LETTER
The New Delhi metallo-β-lactamase (NDM-1) was initially identified in clinical isolates of Escherichia coli and Klebsiella pneumoniae in Sweden from a patient previously hospitalized in India (1). Since then, blaNDM-1 has frequently been reported in Enterobacteriaceae and Acinetobacter spp., with a fast dissemination in the Indian subcontinent, the Balkan countries, China, and the Middle East (2). NDM-1 producers generally associated with Enterobacteriaceae species have also been reported, albeit with much lower frequency, in Latin American countries, including Guatemala, Mexico, Colombia, and Brazil (3). Moreover, production of NDM-1 in this geographic region has also been noted in Acinetobacter baumannii in Honduras and Brazil (3, 4) and in Acinetobacter pittii in Paraguay and Brazil (5, 6). Here, we report the first case of an NDM-1-producing Acinetobacter species in Argentina, an Acinetobacter bereziniae clinical isolate. We describe also the complete sequence of a blaNDM-1-containing plasmid in this strain.
(Part of this work was presented at the 10th International Symposium on the Biology of Acinetobacter 2015, Athens, Greece, 3 to 6 June 2015 [7].)
A. bereziniae HPC229 was isolated on June 2014 from a blood sample of a 53-year-old female patient that underwent chemotherapy due to leukemia at a hospital located in Rosario, Argentina. The patient was treated with ciprofloxacin plus tigecycline, resulting in clinical and microbiological cure as evaluated by negative blood cultures. The patient was readmitted to the hospital 3 months later with symptoms of severe sepsis and died due to septic shock, with positive blood cultures that grew Escherichia coli.
HPC229, originally identified as Acinetobacter lwoffii by the Vitek 2 system (bioMérieux), was reclassified by DNA sequence comparison analyses of its 16S rRNA, gyrB (8), and rpoB (9) genes. The highest identity was found to the corresponding orthologs of the A. bereziniae ATCC 17924 type strain (99.9%, 99.7%, and 99.8%, respectively). The antibiotic susceptibility profile of HPC229 was determined by either the Vitek 2 system or the agar dilution method. The interpretation of the obtained MICs based on CLSI breakpoints (10) indicated resistance to β-lactams, including carbapenems (Table 1).
TABLE 1.
Antimicrobial susceptibilities of A. bereziniae HPC229 and its plasmid-cured HPC229c derivativec
| Antimicrobial(s) | MIC (μg/ml) |
|
|---|---|---|
| HPC229 | HPC229c | |
| Ampicillin-sulbactama | 16 | ≤2 |
| Ceftazidimeb | ≥256 | 8 |
| Cefotaximea | ≥64 | 8 |
| Cefepimeb | ≥256 | 1 |
| Piperacillin-tazobactama | ≥128 | 8 |
| Imipenemb | 128 | 0.125 |
| Meropenema | ≥16 | 1 |
| Colistina | ≤0.5 | ≤0.5 |
| Gentamicina | ≤1 | ≤1 |
| Amikacina | ≤2 | ≤2 |
| Trimethoprim-sulfamethoxazolea | ≤20 | ≤20 |
| Ciprofloxacina | 0.5 | 0.5 |
MICs were determined by the Vitek 2 system.
MICs were determined by the agar dilution method.
HPC229 was resistant to ampicillin-sulbactam, ceftazidime, cefotaxime, cefepime, piperacillin-tazobactam, imipenem, and meropenem, and it was susceptible to colistin, gentamicin, amikacin, trimethoprim-sulfamethoxazole, and ciprofloxacin according to CLSI breakpoints (10).
PCR amplification using specific primers for the blaIMP, blaVIM, blaSPM, blaNDM, blaOXA-23-like, blaOXA-24/40-like, and blaOXA-58-like genes (11–13) and sequencing analysis revealed the presence of blaNDM-1. Evidence that blaNDM-1 was located in a plasmid was first obtained by plasmid curing (14) in which HPC229 was cultured in 5 ml of LB liquid medium containing 0.2 ml of 10% SDS for 48 h at 40°C. This procedure allowed the isolation of HPC229c, which showed a marked increase in susceptibility to β-lactams (Table 1). Susceptibility to aztreonam as judged by the disk diffusion assay, in contrast, showed no differences between HPC229 and HPC229c. All conjugation attempts using HPC229 as the donor and Escherichia coli DH5α, A. baumannii ATCC 17978, or Pseudomonas aeruginosa PAO1 as the recipient (11) were unsuccessful. The genomic relatedness between HPC229 and HPC229c was confirmed using previously described PCR-based procedures (15), and the loss of blaNDM-1 was confirmed by specific PCR analysis.
Analysis of whole-genome sequencing performed with pyrosequencing (454 Genome Sequencer FLX system; Roche Diagnostics) demonstrated that the blaNDM-1 metallo-β-lactamase gene was located in HPC229 in a 44,560-bp plasmid, designated pNDM229. This gene was found located inside a composite transposon bracketed by two copies of ISAba125 and identical to that first described in A. baumannii (16) and designated Tn125 by Poirel et al. (17). The immediate genetic environment of Tn125 in pNDM229 is most similar to that of the conjugative plasmid pNDM-BJ01 (18) present in an A. lwoffii clinical strain (Fig. 1). In both cases, the Tn125 element is located immediately downstream of an aphA6 gene, which is in turn adjacent to an upstream ISAba14 insertion sequence. In the case of pNDM229, however, a second ISAba14 copy was found immediately downstream of the Tn125 element (Fig. 1), thus representing a novel difference from the similar arrangements previously reported (18, 19). This suggests the evolution of a novel composite transposon now bounded by two ISAba14 elements with the potential ability to mobilize as a whole.
FIG 1.
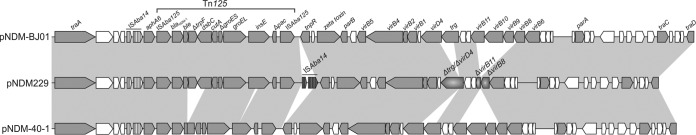
Comparative analysis of Acinetobacter blaNDM-1-harboring plasmids. The linearized genetic maps of pNDM229 from A. bereziniae HPC229 (GenBank accession number KT072713), pNDM-BJ01 from A. lwoffii WJ10621 (NC_019268.1 [18]), and pNDM-40-1 from A. bereziniae CHI-40-1 (KF702385 [19]) are compared. Gray shading indicates more than 99% nucleotide sequence identity. In all plasmids, the ISAba14 element is indicated by black vertical lines. A second copy of ISAba14 present only in pNDM229 is shown in dark gray and exhibits 94.0% nucleotide identity with the upstream ISAba14 element. Partial deletions in conjugative genes in pNDM229 are indicated by radial gray gradients. Arrowheads indicate the direction of transcription, and open reading frames codifying hypothetical proteins are indicated by white arrows.
In summary, we describe here the first case of an NDM-1-producing A. bereziniae isolate in Argentina. To our knowledge, only one other A. bereziniae NDM-1-producing clinical isolate has been recently reported (CHI-40-1), and the blaNDM-1 gene was located in this strain in a conjugative plasmid (19). However, a number of differences were noted between HPC229 and CHI-40-1 and between the plasmids carried by them (Fig. 1). First, comparison analyses of the corresponding 16S rRNA, gyrB, and rpoB genes revealed identities of 99.9%, 99.8%, and 99.6%, respectively, indicating no immediate clonal relationship between these two A. bereziniae strains. Second, comparison of the corresponding blaNDM-1-containing platforms revealed two deletions in pNDM-40-1 which were not observed in pNDM229, one of which removed 1,298 bp from the 3′ end of ble to dsbC and the other of which removed 150 bp within insE (Fig. 1). Third, the above-noted second copy of ISAba14 downstream of Tn125 was present only in pNDM229. Fourth, unlike with other conjugative plasmids, such as pNDM-40-1 and pNDM-BJ01, some conjugation-related genes, including virD4, trg, virB11, virB8, virB10, and virB9, were either truncated or absent in pNDM229 (Fig. 1), thus explaining conjugation failures.
The origin of A. bereziniae HPC229 remains obscure, since no history of overseas travel of the compromised patient or direct contact with foreigners could be identified. The above findings represent a worrisome concern, especially when considering that Acinetobacter species other than A. baumannii can act as reservoirs of blaNDM genes and most probably contribute to their spreading among clinically relevant Enterobacteriaceae species (6, 19).
Nucleotide sequence accession numbers.
The 16S rRNA, gyrB, and rpoB sequences of the HPC229 strain used for comparison with other Acinetobacter species have been released in GenBank under the accession numbers KP765739, KP765740, and KP765741, respectively. The 44,560 bp blaNDM-1-containing pNDM229 plasmid sequence has been released under accession number KT072713.
ACKNOWLEDGMENTS
We thank the anonymous reviewers for their careful reading of our manuscript and their many insightful comments and suggestions for improvement. We are grateful to the Bacteriology Service personnel of the Hospital Provincial del Centenario of Rosario City for kindly providing the HPC229 strain.
P.M.M., G.C., M.R., and A.S.L. are researchers at the National University of Rosario. J.M.-B. and A.M.V. are staff members of CONICET. M.B. and M.C. are fellows of ANPCyT. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina), the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), the Ministerio de Salud and the Secretaría de Ciencia, Tecnología e Innovación, Provincia of Santa Fe, and the Secretaría de Salud Pública of the municipality of Rosario.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TP, Carvalho-Assef AP, Asensi MD. 2014. First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother 58:7592–7594. doi: 10.1128/AAC.03444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterman PE, McGann P, Snesrud E, Robert J, Clifford RJ, Kwak YI, Munoz-Urbizo IP, Tabora-Castellanos J, Milillo M, Preston L, Aviles R, Sutter DE, Leshoa EP. 2013. Bacterial peritonitis due to Acinetobacter baumannii sequence type 25 with plasmid-borne New Delhi metallo-β-lactamase in Honduras. Antimicrob Agents Chemother 57:4584–4586. doi: 10.1128/AAC.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasteran F, Mora MM, Albornoz E, Faccone D, Franco R, Ortellado J, Melgarejo N, Gomez S, Riquelme I, Matheu J, Ramon-Pardo P, Corso A. 2014. Emergence of genetically unrelated NDM-1-producing Acinetobacter pittii strains in Paraguay. J Antimicrob Chemother 69:2575–2578. doi: 10.1093/jac/dku139. [DOI] [PubMed] [Google Scholar]
- 6.Pagano M, Poirel L, Martins AF, Rozales FP, Zavascki AP, Barth AL, Nordmann P. April 2015. Emergence of NDM-1-producing Acinetobacter pittii in Brazil. Int J Antimicrob Agents doi: 10.1016/j.ijantimicag.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Brovedan M, Marchiaro PM, Morán-Barrio J, Cameranesi M, Cera G, Rinaudo M, Viale AM, Limansky AS. 2015. 10th International Symposium on the Biology of Acinetobacter 2015, Athens, Greece, 3 to 6 June 2015, abstr P33A. [Google Scholar]
- 8.Ibrahim A, Gerner-Smidt P, Liesack W. 1997. Phylogenetic relationship of the twenty-one DNA groups of the genus Acinetobacter as revealed by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol 47:837–841. doi: 10.1099/00207713-47-3-837. [DOI] [PubMed] [Google Scholar]
- 9.Bonnin RA, Ocampo-Sosa AA, Poirel L, Guet-Revillet H, Nordmann P. 2012. Biochemical and genetic characterization of carbapenem-hydrolyzing β-lactamase OXA-229 from Acinetobacter bereziniae. Antimicrob Agents Chemother 56:3923–3927. doi: 10.1128/AAC.00257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI). 2014. Performance standards for antimicrobial susceptibility testing. Twenty-third informational supplement. Document M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 11.Marchiaro P, Viale AM, Ballerini V, Rossignol G, Vila AJ, Limansky A. 2010. First report of a Tn402-like class 1 integron carrying blaVIM-2 in Pseudomonas putida from Argentina. J Infect Dev Ctries 4:412–416. [PubMed] [Google Scholar]
- 12.Manchanda V, Rai S, Gupta S, Rautela RS, Chopra R, Rawat DS, Verma N, Singh NP, Kaur IR, Bhalla P. 2011. Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J Med Microbiol 29:249–253. doi: 10.4103/0255-0857.83907. [DOI] [PubMed] [Google Scholar]
- 13.Marchiaro P, Ballerini V, Spalding T, Cera G, Mussi MA, Morán-Barrio J, Vila AJ, Viale AM, Limansky AS. 2008. A convenient microbiological assay employing cell-free extracts for the rapid characterization of Gram-negative carbapenemase producers. J Antimicrob Chemother 62:336–344. doi: 10.1093/jac/dkn185. [DOI] [PubMed] [Google Scholar]
- 14.Lopes BS, Al-Hassan L, Amyes SG. 2012. ISAba825 controls the expression of the chromosomal blaOXA-51-like and the plasmid borne blaOXA-58 gene in clinical isolates of Acinetobacter baumannii isolated from the USA. Clin Microbiol Infect 18:E446–E451. doi: 10.1111/j.1469-0691.2012.03979.x. [DOI] [PubMed] [Google Scholar]
- 15.Limansky AS, Viale AM. 2002. Can composition and structural features of oligonucleotides contribute to their wide-scale applicability as random PCR primers in mapping bacterial genome diversity? J Microbiol Methods 50:291–297. doi: 10.1016/S0167-7012(02)00040-4. [DOI] [PubMed] [Google Scholar]
- 16.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Göttig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Yang X, Xiao X, Luan Ch, Yang Y, Cui Y, Yang R, Gao G, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a blaNDM-1 gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones LS, Carvalho MJ, Toleman MA, White PL, Connor TR, Mushtaq A, Weeks JL, Kumarasamy KK, Raven KE, Török ME, Peacock SJ, Howe RA, Walsh TR. 2015. Characterization of plasmids in extensively drug-resistant Acinetobacter strains isolated in India and Pakistan. Antimicrob Agents Chemother 59:923–929. doi: 10.1128/AAC.03242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]


