Abstract
Chitosan oligosaccharides were modified with N-diazeniumdiolates to yield biocompatible nitric oxide (NO) donor scaffolds. The minimum bactericidal concentrations and MICs of the NO donors against Pseudomonas aeruginosa were compared under aerobic and anaerobic conditions. Differential antibacterial activities were primarily the result of NO scavenging by oxygen under aerobic environments and not changes in bacterial physiology. Bacterial killing was also tested against nonmucoid and mucoid biofilms and compared to that of tobramycin. Smaller NO payloads were required to eradicate P. aeruginosa biofilms under anaerobic versus aerobic conditions. Under oxygen-free environments, the NO treatment was 10-fold more effective at killing biofilms than tobramycin. These results demonstrate the potential utility of NO-releasing chitosan oligosaccharides under both aerobic and anaerobic environments.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic human pathogen that frequently colonizes people with compromised immune systems, such as those with cystic fibrosis (CF) or severe burn wounds (1). The success of P. aeruginosa as a pathogen is related to its multitude of virulence factors, which increase adherence to the host cells, induce inflammation, and disrupt the host immune response (1). Furthermore, P. aeruginosa is intrinsically resistant to many antibiotics due to low membrane permeability and increased expression of β-lactamases and efflux pumps (2–4). In addition to this native resistance, P. aeruginosa readily adapts to antibiotic challenge by acquiring resistance genes (3, 5) and forming protective, cooperative communities known as biofilms (6, 7).
While all antibiotic resistance mechanisms are not fully understood, at least three main factors reduce antibiotic efficacy against bacterial biofilms compared to planktonic cells. First, P. aeruginosa in biofilms secretes a protective layer of exopolysaccharides that prevent the diffusion of antibiotics (7). In the context of cystic fibrosis, biofilm-bound P. aeruginosa exists predominantly as the mucoid phenotype, characterized by a secreted alginate matrix that provides a physical barrier against the host immune response and antibiotics (8). This exopolysaccharide matrix also prevents the diffusion of oxygen into biofilms, causing P. aeruginosa to switch from aerobic to anaerobic respiration (9). The reduced metabolic activity of P. aeruginosa undergoing anaerobic respiration protects the bacterium against traditional antibiotics that are most effective against rapidly dividing cells, including aminoglycosides and β-lactams (10, 11). Finally, biofilms produce persister cells (i.e., dormant bacteria that are highly resistant to chemical disinfectants and exhibit multidrug tolerance) much more frequently than planktonic bacterial cultures (3, 7).
The failure of conventional antibiotics to treat P. aeruginosa biofilms and infections necessitates the development of new antibacterial agents. Nitric oxide (NO), an endogenously produced free radical that can disperse (12, 13) and eradicate (14, 15) biofilms, holds particular promise as an alternative to current antibiotic treatments. Gaseous NO has been repeatedly used to eradicate P. aeruginosa infections in small-animal models with no apparent toxicity (16, 17). Under aerobic environments, NO reacts with molecular oxygen, superoxide, and hydrogen peroxide to form highly reactive intermediates (peroxynitrite, nitrogen dioxide, and dinitrogen trioxide). These molecules exert nitrosative and oxidative stresses, such as DNA deamination, nitrosation of membrane and intracellular proteins, and membrane damage via lipid peroxidation, culminating in bacterial death (18–21). Some of these congener molecules, especially peroxynitrite, are more potent antimicrobials than NO alone (19). In anaerobic environments, NO toxicity is less understood. Ren et al. reported that the bacteriostatic mechanisms included modification of iron-sulfur proteins (22). As these proteins are linked to nearly every cellular process, including metabolism, respiration, RNA modification, and DNA repair and replication, their alteration greatly influences bacterial viability (23). The killing activities of NO are expected to be different under aerobic and anaerobic conditions due to the differences in bactericidal mechanisms, but this hypothesis has yet to be studied systematically. As P. aeruginosa grows rapidly and forms biofilms under both aerobic and anaerobic conditions, understanding the effects of oxygen on the antibacterial activity of NO is essential for developing NO-based therapeutics.
While the administration of exogenous NO holds promise as a therapeutic, treatment of infections or chronic wounds with gaseous NO is impractical, expensive, and potentially dangerous, as NO mediates other physiological processes (e.g., vasodilation and blood clotting) (21, 24–26). Macromolecular scaffolds capable of effectively storing and releasing NO have been developed to enable local delivery (27, 28). The most promising NO release vehicles to date include NO donor-modified N-diazeniumdiolate silica nanoparticles (29–31), dendrimers (32–36), and chitosan (15). While silica nanoparticles (14, 37–39) and dendrimers (32–34) are effective as antimicrobials, they do not easily break down and thus have limited potential as inhaled therapeutics. Chitosan-based oligosaccharides represent attractive scaffolds for NO delivery, as they are biodegradable and have low toxicity to mammalian cells (40, 41). We have previously reported that NO-releasing chitosan oligosaccharides are capable of NO storage/release and of eradicating P. aeruginosa biofilms under aerobic environments at concentrations nontoxic to mammalian cells (15). Here, we evaluate the antibacterial efficacy of NO-releasing chitosan oligosaccharides as a function of oxygen availability using nonmucoid, mucoid, and biofilm P. aeruginosa phenotypes.
MATERIALS AND METHODS
Materials.
Medium-molecular-weight chitosan, 2-methylaziridine, and tobramycin were purchased from Sigma-Aldrich (St. Louis, MO). Methyltrimethoxysilane (MTMOS) was purchased from Fluka (Buchs, Switzerland). (Heptadecafluoro-1,1,2,2-tetrahydrodecyl)trimethoxysilane (17 FTMS) was purchased from Gelest (Morrisville, PA). Nitric oxide gas was purchased from Praxair (Sanford, NC). Standardized NO (26.85 ppm; balance N2), argon (Ar), and nitrogen (N2) gases were purchased from Airgas National Welders (Durham, NC). Sodium methoxide was purchased from Acros Organics (Geel, Belgium). Distilled water was purified using a Millipore Milli-Q UV Gradient A-10 system (Bedford, MA). All common laboratory salts and reagents were purchased from Fisher Scientific (Pittsburgh, PA). All materials were used without further purification unless otherwise specified.
Bacterial strains and media.
The laboratory P. aeruginosa strain used in this study was strain K (PAK). The mucoid phenotype was the mucA22 isogenic mutant of the nonmucoid PAK strain. Both bacterial strains were a gift from Matthew Wolfgang, Department of Microbiology and Immunology, University of North Carolina (UNC) (Chapel Hill, NC). Clinical isolates were collected from patients at the UNC Hospital Clinical Microbiology Laboratory (Chapel Hill, NC). The clinical isolates were screened for tobramycin resistance using the Kirby-Bauer disk diffusion method according to standards published by the Clinical and Laboratory Standards Institute in document M100-S23 (42). All bacteria were grown in Luria Bertani (LB) broth (BD Biosciences, San Jose, CA) with the pH adjusted to 6.5 using 10 mM sodium phosphate. When indicated, potassium nitrate (15 mM) was added to the broth. Phosphate-buffered saline (PBS) was adjusted to pH 6.5 with 10 mM sodium phosphate. All bacterial media were adjusted to pH 6.5 to more accurately mimic the pH of CF mucus (22). Anaerobic media were kept in a Coy anaerobic chamber (Coy Laboratory Products, Ann Arbor, MI) with the lid loosened for 1 week prior to use.
Synthesis of COS.
2-Methylaziridine-modified chitosan oligosaccharides (COS) (specifically chitosan2-5k) were synthesized as previously described (15). Briefly, medium-molecular-weight chitosan (2.5 g) was oxidatively degraded to ∼5 kDa in 15% hydrogen peroxide for 1 h at 85°C. The nondegraded chitosan was removed by filtration. The remaining chitosan oligosaccharides were precipitated from solution with acetone, collected via centrifugation, and dried in vacuo. The ∼5-kDa chitosan oligosaccharides (500 mg) were then dissolved in water (10 ml). Concentrated hydrochloric acid (27.6 μl), water (250 μl), and 2-methylaziridine (356 μl) were then added to this solution. The reaction mixture was stirred for 5 days at 25°C, followed by 24 h at 75°C. The modified chitosan oligosaccharides were again precipitated with acetone, collected via centrifugation, and dried in vacuo. The 1H nuclear magnetic resonance (NMR) data for COS were (400 MHz, D2O, δ): 0.8–1.1 [NH2CH(CH3)CH2NH], 1.9 (C-7: CHNHCOCH3), 2.3–2.9 [NH2CH(CH3)CH2NHCH, C-2: NH2CH(CH3)CH2NHCH], 3.3–4.0 [C-3, C-4, C-5, C-6: OHCH, OCHCH(OH)CH(NH2)CH, OHCH2CH, OHCH2CH], and 4.4 [C-1: OCH(CHNH2)O].
Synthesis of COS-NO.
In order to impart NO storage and release, N-diazeniumdiolates were formed on the secondary amines of COS (15). Briefly, COS (15 mg) was dissolved in a solution of water (300 μl), methanol (700 μl), and 5.4 M sodium methoxide (25 μl) in a 1-dram vial equipped with a stir bar. The open vial was placed in a 160-ml Parr general purpose stainless steel pressure vessel and rigorously stirred. Oxygen was removed from the reaction vessel by purging with argon (10 s; 8 × 105 Pa) 3 times, followed by 3 additional long argon purges (10 min; 8 × 105 Pa). The vessel was then filled with potassium hydroxide-purified NO gas (10 × 105 Pa) for 72 h at room temperature. Afterward, the argon-purging procedure was repeated to remove unreacted NO. The N-diazeniumdolate-modified chitosan oligosaccharides (COS-NO) were precipitated in acetone, collected via centrifugation, dried in vacuo, and stored at −20°C as a yellow powder.
Chemiluminescence detection of NO release.
A Sievers (Boulder, CO) 280i chemiluminescence nitric oxide analyzer was used for chemiluminescence detection of NO from COS-NO (1.0 mg) in 30 ml of deoxygenated PBS (pH 6.5) at 37°C. The released NO was carried by N2 gas to the reaction vessel/detector at a flow rate of 80 ml/min. Additional N2 flow was supplied to the sample flask at 200 ml/min to match the collection rate of the instrument. The analysis was terminated when NO concentrations fell below 10 ppb NO/mg COS-NO. Prior to analysis, the instrument was calibrated with air passed through an NO zero filter (0 ppm NO) and 26.8 ppm of NO standard gas (balance N2).
Electrochemical detection of NO release.
NO-selective electrochemical sensors were fabricated in house as previously reported (43). Briefly, polished polycrystalline Pt disk electrodes (2 mm) sealed in Kel-F (CH Instruments, Austin, TX) were coated with an NO-selective membrane prepared by mixing MTMOS (60 μl), ethanol (300 μl), 17 FTMS (15 μl), water (80 μl), and 0.5 M hydrochloric acid (5 μl) for 1 h at 25°C. The resulting solution was spreadcast over the Pt electrode and dried overnight at room temperature. Amperometric NO measurements followed, using a three-electrode setup with the NO-selective membrane-modified Pt electrode as the working electrode, a Pt-coiled counter electrode, and an Ag/AgCl reference electrode. The applied potential for NO oxidation was +700 mV versus Ag/AgCl. Immediately prior to use, the NO sensors were calibrated by adding a known amount of PBS saturated with NO gas (1.9 mM) to deoxygenated PBS (pH 6.5). Saturated NO solutions were made on the day of use by degassing PBS (pH 6.5) for 30 min with Ar, followed by 20-min purging with NO gas. The sensors were immersed in 10.0 ml of PBS or LB broth (stirred; 37°C) and polarized at +700 mV versus Ag/AgCl until a stable baseline was achieved prior to the addition of COS-NO. The NO oxidation current was measured every 0.1 s and ceased when the current returned to its background value. Measurement of NO release in anaerobic media was carried out in a Coy anaerobic chamber. Total NO for 1.0-mg COS-NO/ml solutions are reported as the average and standard deviation for 4 or more separate measurements.
Planktonic bactericidal assays.
Bacteria were grown as overnight cultures, diluted 1:100 in fresh LB broth (with or without nitrate supplementation), and grown to mid-log phase (2 × 108 CFU/ml). These cultures were centrifuged, resuspended in PBS, and diluted to 2 × 106 CFU/ml in PBS. Each suspension was then added to vials containing 2-fold serial dilutions of COS-NO or COS controls and incubated at 37°C for 4 h with gentle shaking. Following treatment, the bacterial solutions were serially diluted, spiral plated on LB agar, and incubated for 24 h at 37°C. Colonies were enumerated using a Flash and Go colony counter (IUL, Farmingdale, NY). The minimum bactericidal concentration after a 4-h exposure (MBC4 h) was defined as the minimum concentration required to achieve a 3-log-unit reduction in viable bacteria (from 106 to 103 CFU/ml). The plating-counting method employed has a limit of detection of 2.5 × 103 CFU/ml (44). The corresponding NO dose was calculated by multiplying the MBC4 h of COS-NO (in milligrams per milliliter) with the available NO in aerobic and anaerobic PBS (in micromoles NO per milligram COS-NO).
Planktonic inhibition assays.
Bacteria were grown as overnight cultures, diluted 1:100 in fresh LB broth, grown to mid-log phase (2 × 108 CFU/ml), and diluted to 2 × 106 CFU/ml in LB broth. The bacterial cultures were then added to vials containing 2-fold serial dilutions of COS-NO or COS controls and incubated at 37°C for 18 h with gentle shaking. The MIC was determined to be the minimum concentration that prevented visible growth, defined as an optical density at 600 nm of <0.1. All untreated (control) cultures became visibly turbid during the 18-h growth period. The corresponding NO dose was calculated by multiplying the MIC of COS-NO (in milligrams per milliliter) with the available NO in aerobic and anaerobic broth (in micromoles NO per milligram COS-NO). Nitrate-supplemented LB was used for all stages of bacterial growth and exposure. Anaerobic experiments were performed in a Coy anaerobic chamber.
Biofilm eradication assays.
Bacteria were grown as overnight cultures, diluted 1:100 in fresh LB broth, and grown to mid-log phase (2 × 108 CFU/ml). The bacterial cultures were then diluted to 106 CFU/ml in diluted (25%) LB broth supplemented with 15 mM KNO3 (pH 6.5) and grown for 72 h at 37°C with gentle shaking. The viscous microcolony biofilms formed were easily separated from the growth media via pipetting. The biofilms were harvested by placing a pipette tip near the center of the biofilm and applying suction. The biofilms were then washed by injection into PBS and extracted using the same pipetting procedure to remove planktonic or loosely associated bacteria. The freshly washed biofilms (250 μl) were combined with 750 μl of PBS (pH 6.5); added to vials containing COS, COS-NO, or tobramycin; and incubated with gentle shaking for 18 h at 37°C. After treatment, the biofilms were washed via pipetting in PBS to remove excess antibacterial agent, transferred to 750 μl of PBS (pH 6.5), and gently sonicated to disrupt the biofilm matrix. The dispersed biofilms were vortexed, serially diluted, plated, and enumerated on LB agar. The minimum biofilm eradication concentration at 18 h (MBEC18 h) was defined as the concentration that caused a 5-log-unit reduction in viable bacteria (i.e., 108 to 103 CFU/ml) after the 18-h treatment. The corresponding NO dose was calculated by multiplying the MBEC18 h of COS-NO (in milligrams per milliliter) with the available NO in aerobic and anaerobic PBS (in micromoles NO per milligram COS-NO).
Statistical analysis.
All data are expressed as the mean ± 1 standard deviation and were analyzed for significance (P < 0.05) with a two-tailed Student t test.
RESULTS
Nitric oxide release from COS-NO in media.
Nitric oxide release from COS-NO was measured via chemiluminescence in deoxygenated PBS (pH 6.5) at 37°C to yield total NO release payloads of 0.86 ± 0.05 μmol NO/mg, with an overall release duration of 10.2 ± 2.7 h (see Fig. S1 in the supplemental material). While NO release from macromolecular scaffolds is generally measured in deoxygenated medium, measuring the amount of bioavailable (i.e., nonscavenged) NO is critical for elucidating the biocidal dose-response relationship of NO under the intended conditions, as NO is rapidly scavenged by oxygen and proteins in biological media (45). Unfortunately, foaming associated with nutrient-rich medium makes chemiluminescence detection difficult and irreproducible (43). Thus, we turned to amperometric NO detection to carry out NO measurements in broth (LB).
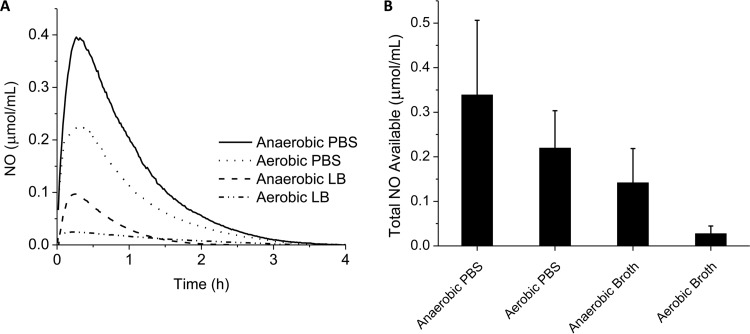
In the absence of NO scavenging (i.e., in deoxygenated PBS), amperometric measurements revealed a total NO payload of 0.34 ± 0.17 μmol/ml from the 1.0-mg/ml solution of COS-NO over 4 h (Fig. 1). As might be expected, both the total NO payload and the release duration of COS-NO measured via amperometry were reduced compared to chemiluminescence detection. These decreases are common for electrochemical sensors that are based on the diffusion of NO to the working electrode and the inherent loss of NO to the ambient atmosphere (43).
FIG 1.
Electrochemical measurements of available NO in media. (A) Representative NO release profiles for 1.0 mg/ml of COS-NO in anaerobic PBS, aerobic PBS, anaerobic LB broth, and aerobic LB broth. (B) The values from panel A were integrated to find the total concentrations of available NO in a 1.0-mg/ml solution of COS-NO in biological media after 4 h. The error bars indicate standard deviations.
Under aerobic conditions, oxygen scavenging reduced the amount of free NO available in PBS by approximately 35% (0.22 ± 0.08 μmol NO/ml). Nutrient broth (LB) further diminished the available NO payload via scavenging of the NO by proteins in the broth. The amount of NO available in anaerobic LB broth was reduced to 0.14 ± 0.08 μmol NO/ml, a 66% reduction relative to anaerobic PBS. Further reductions (0.027 ± 0.017 μmol/ml) were observed in aerobic LB broth due to reaction of NO with oxygen.
Bactericidal action of the COS scaffold.
To confirm that NO and not the scaffold was responsible for the observed bacterial killing, all bacterial assays were performed using NO-releasing and control (i.e., non-NO-releasing) chitosan oligosaccharides. In MBC4 h assays of planktonic cells, COS did not influence bacterial viability at 1× or 10× the MBC4 h of COS-NO, indicating that the chitosan oligosaccharide alone was not bactericidal (see Fig. S2 in the supplemental material). Similarly, bacterial viability was not reduced upon treatment of the biofilms with 4.0 mg COS/ml (1× the MBEC18 h) for 18 h under both aerobic and anaerobic conditions (see Fig. S3 in the supplemental material). Based on these data, the bactericidal activity of COS-NO was attributed solely to the effects of NO and not to toxicity of the COS scaffold.
Effect of oxygen on bactericidal action of NO against planktonic P. aeruginosa.
The biocidal action of NO was evaluated with respect to the oxygen concentration in the treatment medium by exposing planktonic cultures to COS-NO in both aerobic and anaerobic PBS (pH 6.5) (Table 1). When grown under aerobic conditions, the same concentration of COS-NO (100 μg/ml) was required to eradicate both the mucoid and nonmucoid strains. Furthermore, oxygen in the exposure medium did not alter the bactericidal concentration of COS-NO. However, the NO dose delivered was slightly greater under anaerobic exposure conditions due to the decreased reactions between NO and oxygen. For example, the bactericidal NO dose was 0.022 ± 0.008 μmol NO/ml under aerobic conditions versus 0.034 ± 0.017 μmol NO/ml in deoxygenated (anaerobic) medium. This difference in the NO payload was not statistically significant.
TABLE 1.
Effects of oxygen on nongrowing planktonic culturesa
| Strain | Growth medium | Aerobic exposure |
Anaerobic exposure |
||
|---|---|---|---|---|---|
| MBC4 h (μg COS-NO/ml) | NO doseb (μmol NO/ml) | MBC4 h (μg COS-NO/ml) | NO doseb (μmol NO/ml) | ||
| Nonmucoid | Aerobic | 100 | 0.022 ± 0.008 | 100 | 0.034 ± 0.017 |
| Anaerobic | 100 | 0.022 ± 0.008 | 100 | 0.034 ± 0.017 | |
| Mucoid | Aerobic | 100 | 0.022 ± 0.008 | 100 | 0.034 ± 0.017 |
| Anaerobic | 200 | 0.044 ± 0.016 | 200 | 0.068 ± 0.033 | |
P. aeruginosa cultures were grown in LB broth (plus 15 mM KNO3) under aerobic or anaerobic conditions and then exposed to COS-NO in PBS (pH 6.5) for 4 h under aerobic or anaerobic conditions.
Determined via amperometry. The values are presented as means ± standard deviations for 3 or more pooled experiments.
Bacterial cultures were also grown aerobically and anaerobically to determine if oxygen in the growth medium affected P. aeruginosa susceptibility to NO. As anaerobic growth requires nitrate, all assays were carried out using nitrate-supplemented LB medium to enable direct comparison. The absence of oxygen in the growth medium had no effect on the susceptibility of nonmucoid P. aeruginosa to NO. However, strict anaerobic growth of mucoid P. aeruginosa increased the tolerance of the strain for NO by 2-fold (MBC4 h = 0.044 ± 0.016 and 0.022 ± 0.008 μmol NO/ml for anaerobic and aerobic growth conditions, respectively) (Table 1). The increased tolerance for NO was observed under both aerobic and anaerobic exposure conditions.
Inhibition of planktonic P. aeruginosa growth by COS-NO.
MIC assays were performed to evaluate the efficacy of COS-NO during bacterial growth under aerobic and anaerobic environments (Table 2). Under aerobic conditions, the nonmucoid phenotype was more tolerant of COS-NO than the mucoid strain, with inhibitory doses of 800 μg COS-NO/ml (0.022 ± 0.014 μmol NO/ml) versus 400 μg COS-NO/ml (0.011 ± 0.007 μmol NO/ml), respectively. Anaerobic conditions decreased the MIC to 100 μg COS-NO/ml (0.014 μmol NO/ml) for both phenotypes. While the COS-NO concentration required to inhibit growth was reduced in anaerobic environments, the NO dose delivered was not significantly lower, indicating that NO lost to reaction with oxygen accounts for the increased MICs against COS-NO under aerobic conditions.
TABLE 2.
Influence of oxygen on the inhibitory efficacy of COS-NOa
| Growth medium | Nonmucoid strain |
Mucoid strain |
||
|---|---|---|---|---|
| MIC (μg COS-NO/ml) | NO doseb (μmol NO/ml) | MIC (μg COS-NO/ml) | NO doseb (μmol NO/ml) | |
| Aerobic | 800 | 0.022 ± 0.014 | 400 | 0.011 ± 0.007 |
| Anaerobic | 100 | 0.014 ± 0.008 | 100 | 0.014 ± 0.008 |
Bacterial cultures in mid-log-phase growth were diluted to 2 × 106 CFU/ml in LB broth (plus 15 mM KNO3) with COS-NO and grown for 18 h under aerobic or anaerobic conditions. The MIC was determined as the concentration of COS-NO that visibly inhibited growth.
Determined via amperometry. The values are presented as means ± standard deviations for 3 or more pooled experiments.
Inhibition of growth by COS-NO for clinical isolates, including tobramycin-resistant strains.
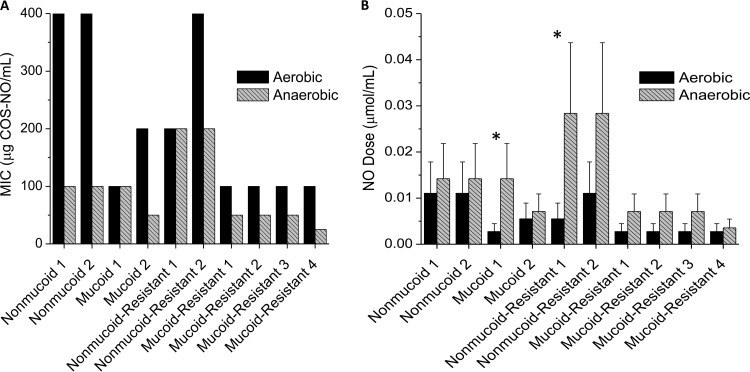
To ensure that the increased inhibition of bacterial growth by COS-NO under anaerobic conditions was not solely a function of the laboratory P. aeruginosa strain used, 10 clinical isolates of P. aeruginosa were tested, including mucoid, nonmucoid, and tobramycin-resistant isolates. The concentrations of COS-NO required to inhibit growth under anaerobic conditions were less than or equal to those under aerobic conditions for all 10 isolates (Fig. 2A). There was no statistical difference in the NO dose required to inhibit growth for most of the strains tested (Fig. 2B). However, two isolates (Fig. 2B, asterisks) showed a statistically significant increase in the NO dose required to inhibit growth under anaerobic conditions relative to aerobic conditions. Overall, COS-NO was more effective at inhibiting growth under anaerobic environments, but the efficacy of NO was unchanged for most of the isolates tested.
FIG 2.
MICs against clinical isolates of P. aeruginosa. (A) Minimum concentrations of COS-NO resulting in no bacterial growth for nonmucoid, mucoid, and tobramycin-resistant isolates. (B) Corresponding NO dose for each isolate under aerobic and anaerobic environments. All bacteria were grown and exposed in LB medium supplemented with nitrate (pH 6.5) for 18 h. Statistically significant differences (P < 0.05) between aerobic and anaerobic NO payloads are indicated by the asterisks. The error bars indicate standard deviations.
As with the laboratory strain, differences were seen between the mucoid and nonmucoid phenotypes (Fig. 2). Mucoid strains were more susceptible to COS-NO, as indicated by the low MIC range (25 to 200 μg COS-NO/ml) relative to nonmucoid strains (MIC range, 100 to 400 μg COS-NO/ml). No apparent differences were observed between tobramycin-susceptible and tobramycin-resistant strains of the same phenotype; however, more isolates would be needed to confirm the statistical significance of these trends.
Biofilm eradication by COS-NO and tobramycin.
As bacterial biofilms exist in both aerobic and anaerobic environments (46), it was important to determine how oxygen concentrations affected the antibiofilm activity of COS-NO. Under aerobic conditions, highly viscous microcolony biofilms were formed (∼250 μl in volume) with bacterial viability of 4.0 ± 0.6 × 108 and 2.5 ± 0.5 × 108 CFU/ml for nonmucoid and mucoid phenotypes, respectively. Of note, nitrate supplementation was required to prevent phenotypic switching from the mucoid to the nonmucoid phenotype (47). Under anaerobic growth conditions, we were unable to cause the bacteria to form robust biofilms even after 7 days of growth.
Bacterial biofilms were exposed to COS-NO for 18 h in PBS (pH 6.5) under aerobic or anaerobic conditions. The MBEC18 h values for the two phenotypes were 4,000 μg COS-NO/ml and 1,000 μg COS-NO/ml (0.88 ± 0.33 and 0.34 ± 0.17 μmol NO/ml) under aerobic and anaerobic conditions, respectively (Table 3). These results indicate that NO released from a scaffold is equally effective at eliminating biofilms derived from nonmucoid and mucoid strains. Moreover, NO is significantly more effective at eliminating biofilms in the absence of oxygen.
TABLE 3.
Bactericidal efficacy of COS-NO against P. aeruginosa biofilmsa
| Strain | Aerobic exposure |
Anaerobic exposure |
||
|---|---|---|---|---|
| MBEC (μg COS-NO/ml) | NO doseb (μmol NO/ml) | MBEC (μg COS-NO/ml) | NO doseb (μmol NO/ml) | |
| Nonmucoid | 4,000 | 0.88 ± 0.33 | 1,000 | 0.34 ± 0.17 |
| Mucoid | 4,000 | 0.88 ± 0.33 | 1,000 | 0.34 ± 0.17 |
Biofilms were exposed to COS-NO in PBS (pH 6.5) for 18 h under aerobic or anaerobic conditions. The MBEC18 h is reported as the concentration of COS-NO required for 5-log-unit reduction in biofilm viability.
Determined via amperometry. The values are presented as means ± standard deviations for 3 or more pooled experiments.
The MBEC18 h of tobramycin against biofilms was determined under the same conditions (i.e., 18-h exposure in PBS) to allow comparison of NO to current antibiotic therapies. Under aerobic environments, the nonmucoid strain was eradicated at lower concentrations of tobramycin than the mucoid strain (200 and 800 μg/ml, respectively) (Table 4). Both strains required greater tobramycin levels (1,600 μg/ml) to eradicate bacterial biofilms under anaerobic conditions.
TABLE 4.
Bactericidal efficacy of tobramycin against P. aeruginosa biofilmsa
| Strain | MBEC [μg/ml (μmol/ml)] |
|
|---|---|---|
| Aerobic exposure | Anaerobic exposure | |
| Nonmucoid | 200 (0.43) | 1,600 (3.42) |
| Mucoid | 800 (1.71) | 1,600 (3.42) |
Biofilms were exposed to tobramycin in PBS (pH 6.5) for 18 h under aerobic or anaerobic conditions. The MBEC18 h is reported as the concentration of COS-NO required for 5-log-unit reduction in biofilm viability.
DISCUSSION
We have previously reported on the antibacterial activity of NO against planktonic and biofilm-based P. aeruginosa (14, 15, 39). However, little is understood regarding how oxygen and the bacterial phenotype impact NO's efficacy. Such knowledge is critical in the development of NO-based therapeutics. Water-soluble NO-releasing chitosan oligosaccharides were used as the NO release scaffold in the studies described here due their biocompatibility and ability to be degraded in vivo (40, 41, 48). Although chitosan is a known bactericidal agent, the reduced molecular mass (to ensure water solubility) and 2-methylaziridine modification (for NO donor addition) resulted in a material with no bactericidal activity (see Fig. S2 and S3 in the supplemental material).
It is well known that NO reacts with oxygen and superoxide to form highly reactive intermediates that facilitate bacterial killing through oxidative and nitrosative stresses (19, 49). As oxygen plays an integral role in the antibacterial action of NO, anaerobic environments may reduce the biocidal efficacy of NO (20, 50). However, NO also reacts with oxygen to form nitrate and nitrite. These seemingly paradoxical roles of oxygen in NO-mediated killing are not fully understood. Therefore, we carried out electrochemical measurements of NO under aerobic and anaerobic conditions to quantify the amounts of bioavailable NO. Under aerobic conditions, the measured NO decreased by 35% compared to anaerobic conditions (Fig. 1). To elucidate the effects of oxygen availability in treatment media, bacteria were first grown aerobically and then exposed to COS-NO in aerobic or anaerobic PBS. Identical concentrations of COS-NO were required to kill P. aeruginosa regardless of the treatment conditions. Due to the reaction of NO with oxygen, the bioavailable concentration of NO (i.e., the NO dose) was slightly, but not significantly, higher under anaerobic conditions (Table 1). As such, oxygen availability in the treatment medium has no statistically significant effect on the biocidal activity of NO released from COS-NO.
While the oxygen concentration in the exposure medium did not alter the bactericidal efficacy of NO, the presence of oxygen during bacterial growth did influence P. aeruginosa susceptibility to NO. Anaerobic growth conditions reduce the efficacy of current antibiotics by altering certain properties of the bacteria, such as alginate production (46) and metabolic rates (10). To separate these factors, MBC4 h assays were performed under nonnutritive conditions to minimize the effects of bacterial metabolism on the bactericidal activity of NO. When bacteria were grown under anaerobic conditions, the efficacy of NO released from the chitosan oligosaccharide scaffold was decreased against the mucoid, but not the nonmucoid, phenotype, indicating that growing mucoid bacteria without oxygen significantly alters their defense against NO (Table 1). Worlitzsch et al. previously reported that P. aeruginosa produces a protective alginate exopolysaccharide that is 50% thicker when grown anaerobically (46). As alginate restricts the diffusion of oxygen, the increased thickness of this protective layer could potentially prevent NO diffusion into the bacteria, therefore requiring a larger NO dose for killing.
To study the role of anaerobic growth on the efficacy of NO and COS-NO, we evaluated the inhibition of P. aeruginosa growth by COS-NO in nutrient-rich medium under both aerobic and anaerobic conditions. In contrast to the static conditions of MBC4 h assays, bacteria are actively growing during inhibition assays. Comparison of MIC values under oxygen and oxygen-free environments showed that the efficacy of COS-NO was enhanced under anaerobic conditions while there was no statistical difference in the corresponding NO dose. This behavior was observed in the laboratory strains (Table 2) and most of the clinical isolates tested (Fig. 2). As the efficacy of NO is not reduced under anaerobic environments, NO-based treatments represent a potential alternative to current antibiotic treatments, including aminoglycosides and β-lactams, which are less effective under anaerobic conditions because their mechanism of action requires actively dividing cells (51, 52).
It is important to characterize the antibiofilm activities of antibacterial agents, as P. aeruginosa exists as biofilms on medical implants (e.g., catheters) (53, 54), on burn wounds (55, 56), and in the airways of patients with cystic fibrosis (57, 58). As shown in Table 4, mucoid biofilms are significantly more resilient against tobramycin than nonmucoid biofilms. Hentzer et al. attributed decreased antibiotic efficacy against mucoid strains to the overproduction of alginate (59). As has been previously reported (60, 61), low-oxygen conditions further decrease the effectiveness of tobramycin (the MBEC18 h is increased to 1,600 μg/ml under anaerobic conditions). While tobramycin is a highly effective antipseudomonal agent, these factors compromise its ability to kill P. aeruginosa biofilms in oxygen-free environments. In contrast, NO released from the chitosan scaffold was equally effective at eradicating mucoid and nonmucoid biofilms. Similarly, the antibiofilm activity was not reduced under anaerobic environments (Table 3). Furthermore, the NO dose required for biofilm eradication under anaerobic conditions is 0.34 ± 0.17 μmol NO/ml, 10-fold lower than that of tobramycin (3.42 μmol/ml) (Tables 3 and 4).
In conclusion, these studies examined the susceptibilities of nonmucoid, mucoid, and biofilm P. aeruginosa phenotypes to NO-releasing chitosan oligosaccharides as a function of oxygen availability. The antibacterial activity of NO-releasing chitosan oligosaccharides was enhanced in oxygen-free environments, despite a concomitant decrease in the number of possible mechanisms available to kill bacteria (i.e., fewer toxic by-products from the reactions of NO and oxygen). Furthermore, the antibiofilm action of NO was more effective than that of tobramycin and was not influenced by the bacterial phenotype. When combined with NO's significant biocidal action against P. aeruginosa, these results suggest that NO-releasing chitosan oligosaccharides may represent a potential alternative to traditional antibiotics, particularly when treating biofilms or in low-oxygen environments. We are currently seeking to enhance the NO payloads and evaluate the effects of NO release kinetics on the antibacterial efficacy of chitosan oligosaccharides.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health (AI 112029).
We thank Matthew Wolfgang of the Department of Microbiology and Immunology at the University of North Carolina at Chapel Hill for providing the PAK and PAK mucA22 laboratory strains. We also thank Peter Gilligan and Anthony Tran of the Clinical Microbiology-Immunology Laboratories at the UNC Hospitals for providing P. aeruginosa clinical isolates.
Mark H. Schoenfisch declares a competing financial interest: he is a cofounder and member of the board of directors and maintains a financial interest in Novan Therapeutics, Inc. Novan Therapeutics, Inc. provided partial funding for this research and is commercializing macromolecular nitric oxide storage and release vehicles for dermatological applications.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01208-15.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.Mesaros N, Nordmann P, Plesiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. [DOI] [PubMed] [Google Scholar]
- 3.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole K. 2002. Outer membranes and efflux: the path to multidrug resistance in Gram-negative bacteria. Curr Pharm Biotechnol 3:77–98. doi: 10.2174/1389201023378454. [DOI] [PubMed] [Google Scholar]
- 5.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Stewart PS, William Costerton J. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 7.Stewart PS. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol 292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 8.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 9.Xu KD, Stewart PS, Xia F, Huang C-T, McFeters GA. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol 64:4035–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Sun Yoon S, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ. 2002. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv Drug Deliv Rev 54:1425–1443. doi: 10.1016/S0169-409X(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 11.Schlessinger D. 1988. Failure of aminoglycoside antibiotics to kill anaerobic, low-pH, and resistant cultures. Clin Microbiol Rev 1:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. 2009. Nitric oxide-mediated dispersal in single-and multi-species biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol 2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hetrick EM, Shin JH, Paul HS, Schoenfisch MH. 2009. Anti-biofilm efficacy of nitric oxide-releasing silica nanoparticles. Biomaterials 30:2782–2789. doi: 10.1016/j.biomaterials.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, Slomberg DL, Schoenfisch MH. 2014. Nitric oxide-releasing chitosan oligosaccharides as antibacterial agents. Biomaterials 35:1716–1724. doi: 10.1016/j.biomaterials.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller CC, Hergott CA, Rohan M, Arsenault-Mehta K, Döring G, Mehta S. 2013. Inhaled nitric oxide decreases the bacterial load in a rat model of Pseudomonas aeruginosa pneumonia. J Cyst Fibros 12:817–820. doi: 10.1016/j.jcf.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Webert KE, Vanderzwan J, Duggan M, Scott JA, McCormack DG, Lewis JF, Mehta S. 2000. Effects of inhaled nitric oxide in a rat model of Pseudomonas aeruginosa pneumonia. Crit Care Med 28:2397–2405. [DOI] [PubMed] [Google Scholar]
- 18.Möller MN, Li Q, Lancaster JR, Denicola A. 2007. Acceleration of nitric oxide autoxidation and nitrosation by membranes. IUBMB Life 59:243–248. doi: 10.1080/15216540701311147. [DOI] [PubMed] [Google Scholar]
- 19.Fang FC. 1997. Perspectives series: host/pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest 99:2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiro S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev 31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 21.Carpenter AW, Schoenfisch MH. 2012. Nitric oxide release. Part II. Therapeutic applications. Chem Soc Rev 41:3742–3752. doi: 10.1039/c2cs15273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren B, Zhang N, Yang J, Ding H. 2008. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol Microbiol 70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 24.Loscalzo J, Welch G. 1995. Nitric oxide and its role in the cardiovascular system. Prog Cardiovasc Dis 38:87–104. doi: 10.1016/S0033-0620(05)80001-5. [DOI] [PubMed] [Google Scholar]
- 25.Samama CM, Diaby M, Fellahi J-L, Mdhafar A, Eyraud D, Arock M, Guillosson J-J, Coriat P, Rouby J-J. 1995. Inhibition of platelet aggregation by inhaled nitric oxide in patients with acute respiratory distress syndrome. Anesthesiology 83:56–65. doi: 10.1097/00000542-199507000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari CKB, França EL, Honorio-França AC. 2009. Nitric oxide, health and disease. J Appl Biomed 7:163–173. [Google Scholar]
- 27.Riccio DA, Schoenfisch MH. 2012. Nitric oxide release. Part I. Macromolecular scaffolds. Chem Soc Rev 41:3731–3741. doi: 10.1039/c2cs15272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seabra AB, Justo GZ, Haddad PS. 28 January 2015. State of the art, challenges and perspectives in the design of nitric oxide-releasing polymeric nanomaterials for biomedical applications. Biotechnol Adv S0734-9750(15)00018-X. doi: 10.1016/j.biotechadv.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Carpenter AW, Reighard KP, Saavedra JE, Schoenfisch MH. 2013. O2-Protected diazeniumdiolate-modified silica nanoparticles for extended nitric oxide release from dental composites. Biomater Sci 1:456–459. doi: 10.1039/c3bm00153a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin JH, Metzger SK, Schoenfisch MH. 2007. Synthesis of nitric oxide-releasing silica nanoparticles. J Am Chem Soc 129:4612–4619. doi: 10.1021/ja0674338. [DOI] [PubMed] [Google Scholar]
- 31.Stevens EV, Carpenter AW, Shin JH, Liu J, Der CJ, Schoenfisch MH. 2010. Nitric oxide-releasing silica nanoparticle inhibition of ovarian cancer cell growth. Mol Pharm 7:775–785. doi: 10.1021/mp9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Slomberg DL, Shah A, Schoenfisch MH. 2013. Nitric oxide-releasing amphiphilic poly(amidoamine) (PAMAM) dendrimers as antibacterial agents. Biomacromolecules 14:3589–3598. doi: 10.1021/bm400961r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun B, Slomberg DL, Chudasama SL, Lu Y, Schoenfisch MH. 2012. Nitric oxide-releasing dendrimers as antibacterial agents. Biomacromolecules 13:3343–3354. doi: 10.1021/bm301109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worley BV, Slomberg DL, Schoenfisch MH. 2014. Nitric oxide-releasing quaternary ammonium-modified poly(amidoamine) dendrimers as dual action antibacterial agents. Bioconjug Chem 25:918–927. doi: 10.1021/bc5000719. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Sun B, Li C, Schoenfisch MH. 2011. Structurally diverse nitric oxide-releasing poly(propylene imine) dendrimers. Chem Mater 23:4227–4233. doi: 10.1021/cm201628z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stasko NA, Schoenfisch MH. 2006. Dendrimers as a scaffold for nitric oxide release. J Am Chem Soc 128:8265–8271. doi: 10.1021/ja060875z. [DOI] [PubMed] [Google Scholar]
- 37.Carpenter AW, Slomberg DL, Rao KS, Schoenfisch MH. 2011. Influence of scaffold size on bactericidal activity of nitric oxide-releasing silica nanoparticles. ACS Nano 5:7235–7244. doi: 10.1021/nn202054f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter AW, Worley BV, Slomberg DL, Schoenfisch MH. 2012. Dual action antimicrobials: nitric oxide release from quaternary ammonium-functionalized silica nanoparticles. Biomacromolecules 13:3334–3342. doi: 10.1021/bm301108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slomberg DL, Lu Y, Broadnax AD, Hunter RA, Carpenter AW, Schoenfisch MH. 2013. Role of size and shape on biofilm eradication for nitric oxide-releasing silica nanoparticles. ACS Appl Mater Interfaces 5:9322–9329. doi: 10.1021/am402618w. [DOI] [PubMed] [Google Scholar]
- 40.Grenha A, Al-Qadi S, Seijo B, Remuñán-López C. 2010. The potential of chitosan for pulmonary drug delivery. J Drug Deliv Sci Technol 20:33–43. doi: 10.1016/S1773-2247(10)50004-2. [DOI] [Google Scholar]
- 41.Kean T, Thanou M. 2010. Biodegradation, biodistribution and toxicity of chitosan. Adv Drug Deliv Rev 62:3–11. doi: 10.1016/j.addr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: 23rd informational supplement M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 43.Hunter RA, Storm WL, Coneski PN, Schoenfisch MH. 2013. Inaccuracies of nitric oxide measurement methods in biological media. Anal Chem 85:1957–1963. doi: 10.1021/ac303787p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breed RS, Dotterrer W. 1916. The number of colonies allowable on satisfactory agar plates. J Bacteriol 1:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coneski PN, Schoenfisch MH. 2012. Nitric oxide release. Part III. Measurement and reporting. Chem Soc Rev 41:3753–3758. doi: 10.1039/c2cs15271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyckoff TJ, Thomas B, Hassett DJ, Wozniak DJ. 2002. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 148:3423–3430. [DOI] [PubMed] [Google Scholar]
- 48.Illum L. 1998. Chitosan and its use as a pharmaceutical excipient. Pharm Res 15:1326–1331. doi: 10.1023/A:1011929016601. [DOI] [PubMed] [Google Scholar]
- 49.Jones ML, Ganopolsky JG, Labbé A, Wahl C, Prakash S. 2010. Antimicrobial properties of nitric oxide and its application in antimicrobial formulations and medical devices. Appl Microbiol Biotechnol 88:401–407. doi: 10.1007/s00253-010-2733-x. [DOI] [PubMed] [Google Scholar]
- 50.Brunelli L, Crow JP, Beckman JS. 1995. The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch Biochem Biophys 316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- 51.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, Elkins M, Thompson B, MacLeod C, Aaron SD. 2005. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol 43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aaron SD, Ferris W, Ramotar K, Vandemheen K, Chan F, Saginur R. 2002. Single and combination antibiotic susceptibilities of planktonic, adherent, and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J Clin Microbiol 40:4172–4179. doi: 10.1128/JCM.40.11.4172-4179.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donlan R. 2008. Biofilms on central venous catheters: is eradication possible? Curr Top Microbiol Immunol 322:133–161. [DOI] [PubMed] [Google Scholar]
- 54.Cole SJ, Records AR, Orr MW, Linden SB, Lee VT. 2014. Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect Immun 82:2048–2058. doi: 10.1128/IAI.01652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison-Balestra C, Cazzaniga AL, Davis SC, Mertz PM. 2003. A wound-isolated Pseudomonas aeruginosa grows a biofilm in vitro within 10 hours and is visualized by light microscopy. Dermatol Surg 29:631–635. [DOI] [PubMed] [Google Scholar]
- 56.Kennedy P, Brammah S, Wills E. 2010. Burns, biofilm and a new appraisal of burn wound sepsis. Burns 36:49–56. doi: 10.1016/j.burns.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 57.Høiby N, Ciofu O, Bjarnsholt T. 2010. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- 58.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock RE, Parsek MR, Noah TL, Boucher RC, Hassett DJ. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3:593–603. doi: 10.1016/S1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 59.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. 2004. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Field T, White A, Elborn J, Tunney M. 2005. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur J Clin Microbiol Infect Dis 24:677–687. doi: 10.1007/s10096-005-0031-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.