Abstract
Candida infection has emerged as a critical health care burden worldwide, owing to the formation of robust biofilms against common antifungals. Recent evidence shows that multidrug-tolerant persisters critically account for biofilm recalcitrance, but their underlying biological mechanisms are poorly understood. Here, we first investigated the phenotypic characteristics of Candida biofilm persisters under consecutive harsh treatments of amphotericin B. The prolonged treatments effectively killed the majority of the cells of biofilms derived from representative strains of Candida albicans, Candida glabrata, and Candida tropicalis but failed to eradicate a small fraction of persisters. Next, we explored the tolerance mechanisms of the persisters through an investigation of the proteomic profiles of C. albicans biofilm persister fractions by liquid chromatography-tandem mass spectrometry. The C. albicans biofilm persisters displayed a specific proteomic signature, with an array of 205 differentially expressed proteins. The crucial enzymes involved in glycolysis, the tricarboxylic acid cycle, and protein synthesis were markedly downregulated, indicating that major metabolic activities are subdued in the persisters. It is noteworthy that certain metabolic pathways, such as the glyoxylate cycle, were able to be activated with significantly increased levels of isocitrate lyase and malate synthase. Moreover, a number of important proteins responsible for Candida growth, virulence, and the stress response were greatly upregulated. Interestingly, the persisters were tolerant to oxidative stress, despite highly induced intracellular superoxide. The current findings suggest that delicate metabolic control and a coordinated stress response may play a crucial role in mediating the survival and antifungal tolerance of Candida biofilm persisters.
INTRODUCTION
Fungal infections are a common and critical problem associated with extremely high morbidity and mortality rates, especially in immunocompromised individuals (1, 2). Candida species are the predominant pathogens in fungal infections, as they are part of normal human microbiota and are ubiquitous in the oral cavity, gastrointestinal tract, and skin of healthy individuals. Under particular conditions, these opportunistic pathogens might contribute to various superficial and even life-threatening systemic infections (3). It has been recognized that biofilm is the preferred mode of growth and existence for microorganisms, including Candida species, and ≥65% of human infections are attributed to biofilm formation and persistence (4). Biofilms attach to surfaces/interfaces and form by embedding themselves in a protective extracellular polymeric matrix. In particular, Candida species are notorious biofilm formers on indwelling medical devices, which is directly linked to therapeutic failure (3, 5). We and other groups have shown that Candida biofilms are highly resistant to antifungals (6, 7). Certain hypotheses have been made to elaborate on the mechanisms of increased antifungal resistance in Candida biofilms, such as the presence of extracellular polymeric substance, the oxidative stress response, and a highly drug-tolerant persister population (8, 9).
Microbial persisters are a subset of microorganisms that have escaped from lethal-dose antimicrobial treatments. Unlike resistant mutants, persisters are multidrug-tolerant phenotypic variants without acquired genetically heritable resistance (10, 11). The concept of persisters was first proposed by Bigger in 1944 (12). It was demonstrated that a growing culture of Staphylococcus pyogenes could not be completely sterilized by penicillin (12). However, the importance of persisters in infections was not well recognized until the recent discovery of persisters in the biofilm community of pathogenic microbes, such as Pseudomonas aeruginosa, Escherichia coli, and Mycobacterium tuberculosis (13–15). Biofilms provide a barrier against the immune system and antimicrobials and prevent persisters contained in the biofilms from being mopped up by immune cells. Once antimicrobial treatment stops, persisters are capable of reviving from the challenges and repopulating the biofilms (10). It is believed that persisters are responsible for the recalcitrance and relapse of many human diseases, such as tuberculosis, cystic fibrosis, and candidiasis (11, 16). Several studies recently reported similar findings in Candida biofilms. Persisters have been detected in Candida albicans biofilms when exposed to high concentrations of antifungals (17, 18), while others are unable to find persisters in biofilms of some Candida strains, including C. albicans SC5314 and certain clinical isolates of C. glabrata and C. tropicalis (19). Therefore, there is still controversy on the presence of persisters in Candida biofilms as a universal trait.
It is evident that persisters have caused a heavy clinical burden, but the biological basis of these tolerant cells remains elusive. Growth arrest triggered by toxin-antitoxin (TA) systems has been the prevailing hypothesis for the survival of microbial persisters, based on the notion that antimicrobials target mainly actively growing cells (20). However, recent studies suggest that dormancy is not the sole mechanism, and some active cellular processes are critical for cellular survival, including active drug efflux and protection against reactive oxygen species (ROS) (11, 20). Nevertheless, our understanding of biofilm persisters is very limited. Currently, the survival mechanisms of Candida biofilm persisters remain unclear. Although one study reported that superoxide dismutase may be associated with C. albicans biofilm persistence against miconazole (18), extensive investigations are urgently needed to elucidate the exact underlying mechanisms.
Given the uncertainty about the presence of persisters in Candida biofilms, here, we first identified the profiles of Candida biofilm persisters in vitro. We then investigated the effects of consecutive antifungal treatments on the persisters. Our data show that Candida biofilms harbor a small fraction of antifungal-tolerant persisters that are unaffected by prolonged courses of treatment. To further explore the survival mechanisms of C. albicans biofilm persisters, we examined their proteome profiles through mass spectrometry (MS)-based shotgun proteomics, and an array of essential proteins for the antifungal tolerance of the persisters was identified. The results indicate that the persisters tend to enter into a metabolically inactive state while exhibiting solid responses to intensive antifungal stress. These novel findings suggest that persisters may be the keystone subpopulation for Candida biofilm resistance and survival, and this study offers new insights into the development of effective antibiofilm therapeutic approaches.
MATERIALS AND METHODS
Candida strains and growth conditions.
A total of six well-defined strains from three representative Candida species were employed in this study, including three reference strains (C. albicans SC5314, C. glabrata ATCC 90030, and C. tropicalis ATCC 13803), and three clinical isolates (C. albicans BF-1, C. glabrata T1570, and C. tropicalis T1427). These species were successfully used in our previous studies (6, 21). The API 32C identification system (bioMérieux, Marcy l'Etoile, France) confirmed the identities of these Candida strains. All Candida strains were streaked on Sabouraud dextrose agar (SDA) (Gibco Ltd., Paisley, United Kingdom) and incubated at 37°C for 24 h. For broth culture, a Candida inoculum was grown overnight in yeast nitrogen base (YNB) medium (Difco, Franklin Lakes, NJ) supplemented with 50 mM glucose in a shaking incubator at 80 rpm. At late-exponential-growth phase, Candida cells were harvested by centrifugation and washed twice with 0.1 M phosphate-buffered saline (PBS) (pH 7.2) for further biofilm experiments. Alternatively, the filamentous growth of C. albicans BF-1 was carried out in Spider medium (1% nutrient broth, 1% mannitol, and 0.2% K2HPO4) (22) and used for subsequent transcriptional analysis.
Biofilm formation.
Candida biofilms were established on presterilized 24-well polystyrene plates (Iwaki, Tokyo, Japan), according to a standard method described previously, with minor modifications (6). Briefly, washed cells were resuspended in YNB medium containing 100 mM glucose, unless otherwise specified, and adjusted to 1 × 107 cells/ml. Next, 1 ml of the standardized suspension was added to the wells of a plate and incubated at 37°C for 1.5 h in a shaking incubator at 80 rpm to allow cell adherence. Following the adhesion stage, nonadherent cells were aspirated, and each well was washed with 1 ml of PBS. Afterwards, 1 ml of new YNB medium containing 100 mM glucose was pipetted into each well. The plates were incubated at 37°C in a shaking incubator at 80 rpm for 48 h to obtain mature biofilms.
Antifungal susceptibility test and XTT reduction assay.
Stock solutions (6.4 mg/ml) of amphotericin B (AMB) (Sigma, St. Louis, MO) were prepared in dimethyl sulfoxide. A 2-fold serial dilution was conducted with YNB medium containing 100 mM glucose to produce final concentrations ranging from 1 to 256 μg/ml. Candida biofilms were washed with 1 ml of PBS and incubated in growth medium with different concentrations of AMB, and growth medium without AMB was used as a control.
After 24 h of incubation at 37°C, the metabolic activity of biofilms was determined by using the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay, as previously described by our group (23). An XTT (Sigma) solution (1 mg/ml in PBS) and a menadione (Sigma) solution (0.4 mM) were prepared and mixed. The final concentration was adjusted with PBS to 0.2 mg/ml for XTT and 0.004 mM for menadione. The biofilms were washed twice with 1 ml of PBS, and 1 ml of the mixed XTT solution was pipetted into each well for incubation in the dark at 37°C for 3 h. The colorimetric changes were measured by transferring 200 μl of the supernatant into a 96-well plate (Iwaki) and reading at 490 nm in a SpectraMax 340 tunable microplate reader (Molecular Devices Ltd., Sunnyvale, CA). Each assay was performed in parallel on three separate occasions.
Consecutive treatments of Candida biofilms and quantitation of persisters.
First, the C. albicans BF-1 biofilms were treated with 1 ml of different concentrations of AMB (1 to 256 μg/ml) for 24 h at 37°C. The biofilms were then washed twice with 1 ml of PBS, and cells were harvested by scraping and vigorous vortexing in 1 ml of PBS. The surviving cells were quantified by serial dilution and plating on SDA for counting of the CFU.
Next, to detect and quantify persisters of all six Candida biofilms, 1 ml of growth medium containing 256 μg/ml AMB was added to the biofilms for 24 h. The number of viable cells was determined, according the same procedure mentioned above. The total cell numbers of untreated biofilms were also recorded. Afterwards, consecutive treatments with AMB were undertaken for another 72 h. The old media were aspirated every 24 h and replaced by 1 ml of fresh ones containing 256 μg/ml AMB, and the number of viable cells was quantified with CFU.
LIVE/DEAD staining and confocal laser scanning microscopy.
The viability of AMB-treated biofilms was further evaluated by LIVE/DEAD staining and confocal laser scanning microscopy (CLSM), as described previously (24). Candida biofilms were formed on polystyrene disks and treated with AMB (256 μg/ml). After 24 h of treatment and 48 h of consecutive treatments, biofilms were stained using the LIVE/DEAD BacLight viability kit (Molecular Probes, Eugene, OR) in the dark for 30 min. Images were obtained by scanning the biofilms in a CLSM system (FluoView FV 1000; Olympus, Tokyo, Japan).
Sample preparation for mass spectrometry.
To investigate the survival mechanisms of Candida biofilm persisters, C. albicans BF-1 biofilms were treated with 256 μg/ml AMB for 24 h. After washing twice with 1 ml of PBS, cells of treated and untreated biofilms were harvested and lysed with yeast protein extraction reagent (Y-PER) (Pierce Biotechnology, Rockford, IL), according to the manufacturer's protocol. The cell lysates were centrifuged at 13,200 rpm for 10 min, and the supernatants were reserved to determine the protein content using the Bradford assay (Bio-Rad, Hercules, CA). Prior to in-solution digestion, the samples were mixed with acetone and incubated at −20°C for 30 min to precipitate proteins. Afterwards, proteins were resuspended with 8 M urea in 0.1 M Tris-HCl, reduced by 20 mM dithiothreitol, and alkylated with 25 mM iodoacetamide. Sequencing-grade trypsin (Promega, Madison, WI) was added in a protease-to-protein ratio of 1:100 (wt/wt) to digest the proteins at 37°C overnight. The output peptides were then desalted and purified using StageTips, as described previously (25). The experiments were independently repeated three times.
Liquid chromatography-tandem MS analysis.
The liquid chromatography-tandem mass spectrometry (LC-MS/MS) experiments were performed on a nanoflow high-performance liquid chromatography (HPLC) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were dried down using SpeedVac (Thermo Fisher Scientific) and redissolved in 0.1% formic acid. Afterwards, the peptide samples were separated by reverse-phase chromatography using an in-house-made PicoTip column (New Objective, Woburn, MA) (360 μm outer diameter, 75 μm inner diameter, 15-μm tip), packed with 8 to 10 cm of octadecyl-silica (ODS)-A C18 5-μm phase (YMC, Allentown, PA). Peptides were rinsed with buffer A (0.1% formic acid) for 5 min and eluted with a 150-min linear gradient from 2 to 35% buffer B (100% acetonitrile, 0.1% formic acid). The LTQ Orbitrap was operated in a data-dependent manner through a full scan, followed by collision-induced dissociation on the 20 most abundant ions.
Data processing and bioinformatics analysis.
The MS data were analyzed with the MaxQuant software (version 1.3.0.5; Max Planck Institute of Biochemistry, Martinsried, Germany) (26). Specifically, the MS/MS spectra were searched by the Andromeda search engine against the reference proteome sequences of C. albicans SC5314 obtained from UniProt release 2014_06. Label-free quantitation algorithms with default parameters were adopted for protein quantification. The precursor mass tolerance was set at 20 ppm and 6 ppm for the first search and main Andromeda search, respectively. The searches included methionine oxidation and N-terminal acetylation as variable modifications and carbamidomethylation as a fixed modification. The minimum peptide length was 7 amino acids, and a maximum of 2 missed cleavages was set. A false discovery rate (FDR) of 0.01 was specified for peptide and protein identifications. If the peptides were shared between multiple proteins, a protein group was reported. The statistical analysis of differential protein expression between the biofilm persisters and controls was performed by a t test, with a permutation-based FDR value of 0.05 using the Perseus software (version 1.4.1.3; Max Planck Institute of Biochemistry) (27). Proteins were grouped by hierarchical clustering according to their expression profiles. The differentially expressed proteins were annotated by Gene Ontology (GO) enrichment analysis with BiNGO (version 2.44) in the categories of biological process, molecular function, and cellular component (28). A GO database of C. albicans SC5314 downloaded from EMBL-EBI in July 2014 was used to map the proteins. Multiple hypothesis testing was corrected with a Benjamini-Hochberg FDR threshold of 0.05.
Measurement of superoxide.
The cytosol and mitochondrion superoxide probes (HKSOX-2 and HKSOX-2m, respectively) for the detection of the endogenous superoxide in live cells were kindly provided by Dan Yang at the Department of Chemistry, Faculty of Science, The University of Hong Kong. For the measurement of superoxide levels in persisters, C. albicans BF-1 biofilms were developed for 48 h on an ibiTreat μ-slide 8 well (ibidi, Martinsried, Germany) and treated with 256 μg/ml AMB for 24 h. For a positive control, biofilms were cultured for 72 h and treated with 20 μM antimycin A for 30 min. Untreated biofilms served as a negative control. The biofilms were washed and incubated with the probes (4 μM HKSOX-2 and 2 μM HKSOX-2m) separately in the dark for 30 min. After incubation, the biofilms were scanned to take images by CLSM.
Quantitative real-time PCR.
To further evaluate and validate the role of the differentially expressed proteins involved in the survival and tolerance of biofilm persisters, the growth and biofilm development of C. albicans BF-1 were determined in Spider medium. The biofilms grown in Spider medium were treated with 256 μg/ml AMB for 24 h and pelleted. Total RNA was extracted using the SV total RNA isolation system (Promega), and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative PCR (qPCR) was performed using the QuantiNova SYBR green PCR kit (Qiagen, Valencia, CA) in the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA), with the following cycling conditions: 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Reactions were carried out in triplicate for three independent experiments. The sequences of the primers used in this experiment are listed in Table S1 in the supplemental material. The data were analyzed with LinRegPCR (version 2015.3) (29, 30), and the relative gene expression levels were normalized to that of the reference gene ACT1. Statistical significance was evaluated by t test, with the significance level set at a P value of <0.05.
RESULTS
Antifungal susceptibility of Candida biofilms.
An XTT reduction assay was employed to determine the susceptibility of Candida biofilms. Overall, AMB displayed potent antifungal effects on the six Candida biofilms. The MIC50 (50% growth reduction) of AMB for these biofilms ranged from 1 to 2 μg/ml. Nevertheless, high doses of AMB were required to inhibit 95% of the growth of the biofilms (Table 1). The dose-dependent killing of these biofilms by AMB exhibited a biphasic trend, with a reduction of live cells at low concentrations and a relative stable subpopulation unaffected at high concentrations. The survival of C. albicans BF-1 biofilm in the presence of AMB is shown in Fig. 1.
TABLE 1.
MICs of AMB against Candida biofilms
| Strain | AMB MICs ([μg/ml]/[μM])a |
|
|---|---|---|
| MIC50 | MIC95 | |
| C. albicans SC5314 | 2.0/2.2 | 64/70.4 |
| C. albicans BF-1 | 2.0/2.2 | 32/35.2 |
| C. glabrata 90030 | 2.0/2.2 | 128/140.8 |
| C. glabrata T1570 | 1.0/1.1 | 16/17.6 |
| C. tropicalis 13803 | 1.0/1.1 | >256/281.6 |
| C. tropicalis T1427 | 1.0/1.1 | 256/281.6 |
The MIC50 and MIC95 are defined as the lowest AMB concentration with a 50% and 95% reduction in absorbance, respectively, compared to the controls assessed by XTT reduction assay.
FIG 1.
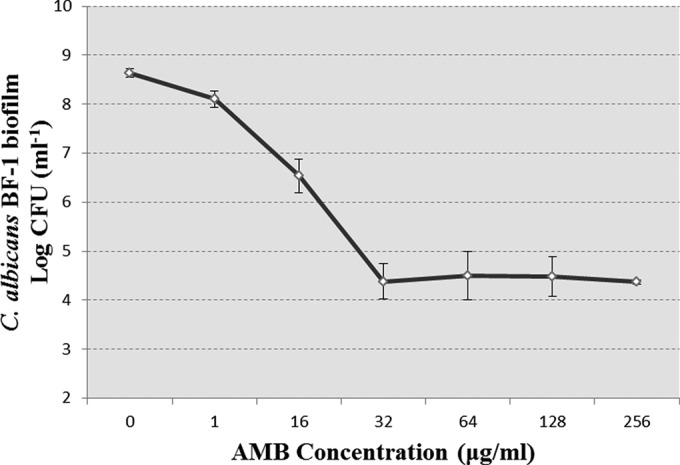
Dose-dependent inhibition of C. albicans BF-1 biofilms by AMB after 24 h treatment. As the concentration of AMB increases, the treatment kills the majority of C. albicans biofilms but fails to eradicate a subpopulation of the biofilms, which remain alive even at lethal doses of AMB. The data represent the means ± SD of three biological replicates from one representative experiment out of two independent experiments with similar results.
Presence of persisters in Candida biofilms.
After 24 h of treatment with AMB at 256 μg/ml, the majority of the six Candida biofilms were killed, while a small amount of persisters survived (Fig. 2). The fractions of biofilm persisters were in a range of 0.01% to 0.5% among all the strains, except C. glabrata T1570. In this case, although the untreated biofilm possessed a large population, the fraction of persisters was extremely low (2.4 × 10−10). A relatively low fraction of persisters was also observed in another C. glabrata strain (ATCC 90030), despite the rapid growth of its biofilm (Fig. 2). The cellular viability of these treated Candida biofilms was then assessed by LIVE/DEAD staining with CLSM, which confirmed the presence of viable persisters in these biofilms (Fig. 3). Taken together, these results demonstrated the presence of persisters in all the Candida biofilms tested after 24 h of treatment with AMB.
FIG 2.
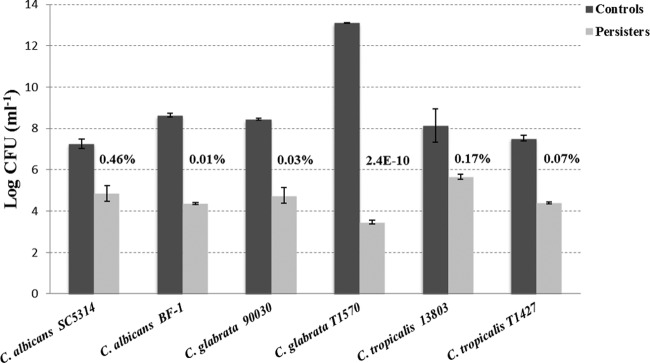
AMB-tolerant persisters in Candida biofilms. The fraction of persisters is defined as the number of persisters accounting for the total cell number of the control biofilm. It is small and varies among the strains tested, ranging from 0.01% to 0.46%, except for C. glabrata T1570 (2.4 ×10−10). The data represent the means ± SD of three biological replicates from one representative experiment out of two independent experiments with similar results.
FIG 3.
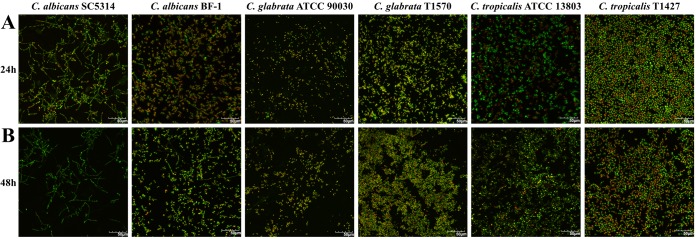
Confocal images (40× magnification) reflecting the viability of Candida biofilms treated with AMB (256 μg/ml). The preformed Candida biofilms on polystyrene disks were exposed to AMB for 24 h (A). Afterwards, fresh AMB medium was added to treat the biofilms until 48 h (B). The treated biofilms were then stained with viability indicators (SYTO 9 and propidium iodide) and scanned with a confocal laser scanning microscope. The bright green cells represent the persisters that stay alive under consecutive AMB treatments. The viability experiments were undertaken twice, and the pictures presented reflect a representative field. Scale bar = 50 μm.
Consecutive treatments of Candida biofilm persisters.
In order to verify that the persisters were not leftover due to insufficient action time and to determine the effect of consecutive antifungal treatments on persisters, the 24-h-treated biofilms were consecutively treated by AMB (256 μg/ml) for another 72 h. Interestingly, consecutive treatments failed to control these biofilms at 48, 72, and 96 h. The quantity of persister cells was maintained at a relatively stable level (Fig. 4). Confocal images of LIVE/DEAD staining confirmed that the persisters remained alive, despite the consecutive AMB treatments (Fig. 3).
FIG 4.
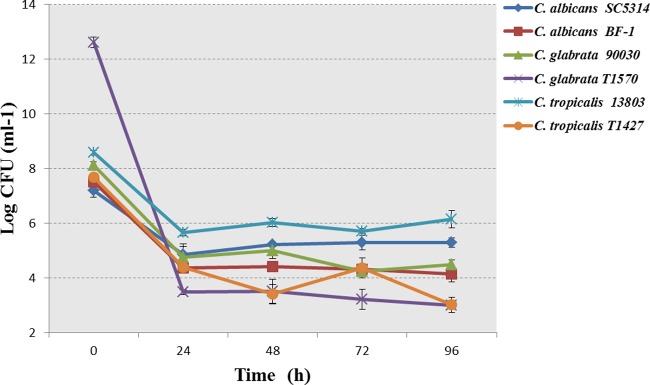
Consecutive treatments of Candida biofilms by AMB (256 μg/ml). The biofilms were treated with AMB consecutively for 96 h. The old media were replaced every 24 h with fresh AMB medium. The consecutive treatments failed to control the biofilm persisters. The data represent the means ± SD of three biological replicates from one representative experiment out of two independent experiments with similar results.
Specific proteomic signature of C. albicans biofilm persisters.
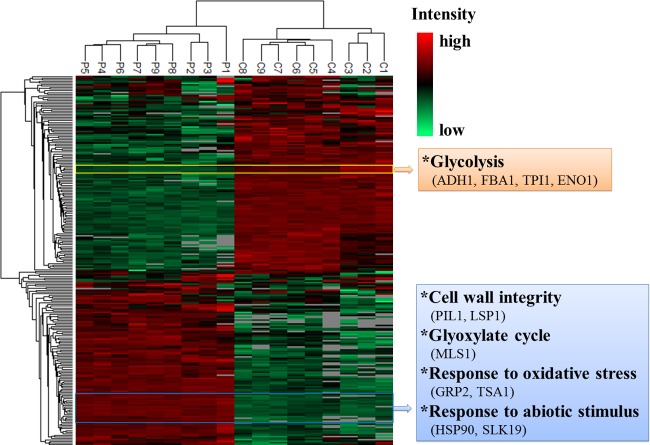
The protein expression profiles of C. albicans biofilm persisters were analyzed using an MS-based shotgun proteomics approach. Digested protein samples from persister fractions and controls of C. albicans BF-1 biofilms, each consisting of 3 biological replicates times 3 technical replicates, were loaded for the identification and quantification of proteins. The relative label-free quantitation of the replicates was highly reproducible, with average Pearson correlation coefficients of 0.952 for the persister samples and 0.954 for the controls. The complete list of peptides identified is presented in Data Set S1 in the supplemental material. Those proteins assembled from shared peptides were pooled into one protein group. A total of 903 proteins/protein groups were identified after excluding potential contaminants and 11 proteins without a valid label-free quantification intensity in both persisters and controls (see Data Set S2 in the supplemental material). Of those, 424 (47.0%) proteins were common to persisters and controls (Fig. 5). For hierarchical clustering and statistical analysis, the data on protein intensities were filtered to ensure that each biological replicate had at least one valid value. The hierarchical clustering showed a clear disparity in protein expression profiles between persisters and controls. The replicates from either persisters or controls were grouped together and demonstrated great similarity (Fig. 6). Compared to the controls, 96 proteins (10.6%) were significantly upregulated (P < 0.05; FDR, 0.05), and 109 proteins (12.1%) were significantly downregulated (P < 0.05; FDR, 0.05) in the persisters (Fig. 5). The differentially expressed proteins were subjected to Gene Ontology (GO) enrichment analysis, which revealed 253 significantly overrepresented biological processes (P < 0.05; FDR, 0.05) (see Data Set S3 in the supplemental material).
FIG 5.
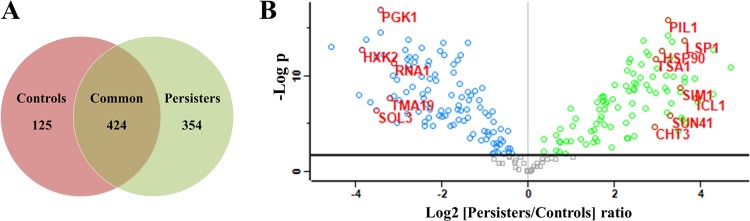
Protein identification and quantification in C. albicans biofilm persisters. (A) Overlap of proteins identified from the persisters (778 proteins) and controls (549 proteins). (B) Volcano plot of quantitative differences in proteins between the persisters and controls. Proteins showing significant differences (P < 0.05; FDR 0.05) are labeled. The green and blue circles represent significantly up- and downregulated proteins in the persisters, respectively.
FIG 6.
Two-way hierarchical clustering (P, persisters; C, controls). The hierarchical clustering analysis groups the samples of persisters and controls separately, and it shows distinct profiles of protein expression between the two clusters. Proteins that reveal the same changing patterns are clustered in the column tree on the left. The proteins listed in the rectangles on the right display similar expression patterns.
Altered metabolic activity in C. albicans biofilm persisters.
We further examined the significant protein hits involved in the cellular metabolism of the persisters (Table 2). Interestingly, the major energy-generating pathways in the C. albicans biofilm persisters were inhibited. Among the significantly changed proteins, the enzymes involved in glycolysis, e.g., phosphoglycerate kinase (PGK1) and hexokinase-2 (HXK2), were all downregulated. Likewise, the dominating enzymes from the tricarboxylic acid (TCA) cycle, including isocitrate dehydrogenase (IDH) and malate dehydrogenase (MDH), which catalyze the generation of NADH, decreased. Persisters also exhibited a lower level of proteins associated with the pentose phosphate pathway, the parallel pathway of glycolysis. In contrast, a few metabolic pathways were activated in the persisters. We observed upregulation of the key enzymes, such as isocitrate lyase (ICL) and malate synthase (MS), in the glyoxylate cycle as an alternative pathway of the TCA cycle. Gluconeogenesis, occurring roughly in reverse order of glycolysis, was likely to be enhanced. The protein that matched the rate-limiting enzyme of gluconeogenesis, namely, fructose-1, 6-bisphosphatase (FBPase), was upregulated.
TABLE 2.
Annotated differentially expressed proteins in C. albicans biofilm persisters
| Proteins by function | UniProt accession no. | Gene | Log2 fold change | P value |
|---|---|---|---|---|
| Glycolysis | ||||
| Fructose-bisphosphate aldolase | Q9URB4 | FBA1 | −2.83 | 3.96E-09 |
| Enolase 1 | P30575 | ENO1 | −2.02 | 3.64E-07 |
| Hexokinase-2 | P83776 | HXK2 | −3.84 | 1.53E-13 |
| Phosphoglycerate kinase | P46273 | PGK1 | −3.42 | 1.11E-17 |
| Phosphoglycerate mutase | P82612 | GPM1 | −0.67 | 0.0013 |
| Triosephosphate isomerase | Q9P940 | TPI1 | −2.63 | 3.02E-12 |
| Hexokinase-like proteina | Q59K93 | GLK2 | −0.84 | 5.13E-07 |
| Likely hexokinasea | Q59TZ8 | GLK1 | ||
| Likely hexokinasea | Q59JX3 | GLK3 | ||
| Likely hexokinasea | Q59RR7 | GLK4 | ||
| Uncharacterized protein | Q5AK23 | ADH1 | −1.39 | 3.24E-08 |
| Pentose phosphate pathway | ||||
| Potential 6-phosphogluconolactonase | Q59PZ6 | SOL3 | −3.53 | 3.21E-07 |
| Transaldolase | Q5A017 | TAL1 | −1.24 | 2.96E-09 |
| 6-Phosphogluconate dehydrogenase, decarboxylating | Q5AKV3 | GND1 | −0.64 | 3.46E-05 |
| Glucose-6-phosphate 1-dehydrogenasea | Q5APL0 | ZWF1 | −1.41 | 2.97E-14 |
| Glucose-6-phosphate 1-dehydrogenasea | Q5AQ54 | |||
| Gluconeogenesis | ||||
| Uncharacterized protein | Q5AAH5 | PCK1 | 0.97 | 0.0006 |
| Uncharacterized protein | G1UB66 | FBP1 | 0.71 | 0.0064 |
| TCA cycle | ||||
| Aconitate hydratase, mitochondrial | P82611 | ACO1 | −1.12 | 1.58E-07 |
| Malate dehydrogenase, cytoplasmic | P83778 | MDH1 | −0.60 | 0.0006 |
| Malate dehydrogenase | Q5AMP4 | MDH1-1 | −0.92 | 1.28E-06 |
| Isocitrate dehydrogenase (NADP) | Q59MF7 | IDP1 | −1.75 | 4.22E-11 |
| Isocitrate dehydrogenase (NADP) | Q59V07 | IDP2 | −1.43 | 2.73E-07 |
| Putative uncharacterized protein IDH2a | Q5A0M1 | IDH2 | 2.70 | 2.46E-07 |
| Putative uncharacterized protein IDH2a | Q5A0T8 | |||
| Citrate synthase | Q59ZZ5 | CIT1 | 2.61 | 4.93E-12 |
| Glyoxylate cycle | ||||
| Isocitrate lyase | Q59RB8 | ICL1 | 3.87 | 1.75E-08 |
| Malate synthase | Q5APD2 | MLS1 | 2.73 | 2.61E-13 |
| Biosynthesis of amino acids | ||||
| Phospho-2-dehydro-3-deoxyheptonate aldolase, tyrosine inhibited | P79023 | ARO4 | −2.32 | 2.44E-09 |
| 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | P82610 | MET6 | −2.17 | 7.23E-11 |
| Uncharacterized protein | Q5A9D9 | LYS12 | −2.48 | 1.17E-13 |
| Homoserine dehydrogenase | Q5AIA2 | HOM6 | −2.99 | 3.17E-08 |
| Uncharacterized protein | Q5ALQ9 | ARO3 | −0.74 | 0.0018 |
| Phosphoserine aminotransferase | Q59P52 | SER1 | −2.29 | 2.97E-07 |
| Branched-chain amino acid aminotransferase | Q59YS9 | BAT22 | −2.87 | 1.80E-13 |
| Branched-chain amino acid aminotransferase | Q5AHJ9 | BAT21 | −2.44 | 1.79E-08 |
| Uncharacterized protein | Q5AAM0 | ARG1 | −0.98 | 0.0004 |
| 3-Isopropylmalate dehydrogenase | Q5AFI8 | LEU2 | −2.33 | 6.64E-12 |
| Likely mitochondrial ketol-acid reductoisomerasea | Q59WW5 | ILV5 | −1.59 | 6.45E-10 |
| Likely mitochondrial ketol-acid reductoisomerasea | Q59XR8 | |||
| Asparagine synthetasea | Q59SH9 | ASN1 | −3.05 | 2.09E-10 |
| Asparagine synthetasea | Q59SE8 | |||
| Translational elongation factor | ||||
| EF-1α1a | P0CY35 | TEF1 | −0.80 | 6.06E-06 |
| EF-1αa | Q59K68 | |||
| EF-1α2a | Q59QD6 | |||
| EF-2 | Q5A0M4 | EFT2 | −1.55 | 1.61E-08 |
| EF-3 | P25997 | CEF3 | 2.34 | 3.09E-09 |
| EF-Tu | Q5ABC3 | TUF1 | −1.09 | 0.0005 |
| Potential translation elongation factor Cam1p | Q59QS2 | CAM1-1 | 2.06 | 1.88E-07 |
| Putative uncharacterized protein EFB1 | Q5A652 | EFB1 | 4.29 | 7.72E-09 |
| Regulation of Ran GTPase activity | ||||
| Uncharacterized proteina | Q5AJS1 | RNA1 | −3.11 | 4.46E-12 |
| Uncharacterized proteina | Q5AJE2 | |||
| Ribosome | ||||
| Likely cytosolic ribosomal protein S3 | Q59N00 | RPS3 | 3.34 | 9.66E-11 |
| Ubiquitin-ribosomal protein fusion S27aa | Q5A109 | UBI3 | −0.81 | 6.65E-05 |
| Uncharacterized proteina | Q5ADS0 | UBI4 | ||
| Likely cytosolic ribosomal acidic protein P0 | Q5AFQ4 | RPP0 | 3.60 | 1.26E-10 |
| Likely cytosolic ribosomal protein S18 | Q5AFQ0 | RPS18 | 1.74 | 1.04E-05 |
| Likely cytosolic ribosomal protein S20 | Q5A389 | RPS20 | 3.30 | 6.08E-08 |
| Likely cytosolic ribosomal protein L12 | Q5AJF7 | RPL12 | 2.30 | 0.0003 |
| Likely cytosolic ribosomal protein S5 | Q5AG43 | RPS5 | 3.60 | 3.38E-10 |
| Likely cytosolic ribosomal protein S7 | Q5AJ93 | RPS7A | 3.32 | 2.42E-14 |
| Likely cytosolic ribosomal protein L4 | Q59ZX4 | RPL4B | 1.58 | 4.61E-08 |
| 40S ribosomal protein S0 | O42817 | RPS0 | 2.41 | 1.86E-09 |
| 40S ribosomal protein S1 | P40910 | RPS1 | 2.72 | 8.23E-10 |
| 40S ribosomal protein S6 | Q5AMI6 | RPS6A | 3.91 | 4.81E-08 |
| 40S ribosomal protein S12 | Q5ADQ6 | RPS12 | 2.40 | 1.13E-06 |
| Heat shock proteins | ||||
| HSP90 homolog | P46598 | HSP90 | 3.09 | 2.26E-13 |
| Small HSP21 | Q5AHH4 | HSP21 | 3.85 | 1.86E-10 |
| Putative uncharacterized protein HSP104 | Q5A376 | HSP104 | 3.56 | 3.55E-06 |
| HSP SSA1 | P41797 | SSA1 | 0.36 | 0.0004 |
| HSP SSC1, mitochondrial | P83784 | SSC1 | 0.96 | 2.23E-07 |
| Likely HSP70/BiP chaperonea | Q5AD54 | KAR2 | 2.25 | 6.29E-06 |
| Uncharacterized proteina | Q5ADI3 | |||
| Response to abiotic stimulus | ||||
| 14-3-3 protein homolog | O42766 | BMH1 | 2.58 | 1.46E-10 |
| Ras-like protein 1 | Q59XU5 | RAS1 | 1.22 | 0.0004 |
| Guanine nucleotide-binding protein subunit β-like protein | P83774 | ASC1 | 1.79 | 7.89E-08 |
| Uncharacterized protein | Q59W54 | SMT3 | 1.53 | 0.0003 |
| Putative uncharacterized protein | Q5ADT0 | SLK19 | 3.27 | 1.25E-12 |
| Adhesion | ||||
| Agglutinin-like protein 3 | Q59L12 | ALS3 | 2.45 | 0.0002 |
| Cell surface mannoprotein MP65 | Q59XX2 | MP65 | 2.41 | 6.67E-08 |
| Cell surface protein | Q5A9D4 | CSP37 | 3.19 | 8.36E-11 |
| Ergosterol biosynthesis | ||||
| NADH-cytochrome b5 reductase 2 | Q59M70 | MCR1 | −1.79 | 8.68E-09 |
| Uncharacterized protein | Q59K50 | ERG10 | −0.75 | 0.001 |
| Uncharacterized protein | Q5A3Z7 | ERG13 | −1.64 | 2.96E-07 |
| Cell wall integrity | ||||
| Glucan 1,3-β-glucosidase | P29717 | XOG1 | 1.32 | 0.0013 |
| Glucan 1,3-β-glucosidase BGL2 | Q5AMT2 | BGL2 | 2.44 | 2.83E-09 |
| Secreted β-glucosidase SUN41 | Q59NP5 | SUN41 | 3.27 | 1.24E-06 |
| Secreted β-glucosidase SIM1 | Q5AKU5 | SIM1 | 3.51 | 1.53E-09 |
| Uncharacterized protein | Q5AAH5 | PCK1 | 0.97 | 0.0006 |
| pH-responsive protein 2 | O13318 | PHR2 | 0.64 | 0.0001 |
| Mannose-1-phosphate guanyltransferase | O93827 | MPG1 | 1.52 | 3.83E-05 |
| Potential cell wall glucanase | Q5AKC7 | SCW11 | 3.51 | 4.75E-09 |
| Uncharacterized protein | Q5AL34 | PSA2 | 0.64 | 0.0061 |
| Possible sphingolipid long-chain-base sensory protein | Q59KV8 | LSP1 | 3.60 | 2.02E-14 |
| Possible sphingolipid long-chain-base sensory protein | Q59VF6 | PIL1 | 3.22 | 2.22E-16 |
| Response to oxidative stress | ||||
| Putative NADPH-dependent methylglyoxal reductase GRP2 | P83775 | GRP2 | 2.41 | 5.27E-11 |
| Peroxiredoxin TSA1 | Q9Y7F0 | TSA1 | 2.95 | 1.44E-12 |
| Potential mitochondrial glutaredoxin | Q5ABB1 | TTR1 | 1.50 | 0.0007 |
Protein group.
In addition, the abundance of enzymes associated with amino acid biosynthesis, such as branched-chain-amino-acid aminotransferase, was lower in the persisters. The expression of a highly conserved protein, TMA19, implicated in protein synthesis and cell growth, was reduced. We found a lower level of RNA1, a protein responsible for RNA processing and export of RNA from the nucleus to the cytosol. The major elongation factors (e.g., EF-1α, EF-2, and EF-Tu) required for translational elongation were downregulated, whereas the fungi-unique elongation factor EF-3 was upregulated, along with several auxiliary components (e.g., EF-1β and EF-Cam1p). Interestingly, we identified a variety of ribosomal proteins (e.g., RPS6A, RPS5, RPP0, RPS3, RPS7A, RPL4B, RPS20, RPS1, RPS0, RPS12, RPL12, and RPS18) and proteins involved in translation regulation (e.g., TIF1, EGD2, PAB1, ASC1, and CAF20) that were enriched in persisters (Table 2).
Enhanced stress response in C. albicans biofilm persisters.
The expression of proteins involved in the stress response revealed considerable changes in the persisters. We found that proteins from heat shock protein (HSP) family, including HSP90, HSP21, HSP104, HSP SSA1, and HSP SSC1, were highly expressed in the persisters (Table 2). The HSP70 molecular chaperone protein KAR2, being essential for viability and the alleviation of the endoplasmic reticulum by regulation of the unfolded protein response, was also induced. Moreover, we identified a high level of proteins accounting for morphogenesis (e.g., BMH1, RAS1, and ASC1), cell cycle control (e.g., BMH1, SMT3, SLK19, and GRP2), and surface adherence (e.g., ALS3, MP65, and CSP37), which are essential for viability, virulence, and response to biotic stimulus of C. albicans (Table 2).
The major mechanism of AMB action is to increase cell permeability through binding to ergosterol present in the cell membrane (31). Interestingly, the proteins associated with ergosterol biosynthesis (e.g., MCR1, ERG10, and ERG13) were downregulated in persisters. Meanwhile, the persisters demonstrated increased expression of cell wall integrity proteins (e.g., XOG1, BGL2, SUN41, PCK1, SIM1, PHR2, MPG1, SCW11, and PSA2). It has been shown that AMB induces oxidative damage in fungal cells to enhance its antifungal effect (32). As expected, we observed the upregulation of some important cellular antioxidants (e.g., TSA1 and TTR1) in the persisters (Table 2). Several aforementioned highly expressed proteins (HSP21, RAS1, and GRP2) also contribute to the antioxidative response. To verify whether persisters are tolerant to oxidative stress, we adopted cytosol and mitochondrion superoxide probes to assess the abundance of the major ROS. As shown in the confocal micrographs, the superoxide level was low in the control biofilms. With reference to the controls, the AMB treatment induced larger amount of superoxide in the persisters. Notably, the majority of superoxide in the cells was located at the mitochondrion instead of the cytosol (Fig. 7).
FIG 7.
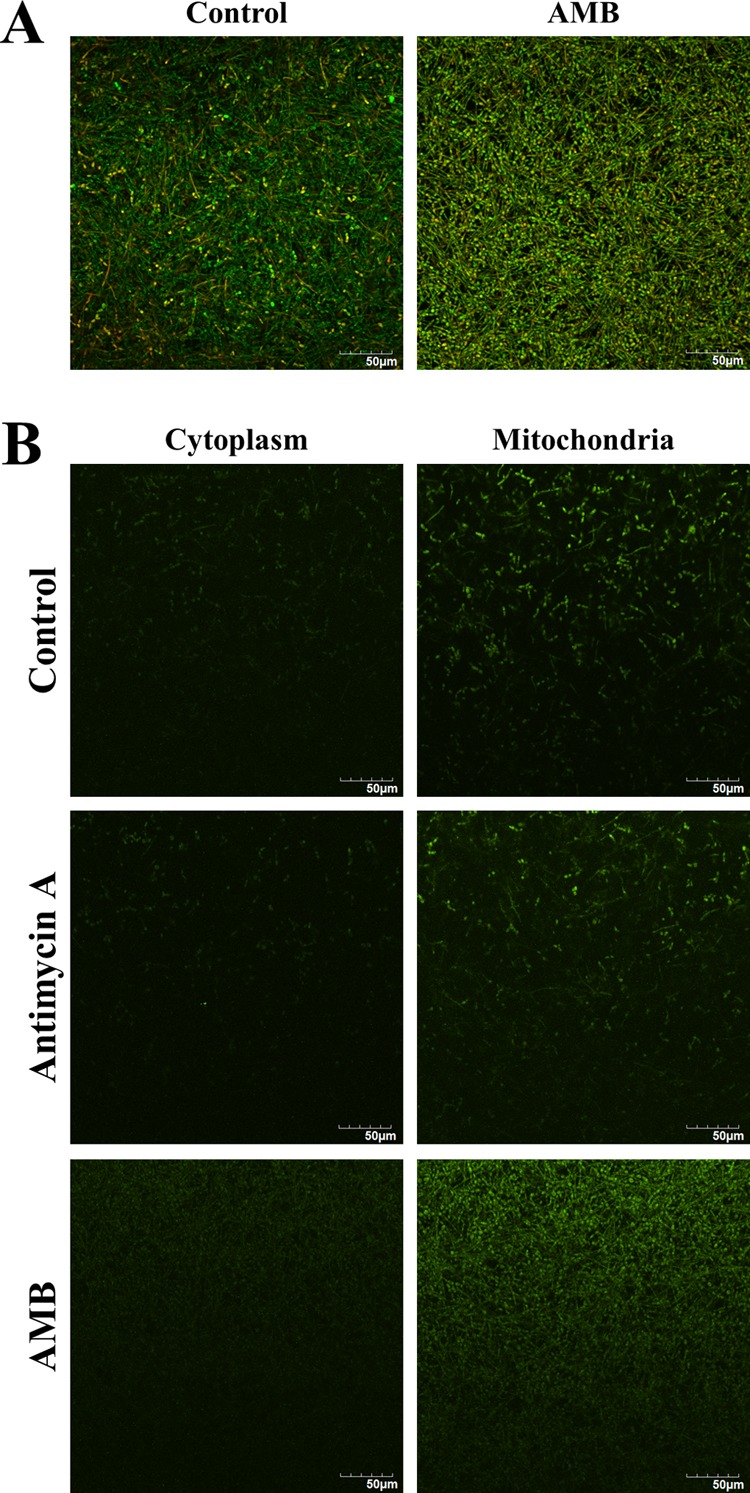
Superoxide levels in C. albicans biofilm persisters. The C. albicans biofilms were formed on ibiTreat μ-slide for 48 h and then exposed to AMB (256 μg/ml) for 24 h. (A) LIVE/DEAD staining (40×) of the control and AMB-treated biofilms. (B) Biofilms were labeled with cytosol and mitochondrion superoxide probes. Those treated with antimycin A (20 μM) for 30 min ahead of probe incubation served as the positive controls. The persisters after AMB treatment exhibited a higher level of superoxide mainly located in the mitochondria with reference to the controls. The experiments were performed twice, and the pictures presented reflect a representative field. Scale bar = 50 μm.
Transcriptional response of C. albicans biofilm persisters in Spider medium.
Under hyphal growth conditions in Spider medium, the AMB treatment gave rise to a smaller fraction (7.34E-06) of persisters in the C. albicans BF-1 biofilms than that in YNB medium. After 24 h treatment, the expression of both PGK1 and HXK2 was markedly downregulated (P < 0.001), while the GRP2 and TSA1 transcripts were significantly enhanced (P < 0.05) in the persisters compared to the controls. No statistically significant differences were observed in the expression of RAS1, ALS3, and BGL2 (Fig. 8).
FIG 8.
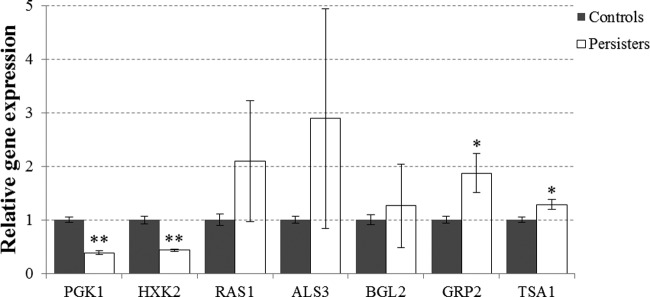
Relative gene expression in C. albicans biofilm persisters under filamentous conditions. The C. albicans biofilms were formed in Spider medium and treated with 256 μg/ml AMB for 24 h. The mRNA expression level was determined by qPCR and normalized against ACT1. The data are presented as the means ± SD from three independent experiments performed in triplicate. *, P < 0.05; **, P < 0.001.
DISCUSSION
The formation and persistence of microbial biofilms have been a leading cause of human infections and failure of antimicrobial therapy worldwide. The underlying mechanisms enabling the survival of biofilms remain unclear. The discovery of persisters in both bacterial and fungal biofilms has expedited the unmasking of this intricate issue. AMB is a potent polyene antifungal commonly used for the treatment of systemic fungal infections through binding to the cell membrane component ergosterol and induction of oxidative stress (31, 32). In this study, although the majority of the six Candida biofilm cells were killed following 24 h of AMB treatment at 256 μg/ml, a small fraction of persisters survived. The level of persisters varied among different strains, indicating the diversity of species susceptibility and their drug tolerance. The results are partly similar to previous observations by other researchers in biofilms of C. albicans and C. glabrata with a higher level of persisters (17, 33). It should be noted that our current data demonstrate the presence of persisters in biofilms of the well-studied reference strain C. albicans SC5314. This differs from a recent study on a reported lack of persisters in biofilms of C. albicans SC5314 treated with 100 μg/ml AMB (19). It has been proposed that the formation of persisters depends on biofilm adherence instead of its complex architecture (17). Notably, here, we show that the clinical isolate C. glabrata T1570 can form thick biofilms with a large population, but interestingly, the fraction of persisters is extremely low. This result might suggest that biofilm-forming ability per se may not account for high drug tolerance.
Currently, the sequential or periodic administration of antimicrobial agents remains the major treatment protocol for controlling infectious diseases in humans (34–36). However, several notorious infections, including candidiasis, are difficult to control despite prolonged courses of antimicrobial therapy and the absence of resistant mutants, which is mainly attributed to drug-tolerant persisters (11). In the present study, we found that sequential AMB treatment fails to eradicate Candida biofilm persisters. This observation indicates that persisters may play a critical role in chronic infections and may account for therapeutic failure even with a prolonged antifungal remedy in clinical practice. Recently, emerging evidence has provided a link between persisters and refractory infectious diseases. One whole-genome comparison of Burkholderia pseudomallei isolated from patients with recurrent infections revealed almost identical genotypes, suggesting that relapse results from recovery of the initial infecting strain instead of a reinfection (37). In cystic fibrosis patients with protracted antibiotic therapy, elevated levels of P. aeruginosa persisters have been identified from the isolates obtained late in the course of airway infection (38). Moreover, an analysis of clinical isolates from patients undergoing chemotherapy reveals that those with long-term oral carriage of C. albicans carry high-persister mutants (33). Therefore, novel therapeutic strategies are urgently needed to target persisters. However, there is limited information on the survival mechanisms of persisters, hampering the course of drug discovery. The present study has secured a reliable in vitro model for the investigation of Candida biofilm persisters, as consecutive AMB treatments are unable to eradicate persisters. With this model, we identified the proteomic profiles of C. albicans BF-1 biofilm persister fractions via MS-based shotgun proteomics, the most used tool for the global identification and quantification of proteins. The Candida biofilm persisters exhibit a specific proteomic signature distinct from that of the controls. The catalyzing enzymes from glycolysis and the TCA cycle are curbed, demonstrating that major energy-generating pathways are inhibited in the persisters. Allison et al. (39) reported that the addition of glycolysis intermediates enables the killing of bacterial persisters induced by aminoglycosides, providing the link between glycolysis inactivation and biofilm persistence. It has been demonstrated that the deletion of the protein TMA19 in yeast might result in the compromise of cell cycle progression and elongation of the cell life span (40, 41); notably, the present study shows that this key protein is downregulated in the C. albicans biofilm persisters. We also found that protein synthesis is downregulated, resulting from the control of regulatory proteins involved in the translation and downregulation of enzymes for amino acid biosynthesis. This differs from a transcriptional study demonstrating that genes involved in amino acid biosynthesis are overexpressed in AMB-treated C. albicans biofilms (42). However, this study used a sublethal concentration of AMB to treat the biofilms, which retained >80% of the metabolic activity of the controls. Kwan and coworkers (43) recently showed that downregulated protein synthesis in E. coli might account for the induction of higher levels of persisters. It is generally assumed that a dormant state triggered by TA systems is responsible for the tolerance of persisters. Balaban et al. (44) adopted a single-cell model and revealed that persisters of the hipA7 mutant E. coli are nongrowing cells, which is the first direct evidence correlating persistence and dormancy. The HipA toxin belongs to the first identified TA system that is linked to persistence and works by phosphorylating the elongation factor EF-Tu, leading to perturbed translation and persister formation (45). The downregulation of major elongation factors (e.g., EF-1α, EF-2, and EF-Tu) may therefore be critical for Candida biofilm persistence. Collectively, these data suggest that Candida biofilm persisters appear to enter into a metabolically inactive state for survival.
Despite the inhibition of major metabolic activities, the Candida biofilm persisters maintain important cellular circuitries and thereby initiate the active stress response. The glyoxylate cycle bypasses the decarboxylation steps of the TCA cycle and converts C2 compounds to C4 precursors for gluconeogenesis and biosynthesis (46). As shown in our study, the unique enzymes from this route, ICL and MS, are significantly enriched in the persisters. The glyoxylate cycle is required for Candida virulence, and its absence in humans makes it a prime target for antifungal agents (47). The use of the ICL inhibitor itaconate reduces the fraction of Burkholderia cenocepacia biofilm persisters, corroborating the importance of the glyoxylate cycle in the survival of persisters (48). Notably, the substantial increase in abundance of many proteins essential for the stress response suggests that the Candida biofilm persisters may adopt an active defense strategy in response to extreme antifungal stimulus. HSPs are a family of cytoprotective proteins ubiquitous within cells, and they are crucial for cellular stress responses and protein folding and translocation (49). We have found a series of HSPs that display tremendously higher expression levels, such as HSP90, HSP21, and HSP104. HSP90 is a key regulator of cell circuitry and plays an important role in potentiating fungal resistance (50, 51). HSP21 mediates adaptation to thermal and oxidative stress and contributes to antifungal drug tolerance in C. albicans (52, 53). Of the HSPs, HSP104 is not found in animal cells, and it governs the thermotolerance and virulence of C. albicans (54). In addition, our results show that persisters increase the expression levels of various proteins that dominate growth, phenotypic switching, surface adherence, quorum sensing, and cell cycle control. Of them, ALS3 is an important adhesion protein that mediates Candida adherence to host tissues and its biofilm formation (55). A previous study reported that the exposure of C. albicans biofilm to caspofungin is associated with the enhanced expression of the ALS3 gene, while AMB-exposed biofilms show upregulation of the ALS1 gene (42). The activation of these regulatory proteins indicates that Candida biofilm persisters probably prepare themselves for a self-protection mode by adjusting their growth and interactions with other cells. The enrichment of an array of ribosomal proteins may be important for correct assembly of crucial proteins under stressed conditions.
As AMB causes lethal effects on Candida by prompting cell membrane rupture and oxidative stress, we further examined the specific cellular pathways in Candida biofilm persisters in response to AMB treatment. It has been reported that C. albicans biofilms contain a lower level of ergosterol than that of planktonic cells, and mutants with impaired ergosterol synthesis exhibit increased resistance to AMB (56). Notably, the biosynthesis of ergosterol, the AMB target, is subdued, indicating that the persisters attempt to lower cell membrane stress. This is consistent with the hypothesis that persisters represent an evolutionary reservoir that gives rise to resistant organisms (11). Interestingly, we detected a number of upregulated cell wall integrity proteins responsible for cell wall biosynthesis and maintenance in persisters. A previous report showed the upregulation of genes associated with β-1,6-glucan biosynthesis in C. albicans biofilms treated with high doses of AMB (57). Taken together, these findings suggest that persisters may enhance cell wall integrity as a compensatory mechanism for the impairment of the cell membrane. The control of oxidative stress may be a common mechanism for bacterial tolerance (58). It has been reported that targeting of the oxidative stress response of Candida is able to enhance the efficacy of AMB treatment (59). Exposure of C. albicans planktonic cultures to AMB at a concentration equivalent to the MIC might result in the upregulation of multiple proteins involved in oxidative stress adaptation (60). Through probing of intracellular superoxide, we further showed that persisters might undergo increased oxidative stress compared to that of control biofilms, suggesting that the survivors are tolerant to oxidative killing. Several important cellular antioxidants and proteins required for adaptation to oxidative stress were found to be increased in persisters. Moreover, the inhibition of glycolysis and the TCA cycle results in decreased production of NADH, and its oxidation via the electron transport chain stimulates the production of superoxide and downstream ROS. The protection against ROS by downregulation of the TCA cycle has been shown to promote the survival of tobramycin-treated biofilms (48). We have previously demonstrated that C. albicans biofilms increase antioxidative capacities with reference to the planktonic counterparts (24). The present study further implies that persisters may be the responsible population for the increased antioxidative capacities of C. albicans biofilms.
Based on the proteomic analysis, we further investigated the transcriptional regulation of C. albicans biofilm persisters in response to AMB treatment under filamentous conditions. Seven significant changing proteins were selected and subjected to qPCR analysis. Consistent with the proteomic analysis, the persisters in Spider medium demonstrate downregulation of the key genes (PGK1and HXK2) from glycolysis and overexpression of the genes (GRP2 and TSA1) responsible for the antioxidative response. These findings further indicate the importance of metabolic and stress regulation in persister survival. The gene expression of RAS1, ALS3, and BGL2 reveals no significant differences, while displaying a similar trend with the proteomics results. As gene expression precedes protein synthesis, this may lead to some discrepancy between the proteomic and transcriptional data measured at the same time point. Additionally, a much lower fraction of persisters was identified under filamentous conditions, suggesting that the environmental conditions may affect the antifungal tolerance of persisters. Due to the complexity of gene regulation in persisters and the current lack of reliable techniques for persister isolation, further studies are highly warranted to confirm the current findings.
In summary, this study has demonstrated that a small fraction of Candida biofilm persisters can withstand consecutive treatments of lethal doses of AMB, and a specific proteomic signature of the persisters has been identified. To our knowledge, this is probably the first study on a concurrent analysis of both the gene and protein expression profiles of Candida biofilm persisters. The persisters could downregulate major energy-generating pathways and protein synthesis and enhance imperative metabolic activities and protein expression associated with the stress response. Therefore, the antifungal tolerance of Candida biofilm persisters is probably triggered by delicate metabolic regulation and coordinated stress adaptation. It is noteworthy that Candida biofilm persisters may have the capacity to invoke a specific adaptive strategy against antifungals. Here, we identified an array of crucial tolerance-related proteins in the persisters. The unmasking of the survival mechanisms of Candida biofilm persisters may contribute to the development of novel antifungal agents for the effective control and management of Candida infections.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Health & Medical Research Fund (project no. 12110752) from the Hong Kong SAR Government and the Modern Dental Laboratory/HKU Endowment Fund to L.J.
J.A.V. is supported by The Wellcome Trust (grant WT101477MA) and European Union FP7 grant PRIME-XS (no. 262067).
We thank Eva Fung and Naikei Wong from the Department of Chemistry, Faculty of Science, The University of Hong Kong (HKU), for excellent technical assistance and Dan Yang from the same department for providing superoxide probes. We also thank Sarah Wong from the HKU Faculty of Dentistry for her contribution to the proteomics data analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00543-15.
REFERENCES
- 1.Brown GD, Denning DW, Levitz SM. 2012. Tackling human fungal infections. Science 336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 2.Goffeau A. 2008. Drug resistance: the fight against fungi. Nature 452:541–542. doi: 10.1038/452541a. [DOI] [PubMed] [Google Scholar]
- 3.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Fanning S, Mitchell AP. 2012. Fungal biofilms. PLoS Pathog 8:e1002585. doi: 10.1371/journal.ppat.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seneviratne CJ, Jin LJ, Samaranayake YH, Samaranayake LP. 2008. Cell density and cell aging as factors modulating antifungal resistance of Candida albicans biofilms. Antimicrob Agents Chemother 52:3259–3266. doi: 10.1128/AAC.00541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seneviratne CJ, Wang Y, Jin L, Wong SS, Herath TD, Samaranayake LP. 2012. Unraveling the resistance of microbial biofilms: has proteomics been helpful? Proteomics 12:651–665. doi: 10.1002/pmic.201100356. [DOI] [PubMed] [Google Scholar]
- 9.Taff HT, Mitchell KF, Edward JA, Andes DR. 2013. Mechanisms of Candida biofilm drug resistance. Future Microbiol 8:1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 11.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin. Lancet 244:497–500. [Google Scholar]
- 13.Brooun A, Liu S, Lewis K. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison JJ, Ceri H, Roper NJ, Badry EA, Sproule KM, Turner RJ. 2005. Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 151:3181–3195. doi: 10.1099/mic.0.27794-0. [DOI] [PubMed] [Google Scholar]
- 15.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol 69:164–174. doi: 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 17.LaFleur MD, Kumamoto CA, Lewis K. 2006. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Chemother 50:3839–3846. doi: 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bink A, Vandenbosch D, Coenye T, Nelis H, Cammue BP, Thevissen K. 2011. Superoxide dismutases are involved in Candida albicans biofilm persistence against miconazole. Antimicrob Agents Chemother 55:4033–4037. doi: 10.1128/AAC.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Dhaheri RS, Douglas LJ. 2008. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob Agents Chemother 52:1884–1887. doi: 10.1128/AAC.01473-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balaban NQ, Gerdes K, Lewis K, McKinney JD. 2013. A problem of persistence: still more questions than answers? Nat Rev Microbiol 11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 21.Seneviratne CJ, Wong SS, Yuen KY, Meurman JH, Pärnänen P, Vaara M, Samaranayake LP. 2011. Antifungal susceptibility and virulence attributes of bloodstream isolates of Candida from Hong Kong and Finland. Mycopathologia 172:389–395. doi: 10.1007/s11046-011-9444-4. [DOI] [PubMed] [Google Scholar]
- 22.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seneviratne CJ, Silva WJ, Jin LJ, Samaranayake YH, Samaranayake LP. 2009. Architectural analysis, viability assessment and growth kinetics of Candida albicans and Candida glabrata biofilms. Arch Oral Biol 54:1052–1060. doi: 10.1016/j.archoralbio.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Seneviratne CJ, Wang Y, Jin L, Abiko Y, Samaranayake LP. 2008. Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8:2936–2947. doi: 10.1002/pmic.200701097. [DOI] [PubMed] [Google Scholar]
- 25.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Cox J, Mann M. 2012. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13(Suppl 16):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 29.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 30.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baginski M, Sternal K, Czub J, Borowski E. 2005. Molecular modelling of membrane activity of amphotericin B, a polyene macrolide antifungal antibiotic. Acta Biochim Pol 52:655–658. [PubMed] [Google Scholar]
- 32.Sokol-Anderson ML, Brajtburg J, Medoff G. 1986. Amphotericin B-induced oxidative damage and killing of Candida albicans. J Infect Dis 154:76–83. doi: 10.1093/infdis/154.1.76. [DOI] [PubMed] [Google Scholar]
- 33.LaFleur MD, Qi Q, Lewis K. 2010. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother 54:39–44. doi: 10.1128/AAC.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlow GD, Nathwani D. 2000. Sequential antibiotic therapy. Curr Opin Infect Dis 13:599–607. doi: 10.1097/00001432-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Taccetti G, Campana S, Neri AS, Boni V, Festini F. 2008. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis. J Chemother 20:166–169. doi: 10.1179/joc.2008.20.2.166. [DOI] [PubMed] [Google Scholar]
- 36.Chen F, Zheng N, Wang Y, Wen JL, Tu WF, Du YQ, Lin JM. 2013. Sequential intravenous/oral moxifloxacin monotherapy for complicated skin and skin structure infections: a meta-analysis of randomised controlled trials. Int J Clin Pract 67:834–842. doi: 10.1111/ijcp.12174. [DOI] [PubMed] [Google Scholar]
- 37.Hayden HS, Lim R, Brittnacher MJ, Sims EH, Ramage ER, Fong C, Wu Z, Crist E, Chang J, Zhou Y, Radey M, Rohmer L, Haugen E, Gillett W, Wuthiekanun V, Peacock SJ, Kaul R, Miller SI, Manoil C, Jacobs MA. 2012. Evolution of Burkholderia pseudomallei in recurrent melioidosis. PLoS One 7:e36507. doi: 10.1371/journal.pone.0036507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinnerthaler M, Jarolim S, Heeren G, Palle E, Perju S, Klinger H, Bogengruber E, Madeo F, Braun RJ, Breitenbach-Koller L, Breitenbach M, Laun P. 2006. MMI1 (YKL056c, TMA19), the yeast orthologue of the translationally controlled tumor protein (TCTP) has apoptotic functions and interacts with both microtubules and mitochondria. Biochim Biophys Acta 1757:631–638. doi: 10.1016/j.bbabio.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Bommer UA, Thiele BJ. 2004. The translationally controlled tumour protein (TCTP). Int J Biochem Cell Biol 36:379–385. doi: 10.1016/S1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 42.Vediyappan G, Rossignol T, d'Enfert C. 2010. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother 54:2096–2111. doi: 10.1128/AAC.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn MF, Ramirez-Trujillo JA, Hernandez-Lucas I. 2009. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155:3166–3175. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 47.Lorenz MC, Fink GR. 2001. The glyoxylate cycle is required for fungal virulence. Nature 412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- 48.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santoro MG. 2000. Heat shock factors and the control of the stress response. Biochem Pharmacol 59:55–63. doi: 10.1016/S0006-2952(99)00299-3. [DOI] [PubMed] [Google Scholar]
- 50.Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 51.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog 5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer FL, Wilson D, Jacobsen ID, Miramon P, Slesiona S, Bohovych IM, Brown AJ, Hube B. 2012. Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS One 7:e38584. doi: 10.1371/journal.pone.0038584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayer FL, Wilson D, Hube B. 2013. Hsp21 potentiates antifungal drug tolerance in Candida albicans. PLoS One 8:e60417. doi: 10.1371/journal.pone.0060417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiori A, Kucharikova S, Govaert G, Cammue BP, Thevissen K, Van Dijck P. 2012. The heat-induced molecular disaggregase Hsp104 of Candida albicans plays a role in biofilm formation and pathogenicity in a worm infection model. Eukaryot Cell 11:1012–1020. doi: 10.1128/EC.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nailis H, Vandenbosch D, Deforce D, Nelis HJ, Coenye T. 2010. Transcriptional response to fluconazole and amphotericin B in Candida albicans biofilms. Res Microbiol 161:284–292. doi: 10.1016/j.resmic.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JH, Faria NC, Martins Mde L, Chan KL, Campbell BC. 2012. Enhancement of antimycotic activity of amphotericin B by targeting the oxidative stress response of Candida and cryptococcus with natural dihydroxybenzaldehydes. Front Microbiol 3:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoehamer CF, Cummings ED, Hilliard GM, Rogers PD. 2010. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob Agents Chemother 54:1655–1664. doi: 10.1128/AAC.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.