Abstract
Enterobacter cloacae is among the most important pathogens responsible for nosocomial infections and outbreaks. In this study, 77 Enterobacter isolates were collected: 27 isolates from Algerian hospitals (in Constantine, Annaba, and Skikda) and 50 isolates from Marseille, France. All strains were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS). Antibiotic susceptibility testing was performed by the disk diffusion method. PCR was used to detect extended-spectrum-beta-lactamase (ESBL)-encoding, fluoroquinolone resistance-encoding, and aminoglycoside-modifying enzyme (AME) genes. Epidemiological typing was performed using MALDI-TOF MS with data mining approaches, along with multilocus sequence typing (MLST). Sixty-eight isolates (27 from Algeria, 41 from Marseille) were identified by MALDI-TOF MS as E. cloacae. Resistance to antibiotics in the Algerian isolates was significantly higher than that in the strains from Marseille, especially for beta-lactams and aminoglycosides. Eighteen of the 27 Algerian isolates and 11 of the 41 Marseille isolates possessed at least one ESBL-encoding gene: blaCTX-M and/or blaTEM. AME genes were detected in 20 of the 27 Algerian isolates and 8 of the 41 Marseille isolates [ant(2″)-Ia, aac(6′)-Ib-cr, aadA1, aadA2, and armA]. Conjugation experiments showed that armA was carried on a transferable plasmid. MALDI-TOF typing showed three separate clusters according to the geographical distribution and species level. An MLST-based phylogenetic tree showed a clade of 14 E. cloacae isolates from a urology unit clustering together in the MALDI-TOF dendrogram, suggesting the occurrence of an outbreak in this unit. In conclusion, the ability of MALDI-TOF to biotype strains was confirmed, and surveillance measures should be implemented, especially for Algerian patients hospitalized in France.
INTRODUCTION
Enterobacter cloacae is an opportunistic pathogen that can cause several type of infections in the lower respiratory tract, surgical sites, urinary tract (1), and central nervous system (2); moreover, it is frequently associated with nosocomial infections in outbreaks, and thus there is a need for rapid detection and typing of such strains. Although multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) are good approaches to identify the spread of a given clone in an outbreak, these techniques remain time-consuming with a substantial cost. Recently, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been introduced in clinical microbiology as a routine tool for rapid identification of bacteria at the species level (3), but it also could be used as a simple tool for typing in nosocomial outbreaks of infections with bacteria such as Acinetobacter baumannii (4). Antibiotic resistance in E. cloacae is being increasingly reported, with the acquisition of genes encoding extended-spectrum beta-lactamases (ESBLs) (5) and aminoglycoside-modifying enzymes (AMEs) being the most critical factor in antibiotic resistance (6). Therefore, epidemiological plans of antibiotic resistance surveillance have been deployed for several years to measure the evolution of the antibiotic resistance phenomenon and to detect outbreaks (7, 8). Some recent investigations reported in Algeria have revealed an increase in ESBL-producing E. cloacae, with blaCTX-M1, blaCTX-M3, blaCTX-M15, blaVEB-1, blaSHV-12, and blaTEM (9, 10) being the most frequent genes detected. However, the epidemiology and molecular support of resistance to aminoglycosides due to AMEs in E. cloacae have never been studied in Algeria, even though they have been reported recently in Acinetobacter baumannii and Klebsiella pneumoniae (11, 12). Indeed, we recently reported an outbreak in a pediatric Algerian hospital of K. pneumoniae infections with strains harboring both genes encoding ESBLs and the armA 16S rRNA methyltransferase gene. Here, we investigate and compare the prevalences and molecular support of antibiotic resistance to aminoglycosides, beta-lactams, and fluoroquinolones in a series of E. cloacae clinical isolates from Constantine, Annaba, and Skikda in Algeria and from Marseille in France. Moreover, we used MALDI-TOF MS and data mining approaches, along with MLST, to study relationships between these isolates to assess whether MALDI-TOF can be used for real-time detection of multidrug-resistant (MDR) E. cloacae during an outbreak.
MATERIALS AND METHODS
Bacterial strains.
We collected consecutive and nonredundant clinical isolates of Enterobacter spp. from several clinical samples from eastern Algerian hospitals (Constantine, Annaba, and Skikda) between March 2012 and March 2013, as well as clinical isolates from Marseille, France, between October 2013 and December 2013.
MALDI-TOF MS identification and clustering.
The Enterobacter isolates were plated on MacConkey agar (bioMérieux, Marcy l'Étoile, France) and incubated for 18 h to 24 h at 37°C. Isolated colonies of each strain were selected and used for MALDI-TOF MS identification using the Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany), as previously described (13). The obtained spectra were downloaded into a MALDI Biotyper 3.0 system (Bruker Daltonics) and used to create a single main spectrum for each Enterobacter isolate. Subsequently, an MSP (main spectrum) dendrogram was constructed using MALDI Biotyper 3.0, and clusters were then analyzed according to the arbitrary distance level.
Antibacterial susceptibility testing.
Antibiotic susceptibility testing was performed by the disk diffusion method on Mueller-Hinton agar (Becton, Dickinson and Company, France), as recommended by the Antibiogram Committee of the French Society for Microbiology (CA-SFM 2013 Ver. June [http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM2013vjuin.pdf]). Phenotypic identification of ESBLs was determined in a systematic manner using a double-disk synergy test by placing disks of ceftazidime, cefotaxime, cefepime, and aztreonam equidistant from a disk with a combination of amoxicillin and clavulanic acid (12). The test was considered positive when a “champagne cork” aspect was observed.
Molecular characterization of antibiotic resistance-encoding genes.
DNA extraction from all isolates was performed using EZ1 DNA extraction kits (Qiagen, Courtaboeuf, France) with the EZ1 Advanced XL bio-robot according to the manufacturer's instructions. ESBL-encoding genes (blaCTX-M, blaTEM, blaSHV, blaVEB, blaGES, blaPER) (14), quinolone antibiotic resistance determinants (qnrA, qnrB) (15), and AME-encoding genes [armA, aad, rmt, aph, ant2, aac(6)-Ib] (16) in both the Algerian and Marseille isolates were detected by PCR, as previously described (16). Positive PCR products were purified and sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). The analysis was performed using an ABI 3130 automated sequencer (Applied Biosystems, Foster City, CA, USA). The obtained sequences were analyzed using CodonCode Aligner software and then aligned with those from the National Center for Biotechnology Information and ARG-ANNOT sequence databases (17) using the BLAST program.
Conjugation and resistance transfer.
The transfer of antibiotic resistance by conjugation was performed using five E. cloacae isolates as donors (three from Algeria and two from Marseille) and E. coli J53, which is resistant to sodium azide, as the recipient strain (12). Both the recipient and donor strains were cultivated in tryptic soy broth (Becton, Dickinson and Company, France) at 37°C for 24 h, mixed at a 1:10 donor-to-recipient ratio, and incubated overnight at 37°C with shaking. The transconjugants were selected by plating 10 μl of the mixture on Luria-Bertani agar (Becton, Dickinson and Company, France) containing 200 mg/liter of sodium azide and 20 mg/liter of ceftazidime. Antibacterial susceptibility tests and PCR were performed on the transconjugants to confirm the transfer of antibiotic resistance genes.
Plasmid extraction.
Plasmid extraction was performed using Qiaprep Spin Miniprep kits (Qiagen Inc., Valencia, CA, USA) according to the manufacturer's instructions. The molecular weight of the plasmids was assessed on a 1% agarose gel.
MLST.
The MLST method based on allelic profiles was used to estimate the evolutionary relationship between E. cloacae isolates and performed using seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB), as previously described (1). Alleles of the seven genes were analyzed at the MLST website (http://pubmlst.org/ecloacae/) to provide the sequence type (ST) for each isolate.
Statistical analysis.
A statistical analysis was performed using Epi Info version 7 software according to recommendations of the CDC. Differences were considered significant at a P of <0.05.
RESULTS
Source of samples and units.
Among our 77 clinical isolates of Enterobacter spp., 27 were collected from Algerian patients with a mean age of 35 years (range, 8 to 62 years), and 50 isolates originated from patients in Marseille, France, with a mean age of 40.7 years (range, 1 month to 89 years). The most common specimen was urine for both the Algerian and Marseille isolates, at 60% and 26%, respectively, followed by pus for the Algerian isolates (22%) and sputum for the Marseille isolates (24%).
MALDI-TOF MS identification and clustering.
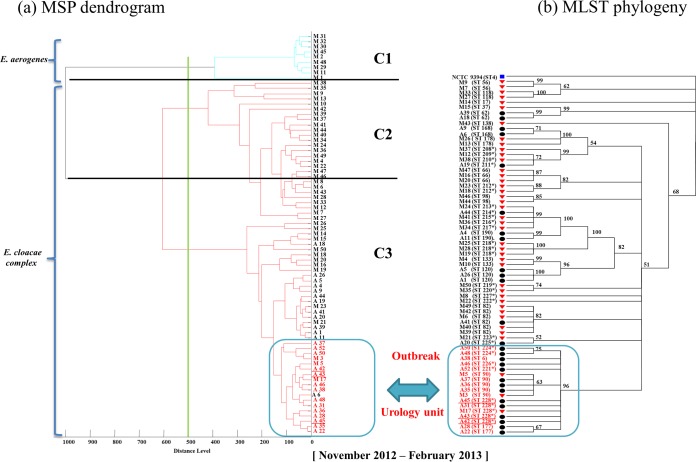
All Enterobacter species isolates were correctly identified at the species level by MALDI-TOF MS (Bruker Daltonics), with a score value of >1.9, as follows: 68 E. cloacae complex isolates (63 E. cloacae, 3 E. asburiae, 1 E. ludwigii, and 1 E. cancerogenus isolate) and 9 E. aerogenes isolates. According to an arbitrary cutoff set up at a distance level of 500, the MSP dendrogram revealed 3 clusters: cluster C1 contained only the 9 E. aerogenes isolates from Marseille, cluster C2 contained 29 E. cloacae isolates from Marseille, and cluster C3 contained 12 E. cloacae complex isolates from Marseille and 27 E. cloacae complex isolates from Algeria. Thus, cluster C2 was significantly associated with the Marseille isolates (P < 10−9) and C3 with the Algerian isolates (P < 10−6) (Fig. 1).
FIG 1.
E. cloacae isolate typing. (a) MSP dendrogram of Enterobacter spp.; (b) phylogenetic tree of multilocus sequence typing (MLST) of E. cloacae isolates using concatenated housekeeping gene sequences. Red triangles, Marseille isolates; black circles, Algerian isolates; blue square, reference strain. Asterisks indicate new STs found in this study. C1, E. aerogenes cluster; C2, E. cloacae isolates from the Marseille cluster; C3, E. cloacae isolates from the Algerian cluster.
Antibacterial susceptibility testing.
Disk diffusion susceptibility testing revealed that all E. cloacae strains were susceptible to imipenem and colistin. However, the Algerian isolates had significantly higher percentages of resistance to third-generation cephalosporins than the Marseille isolates (18/27 versus 11/41, P = 0.002). This was also the case for resistance to fluoroquinolones in the Algerian strains compared to those from Marseille (10/27 versus 5/41, P = 0.03), as well as for aminoglycoside resistance (20/27 in the Algerian isolates versus 8/41 in the Marseille isolates) (P < 10−5) (Table 1).
TABLE 1.
Results of antibiotic susceptibility testing of E. cloacae isolates
| Antibiotica | No. (%) of resistant E. cloacae isolates from: |
P value | |
|---|---|---|---|
| Algeria (n = 27) | Marseille (n = 41) | ||
| CRO | 18 (66.6) | 11 (26.8) | 0.002 |
| CTX | 18 (66.6) | 11 (26.8) | 0.002 |
| CAZ | 17 (62.9) | 10 (24.3) | 0.003 |
| ATM | 17 (62.9) | 10 (24.3) | 0.003 |
| FEP | 17 (62.9) | 10 (24.3) | 0.003 |
| IPM | 0 (0.0) | 0 (0.0) | |
| CIP | 10 (33.3) | 5 (12.1) | 0.03 |
| OFX | 10 (37.0) | 5 (12.1) | 0.03 |
| AMK | 9 (33.3) | 1 (2.4) | 0.001 |
| TOB | 20 (74.0) | 8 (19.5) | <10−5 |
| GAT | 14 (51.8) | 8 (19.5) | 0.01 |
| SXT | 18 (66.6) | 7 (17) | <10−5 |
| NIT | 16 (59.2) | 12 (29.2) | 0.02 |
| CST | 0 (0.0) | 0 (0.0) | |
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; GAT, gentamicin; CRO, ceftriaxone; CST, colistin; CTX, cefotaxime; NIT, nitrofurantoin; FEP, cefipime; FOX, cefoxitin; IPM, imipenem; OFX, ofloxacin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin.
Molecular characterization of antibiotic resistance.
Among the 18 Algerian E. cloacae isolates that showed an ESBL phenotype, 13 (72.2%) isolates had both CTX-M15 and TEM-1, whereas 3 (16.6%) and 2 (11.1%) isolates carried only CTX-M15 or TEM-1, respectively. However, 11 isolates from Marseille were found to be ESBL-producing E. cloacae, with 6 (54.5%) isolates harboring CTX-M15 and 1 (9%) harboring TEM-1; 4 (36.4%) carried both CTX-M15 and TEM-1 (P = 0.002). The fluoroquinolone resistance gene detection results were as follows: in the Algerian isolates, we found qnrB42 in 8 strains along with aac(6′)-Ib-cr (80%), qnrB1 in one strain (10%), and aac(6′)-Ib-cr alone in one strain (10%); in the Marseille isolates, we detected qnrB42 in 2 strains (40%), qnrB1 in one strain (20%), and qnrA1 in 2 strains (40%) (P = 0.03). The aminoglycoside resistance was due to the following AME-encoding genes: aac(6′)-Ib-cr in 9 Algerian strains (45%), in association with ant(2″)-Ia and aadA2 in one Algerian strain and with aadA2 and armA in 3 Algerian strains. Eleven (55%) Algerian strains were found to harbor aadA2. In the Marseille strains, aac(6′)-Ib-cr was found in one strain along with aadA1 (12.5%), and aadA1 was present separately in 3 strains (37.5%) and ant(2″)-Ia in 4 strains (50%) (P < 10−5).
The epidemiology of the Algerian isolates is compared to previous epidemiological studies of E. cloacae in Algeria in Table 2. The antibiotic resistance results are summarized in Table 3.
TABLE 2.
E. cloacae epidemiology in Algeria (2004–2013)a
| Region | Yr(s) of study | β-Lactams |
Quinolones |
Aminoglycosides |
Reference | |||
|---|---|---|---|---|---|---|---|---|
| No. of isolates | Gene(s) detected | No. of isolates | Gene(s) detected | No. of isolates | Genes detected | |||
| Bejaia | 2004–2005 | 1/44 | blaCTX-M3 | ND | 19 | |||
| 2007 | 2/2 | blaCTX-M15 | 2/2 | qnrB | ND | 15 | ||
| Algiers | 2003–2007 | 25/141 | blaCTX-M15, blaCTX-M3, blaVEB-1, blaSHV-12 | 5/141 | qnrB1, qnrB4, qnrS1 | ND | 20 | |
| Tlemcen | 2008 | 2/NM | blaCTX-M15 | ND | 10 | |||
| Annaba | 2009 | 13/65 | blaCTX-M1 | ND | 21 | |||
| Tlemcen | 2008–2010 | 4/4 | blaCTX-M15 | ND | 9 | |||
| Annaba | 2009 | 30/63 | blaCTX-M, blaTEM | ND | 18 | |||
| Eastern Algeria: Constantine, Annaba, Skikda | 2013 | 18/27 | blaCTX-M15, blaTEM-1 | 10/27 | qnrB1, qnrB42 | 20/27 | aadA2, ant2, armA, aac(6′)-Ib-cr | Present study |
ND, not done; NM, not mentioned.
TABLE 3.
Phenotypic and genotypic results of resistant isolates of E. cloacaea
| Region | No. of isolates | β-Lactam resistance | ESBL gene(s) | Quinolone resistance | Quinolone gene | Aminoglycoside resistance | Aminoglycoside gene(s) |
|---|---|---|---|---|---|---|---|
| Algeria | 5 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | TOB-GAT | aadA2 | ||
| 3 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrB42 | AMK-TOB | aac(6′)-Ib-cr | |
| 3 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrB42 | AMK-TOB-GAT | aac(6′)-Ib-cr, aadA2, armA | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrB1 | TOB-GAT | aadA2 | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrB42 | AMK-TOB | aac(6′)-Ib-cr | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | TOB-GAT | aadA2 | |||
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | |||||
| 1 | CRO-CTX | blaCTX-M15 | CIP-OFX | AMK-TOB-GAT | aadA2, ant(2″)-Ia, aac(6′)-Ib-cr | ||
| 1 | CRO-CTX-CAZ-ATM-FEP | blaTEM-1 | TOB-GAT | aadA2 | |||
| 1 | CRO-CTX-CAZ-ATM | blaTEM-1 | TOB-GAT | aadA2 | |||
| 1 | CIP-OFX | qnrB42 | AMK-TOB | aac(6′)-Ib-cr | |||
| 2 | TOB-GAT | aadA2 | |||||
| Marseille | 2 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | TOB-GAT | aadA1 | ||
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrB1 | TOB-GAT | ant(2″)-Ia | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15, blaTEM-1 | CIP-OFX | qnrA1 | TOB-GAT | ant(2″)-Ia | |
| 2 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | |||||
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | CIP-OFX | qnrB42 | TOB-GAT | aadA1 | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | CIP-OFX | qnrB42 | AMK-TOB-GAT | aac(6′)-Ib-cr, aadA1 | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | CIP-OFX | qnrA1 | TOB-GAT | ant(2″)-Ia | |
| 1 | CRO-CTX-CAZ-ATM-FEP | blaCTX-M15 | TOB-GAT | ant(2″)-Ia | |||
| 1 | CRO-CTX-CAZ-ATM-FEP | blaTEM-1 |
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CRO, ceftriaxone; CTX, cefotaxime; CIP, ciprofloxacin; FEP, cefepime; GAT, gentamicin; OFX, ofloxacin; TOB, tobramycin.
Conjugation and resistance transfer.
Conjugation experiments were successful for the five strains tested: three Algerian strains harboring CTX-M15, TEM-1, aadA1, and armA and two isolates from Marseille harboring CTX-M15, TEM-1, and aadA1. E. coli J53 transconjugants showed a phenotype of antibiotic resistance similar to that of the donor strain. The PCR results revealed the presence of donor strain genes.
Plasmid extraction.
The same plasmid, of approximately 9.4 kb molecular size, was extracted from the three Algerian isolates and contained aadA2, armA, and the genes encoding CTX-M15 and TEM-1. Conversely, the two Marseille isolates had one plasmid of approximately 7 kb in size that contained the genes encoding CTX-M15 and TEM-1 and aadA1.
MLST.
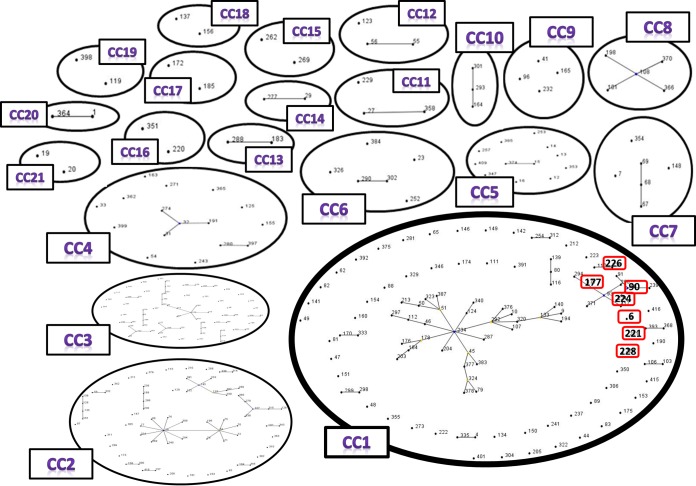
The obtained sequences of the seven loci of the 68 E. cloacae isolates were concatenated and then used to build a phylogenetic tree. E. cloacae NCTC 9394 was used as a reference strain (Fig. 1). The MLST-based phylogenetic tree revealed the presence of 37 STs containing 21 new STs described for the first time in this study. However, within these 37 STs, we identified a small cluster of 7 STs belonging to clonal complex 1 (ST6, ST90, ST177, ST221, ST224, ST226, ST228) (Fig. 2). This cluster contains isolates from the same ward (urology unit) that harbored the armA gene on a plasmid, suggesting an outbreak (Fig. 1).
FIG 2.
eBurst analysis of the E. cloacae MLST database. CC, clonal complex; CC1, the clonal complex that contains the outbreak STs. Outbreak STs are outlined in red.
DISCUSSION
Enterobacter cloacae strains are frequently responsible for nosocomial infections and outbreaks, including urinary tract infections, pneumonia, and bloodstream infections (18). The increase in resistance and the emergence of MDR strains in both Algerian and French hospitals necessitate the implementation of epidemiological surveillance. This study showed a rate of ESBL-producing E. cloacae isolates from eastern Algeria in 2013 of 67%, which is higher than the ESBL production reported in previous epidemiological studies in Algeria: 2.3% in 2004 (19), 17.7% in 2007 (20), and 47% in 2009 (18). The value obtained is also higher than the rate of ESBL production by E. cloacae isolated in Marseille (30%). This can be explained by the overuse of antibiotics in Algerian hospitals (21). The observed resistance is due to the presence of worldwide-spread CTX-M15 and TEM-1 genes in these Algerian and Marseille isolates, which have been described in previous studies (18). Our study reports the first identification of qnrB42 in E. cloacae isolates from both Algeria and France, although it was previously detected in one K. pneumoniae isolate from Algeria (12) and in France (22). qnrB1 was previously described in Algerian E. cloacae strains (20) but not in France; we also report qnrA1 in Marseille isolates, as was detected in a previous study (22). Fluoroquinolone resistance was also associated with the presence of aac(6′)-Ib-cr, which also confers resistance to aminoglycosides, in both the Algerian and Marseille strains. This study presents the first report of the armA 16S rRNA methyltransferase gene in E. cloacae in Algeria and Africa in three strains; it has been reported in an Algerian K. pneumoniae isolate (12) and in two E. cloacae strains in Belgium (23). None of our Marseille strains carried the armA gene; however, it has been detected in one E. cloacae strain of 12 clinical isolates of Enterobacteriaceae in France (24). The presence of aadA1, aadA2, and ant2 in E. cloacae is exclusively described in this study. aadA2 and aadA1 were recently described in K. pneumoniae (12) in Algeria, and another variant (aadA6) was detected in Pseudomonas aeruginosa strains in Korea (25).
MALDI-TOF MS has been successfully used for the identification of E. cloacae (26). The MSP dendrogram clustering of isolates showed significant clusters based on discrimination between Enterobacter species (E. aerogenes and E. cloacae complex). The isolates were also separated according to their geographical origin (Marseille or Algeria), which can be important for rapidly detecting the sources of pathological isolates and is supported by Berrazeg et al. (27) in their study of 535 strains of K. pneumoniae. According to the MLST analysis, among the 37 STs found in our study, 4 STs were common in the Algerian and Marseille isolates, with the remaining strains being heterogeneous, with 21 new STs. Moreover, we identified a specific clade that includes 7 STs (ST6, ST90, ST177, ST221, ST224, ST226, ST228) belonging to the same clonal complex (CC1) (Fig. 2), with 14 Algerian isolates emerging in a uro-nephrology unit. Thus, the isolates were clustered together in the MALDI-TOF MS dendrogram as well as in the MLST-based phylogenetic tree, confirming the suitability of MALDI-TOF MS for biotyping (27) and outbreak detection (Fig. 1) (28, 29). Indeed, this clade suggests the occurrence of an outbreak in this uro-nephrology unit within a period of 4 months, between November 2012 and February 2013, with an unknown common source of transmission. The transmission could be due to hand carriage by paramedical personnel (12, 21) or a contaminated cytoscope, which is typically used to place double J catheters, especially in urology clinics (21).
This outbreak was associated with the presence of the armA 16S rRNA methyltransferase gene in three of the E. cloacae Algerian isolates, and this gene was carried on a conjugative plasmid along with aadA2, blaCTX-M15, and blaTEM-1. This situation can promote the rapid dissemination of armA in various members of the Enterobacteriaceae family (23) (K. pneumoniae in Algeria [12], Escherichia coli, A. baumannii, and Serratia in China [30]).
The armA 16S rRNA methyltransferase gene underlies strong resistance to all aminoglycosides, which usually have a broad antimicrobial spectrum and efficiency, and the cooccurrence of this gene with ESBL-encoding genes presents a real clinical concern. Indeed, this may be a major therapeutic threat in the future (24), as aminoglycoside and beta-lactam combinations, which are widely used by clinicians to treat severe bacterial infections based on their synergistic effects, are no longer a solution (30). Furthermore, with the rapid increase in ESBL-producing strains, imipenem has been massively used and in turn has induced the emergence of carbapenem resistance genes, especially in Gram-negative bacteria, as reported in many epidemiological studies from around the world, including Algeria, where oxacillinases and NDM-1 were detected in A. baumannii (11) and VIM-2 in P. aeruginosa (31).
Hence, to reduce the frequency of MDR bacteria, clinicians and health professionals, particularly in Algeria, should pay attention to the uncontrolled use of antibiotics and should refer to antibiotic susceptibility testing methods for the choice of an effective treatment (21).
Our study demonstrates in a very significant way the high level of strains resistant to the most common antibiotic families in Algeria compared to Marseille, France. Surveillance measures should be systematically implemented when treating an Algerian patient hospitalized in France.
ACKNOWLEDGMENTS
We are very grateful to Linda Hadjadj for technical assistance. We thank Tohru Miyoshi-Akiyama for new ST submissions to the Enterobacter cloacae MLST database. We thank American Journal Experts for English corrections.
We have no conflicts of interest to declare.
This work was partly funded by CNRS and IHU Méditérranée Infection.
REFERENCES
- 1.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagenlehner FM, MacKenzie FM, Forbes KJ, Gould IM. 2002. Molecular epidemiology and antibiotic resistance of Enterobacter spp. from three distinct populations in Grampian, UK. Int J Antimicrob Agents 20:419–425. doi: 10.1016/S0924-8579(02)00179-6. [DOI] [PubMed] [Google Scholar]
- 3.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. On-going revolution in bacteriology: routine identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 4.Mencacci A, Monari C, Leli C, Merlini L, De Carolis E, Vella A, Cacioni M, Buzi S, Nardelli E, Bistoni F, Sanguinetti M, Vecchiarelli A. 2013. Typing of nosocomial outbreaks of Acinetobacter baumannii by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51:603–606. doi: 10.1128/JCM.01811-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres E, Lopez-Cerero L, Del Toro MD, Pascual A. 2013. First detection and characterization of an OXA-48-producing Enterobacter aerogenes isolate. Enferm Infecc Microbiol Clin 32:469–470. doi: 10.1016/j.eimc.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Huang S, Dai W, Sun S, Zhang X, Zhang L. 2012. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PLoS One 7:e47636. doi: 10.1371/journal.pone.0047636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove SE, Kaye KS, Eliopoulous GM, Carmeli Y. 2002. Health and economic outcomes of the emergence of third-generation cephalosporin resistance in Enterobacter species. Arch Intern Med 162:185–190. doi: 10.1001/archinte.162.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Khajuria A, Praharaj AK, Grover N, Kumar M. 2013. First report of an Enterobacter ludwigii isolate coharboring NDM-1 and OXA-48 carbapenemases. Antimicrob Agents Chemother 57:5189–5190. doi: 10.1128/AAC.00789-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba Ahmed-Kazi Tani Z, Decre D, Genel N, Boucherit-Otmani Z, Arlet G, Drissi M. 2013. Molecular and epidemiological characterization of enterobacterial multidrug-resistant strains in Tlemcen Hospital (Algeria) (2008-2010). Microb Drug Resist 19:185–190. doi: 10.1089/mdr.2012.0161. [DOI] [PubMed] [Google Scholar]
- 10.Ahmed ZB, Ayad A, Mesli E, Messai Y, Bakour R, Drissi M. 2012. CTX-M-15 extended-spectrum beta-lactamases in Enterobacteriaceae in the intensive care unit of Tlemcen Hospital, Algeria. East Mediterr Health J 18:382–386. [DOI] [PubMed] [Google Scholar]
- 11.Bakour S, Kempf M, Touati A, Ait AA, Haouchine D, Sahli F, Rolain JM. 2012. Carbapenemase-producing Acinetobacter baumannii in two university hospitals in Algeria. J Med Microbiol 61:1341–1343. doi: 10.1099/jmm.0.045807-0. [DOI] [PubMed] [Google Scholar]
- 12.Belbel Z, Chettibi H, Dekhil M, Ladjama A, Nedjai S, Rolain JM. 2014. Outbreak of an armA methyltransferase-producing ST39 Klebsiella pneumoniae clone in a pediatric Algerian hospital. Microb Drug Resist 20:310–315. doi: 10.1089/mdr.2013.0193. [DOI] [PubMed] [Google Scholar]
- 13.Seng P, Rolain JM, Fournier PE, La SB, Drancourt M, Raoult D. 2010. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol 5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 14.Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, Song EH, Jeong SH. 2005. Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother 56:698–702. doi: 10.1093/jac/dki324. [DOI] [PubMed] [Google Scholar]
- 15.Touati A, Brasme L, Benallaoua S, Gharout A, Madoux J, de Champs C. 2008. First report of qnrB-producing Enterobacter cloacae and qnrA-producing Acinetobacter baumannii recovered from Algerian hospitals. Diagn Microbiol Infect Dis 60:287–290. doi: 10.1016/j.diagmicrobio.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Kim JY, Park YJ, Kwon HJ, Han K, Kang MW, Woo GJ. 2008. Occurrence and mechanisms of amikacin resistance and its association with beta-lactamases in Pseudomonas aeruginosa: a Korean nationwide study. J Antimicrob Chemother 62:479–483. doi: 10.1093/jac/dkn244. [DOI] [PubMed] [Google Scholar]
- 17.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2013. ARG-ANNOT (Antibiotic Resistance Gene-ANNOTation), a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedjai S, Barguigua A, Djahmi N, Jamali L, Zerouali K, Dekhil M, Timinouni M. 2013. Prevalence and characterization of extended spectrum beta-lactamase-producing Enterobacter cloacae strains in Algeria. J Infect Dev Ctries 7:804–811. doi: 10.3855/jidc.3127. [DOI] [PubMed] [Google Scholar]
- 19.Touati A, Benallaoua S, Forte D, Madoux J, Brasme L, de Champs C. 2006. First report of CTX-M-15 and CTX-M-3 beta-lactamases among clinical isolates of Enterobacteriaceae in Bejaia, Algeria. Int J Antimicrob Agents 27:397–402. doi: 10.1016/j.ijantimicag.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Iabadene H, Messai Y, Ammari H, Ramdani-Bouguessa N, Lounes S, Bakour R, Arlet G. 2008. Dissemination of ESBL and Qnr determinants in Enterobacter cloacae in Algeria. J Antimicrob Chemother 62:133–136. doi: 10.1093/jac/dkn145. [DOI] [PubMed] [Google Scholar]
- 21.Nedjai S, Barguigua A, Djahmi N, Jamali L, Zerouali K, Dekhil M, Timinouni M. 2012. Prevalence and characterization of extended spectrum beta-lactamases in Klebsiella-Enterobacter-Serratia group bacteria, in Algeria. Med Mal Infect 42:20–29. doi: 10.1016/j.medmal.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Guillard T, de Champs C, Moret H, Bertrand X, Scheftel JM, Cambau E. 2012. High-resolution melting analysis for rapid characterization of qnr alleles in clinical isolates and detection of two novel alleles, qnrB25 and qnrB42. J Antimicrob Chemother 67:2635–2639. doi: 10.1093/jac/dks292. [DOI] [PubMed] [Google Scholar]
- 23.Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, De Mendonca R, Rodriguez-Villalobos H, Struelens M, Glupczynski Y. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother 59:459–464. doi: 10.1093/jac/dkl527. [DOI] [PubMed] [Google Scholar]
- 24.Galimand M, Sabtcheva S, Courvalin P, Lambert T. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob Agents Chemother 49:2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiddee A, Henghiranyawong K, Yimsabai J, Tiloklurs M, Niumsup PR. 2013. Nosocomial spread of class 1 integron-carrying extensively drug-resistant Pseudomonas aeruginosa isolates in a Thai hospital. Int J Antimicrob Agents 42:301–306. doi: 10.1016/j.ijantimicag.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Stets MI, Pinto AS Jr, Huergo LF, de Souza EM, Guimaraes VF, Alves AC, Steffens MB, Monteiro RA, Pedrosa Fde O, Cruz LM. 2013. Rapid identification of bacterial isolates from wheat roots by high resolution whole cell MALDI-TOF MS analysis. J Biotechnol 165:167–174. doi: 10.1016/j.jbiotec.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Berrazeg M, Diene SM, Drissi M, Kempf M, Richet H, Landraud L, Rolain JM. 2013. Biotyping of multidrug-resistant Klebsiella pneumoniae clinical isolates from France and Algeria using MALDI-TOF MS. PLoS One 8:e61428. doi: 10.1371/journal.pone.0061428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christner M, Trusch M, Rohde H, Kwiatkowski M, Schlüter H, Wolters, Aepfelbacher M, Hentschke M. 2014. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-toxigenic Escherichia coli. PLoS One 9:e101924. doi: 10.1371/journal.pone.0101924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batah R, Loucif L, Olaitan AO, Boutefnouchet N, Allag H, Rolain JM. 17 April 2015. Outbreak of Serratia marcescens coproducing ArmA and CTX-M-15 mediated high levels of resistance to aminoglycoside and extended-spectrum beta-lactamases, Algeria. Microb Drug Resist doi: 10.1089/mdr.2014.0240. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q, Zhang Y, Han L, Sun J, Ni Y. 2009. Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob Agents Chemother 53:271–272. doi: 10.1128/AAC.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touati M, Diene SM, Dekhil M, Djahoudi A, Racherache A, Rolain JM. 2013. Dissemination of a class I integron carrying VIM-2 carbapenemase in Pseudomonas aeruginosa clinical isolates from a hospital intensive care unit in Annaba, Algeria. Antimicrob Agents Chemother 57:2426–2427. doi: 10.1128/AAC.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]