Abstract
Tenofovir alafenamide (TAF) is an investigational prodrug of the HIV-1 nucleotide reverse transcriptase (RT) inhibitor (NtRTI) tenofovir (TFV), with improved potency and drug delivery properties over the current prodrug, tenofovir disoproxil fumarate (TDF). TAF is currently in phase 3 clinical studies for the treatment of HIV-1 infection, in combination with other antiretroviral agents. Phase 1 and 2 studies have shown that TAF was associated with increased peripheral blood mononuclear cell (PBMC) drug loading and increased suppression of HIV-1 replication compared to treatment with TDF. In this study, selection of in vitro resistance to both TAF and the parent compound, TFV, led to the emergence of HIV-1 with the K65R amino acid substitution in RT with 6.5-fold-reduced susceptibility to TAF. Although TAF is more potent than TFV in vitro, the antiviral susceptibilities to TAF and TFV of a large panel of nucleoside/nucleotide RT inhibitor (NRTI)-resistant mutants were highly correlated (R2 = 0.97), indicating that the two compounds have virtually the same resistance profile when assessed as fold change from the wild type. TAF showed full antiviral activity in PBMCs against primary HIV-1 isolates with protease inhibitor, nonnucleoside RT inhibitor (NNRTI), or integrase strand transfer inhibitor resistance but reduced activity against isolates with extensive NRTI resistance amino acid substitutions. However, the increased cell loading of TFV with TAF versus TDF observed in vivo suggests that TAF may retain activity against TDF-resistant mutant viruses.
INTRODUCTION
Tenofovir alafenamide (TAF) is an investigational prodrug of the nucleotide analog phosphonate tenofovir (TFV) (Fig. 1). The current prodrug of tenofovir is tenofovir disoproxil fumarate (TDF) (1). TAF has shown improved pharmacokinetic properties and more potent HIV-1 suppression than TDF in phase 1 and phase 2 clinical trials (2, 3) and is currently being studied in clinical trials for the treatment of HIV-1 infection in patients ≥12 years old, in combination with other antiretroviral agents. Both TAF and TDF prodrugs ultimately led to the delivery of TFV to the target cells; however, TAF showed greater distribution to lymphoid tissues than TDF in nonclinical studies (4). In a phase 1 clinical study, monotherapy with 25 mg TAF achieved a median 1.46-log10-unit decrease in plasma HIV-1 RNA at day 10 compared to 0.97 log10 unit for 300 mg TDF while reducing the systemic exposure of TFV by about 86% and increasing the concentration of the active moiety, tenofovir diphosphate (TFV-DP), in peripheral blood mononuclear cells (PBMCs) by 5- to 7-fold (2). The 25-mg dose of TAF achieves higher intracellular TFV-DP concentrations than 300 mg TDF due to the greater plasma stability of TAF than of TDF and the subsequent intracellular conversion of TAF to TFV. In vitro studies have demonstrated consistent conversion of TAF to TFV in PBMCs from a diverse set of donors (5). (In an accompanying paper [6], we evaluate the in vitro virology profile of TAF and compare it to that of TDF.)
FIG 1.

Structures of the 2 prodrugs of TFV—TAF and TDF—and conversion leading to the active inhibitor of HIV-1 RT, TFV-DP. The prodrugs TAF and TDF are metabolized to give rise to the nucleotide TFV, which is phosphorylated by cellular kinases to the active moiety, TFV-DP. *, fumarate is not represented.
Resistance to TFV has been extensively characterized both in vivo and in vitro (reviewed in reference 7). In vitro resistance selection experiments have established the reverse transcriptase (RT) amino acid substitution K65R as the primary TFV resistance mutation (8), often found in association with a substitution at position S68 (S68N or S68K) (9). The levels of phenotypic resistance in these mutants were within 2- to 5-fold that of the wild-type reference. The RT amino acid substitution K70E was also observed in a resistance selection experiment with TFV and resulted in 3-fold-reduced susceptibility to TFV (10). Analyses of the development of resistance in antiretroviral (ARV) treatment-naive patients experiencing virologic failure further confirmed the role of the K65R mutation and coselected S68G and A62V amino acid substitutions as the main resistance pathway for TDF in vivo (11). The secondary resistance pathway involving the RT amino acid substitution K70E has been infrequently observed in vivo (11, 12). In ARV treatment-experienced patients, development of TDF resistance has been similarly characterized by a K65R (with or without S68G) substitution (13); however, treatment in these populations has also shown that preexisting thymidine analog-associated mutations (TAMs) (M41L, D67N, K70R, L210W, T215Y/F, and K219Q/E/N/R in RT) play a key role in resistance to TDF. The presence of 3 or more TAMs, including either the M41L or the L210W amino acid substitution at baseline in subjects treated with TDF, was found to be associated with reduced response to TDF (14) and in vitro TFV phenotypic resistance in site-directed mutant analyses (15). The presence of a double serine insertion after RT residue 69 further decreased TFV susceptibility in the presence of TAMs (16). Conversely, the presence of the M184V/I amino acid substitution, which is the hallmark of lamivudine (3TC) and emtricitabine (FTC) resistance, is associated with increased response to TDF and hypersusceptibility to TFV in phenotypic assays (15). Mechanistically, resistance to TFV by HIV-1 RT containing the K65R mutation is driven by the ability of the mutated RT enzyme to discriminate against the inhibitor while still recognizing the natural substrate (17). The presence of the K65R substitution in viral isolates is associated with decreased replication (18), in addition to the low-level phenotypic resistance described above (2- to 5-fold that of the wild type). This ability of the mutated HIV-1 RT enzyme to favor the natural substrate over the inhibitor is also responsible for the resistance to 3TC by HIV-1 RT containing the M184V amino acid substitution (19–21). While the M184V/I substitution leads to a complete loss of activity for 3TC and FTC (phenotypic resistance above the limit of detection), it is also associated with overall decreased viral fitness (19, 22) and partial restoration of susceptibility and/or hypersusceptibility to other nucleoside reverse transcriptase inhibitors (NRTIs), such as TFV and zidovudine (AZT) (15, 23–25). Resistance to AZT by HIV-1 RT containing TAMs results from a different mechanism, in which the mutated RT enzyme is able to excise newly incorporated AZT-monophosphate from the growing viral DNA strands (26, 27), thus allowing viral DNA synthesis to resume with the incorporation of natural nucleotides. The presence of multiple TAMs in viral isolates has been associated with high levels of phenotypic resistance to AZT (often >100-fold that of the wild type), as well as low-level cross-resistance to TFV and other NRTIs (15, 25, 28).
Unlike NRTIs, such as 3TC and AZT, that completely lose their anti-HIV activity in vitro in the presence of resistance-associated amino acid substitutions (M184V/I and TAMs, respectively), TFV retains some level of anti-HIV activity in vitro even in mutants with a K65R mutation or multiple TAMs (phenotypic resistance is typically <8-fold that of the wild type) (15, 25). As treatment with TAF is associated with a 5-fold increase in the quantity of TFV-DP delivered to HIV-infected target cells compared to treatment with TDF (2), some of the resistance observed in patients treated with TDF could be overcome when they are treated with TAF. The aim of the studies described in this paper was to characterize the resistance profile of TAF against HIV-1. First, resistance selection experiments comparing TAF and TFV were carried out in parallel, and the selected viruses were analyzed for genotypic and phenotypic resistance. Next, phenotypic analyses were conducted using the HIV single-cycle PhenoSense assay (Monogram Biosciences, South San Francisco, CA) for both TAF and TFV using a large panel of viruses with diverse resistance patterns. Finally, the antiviral activity of TAF was tested in PBMCs against a panel of HIV-1 primary isolates with resistance to various classes of antiretroviral drugs.
MATERIALS AND METHODS
Reagents and cell lines.
TAF, TFV, FTC, elvitegravir (EVG), and efavirenz (EFV) were synthesized at Gilead Sciences (Foster City, CA, USA). AZT was purchased from Sigma-Aldrich (St. Louis, MO, USA). MT-2 cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Germantown, MD, USA). PBMCs were prepared from whole blood as indicated below. The wild-type virus stock HIV-1IIIB was purchased from Advanced Biotechnologies, Inc. (Columbia, MD, USA).
Resistance selections.
The resistance selection experiments were set up in 6-well plates with 1 million MT-2 cells per well in a final volume of 4 ml. At the start of the experiments, cells were infected with 8 μl of a 1:100 dilution of HIV-1IIIB viral stock, corresponding to a multiplicity of infection (MOI) of around 0.01. HIV-1IIIB induces a cytopathic effect (CPE) in MT-2 cells. Two selection experiments were conducted using both TAF and TFV in parallel. The starting drug concentrations were below the effective concentration to inhibit 50% of viral replication (EC50) in the first experiment (8 nM and 3 μM for TAF and TFV, respectively, compared to their respective HIV-1IIIB EC50s of 14 nM and 3.5 μM) and were set at 2-fold the EC50 in the second experiment (28 nM and 7 μM, respectively). One infected culture without drug was maintained throughout the experiments to control the natural evolution of the virus in tissue culture. The cultures were incubated at 37°C and split 1:5 every 6 to 7 days, depending on the growth status of the cells and the extent of the CPE observed in the cultures. CPE in the form of cellular syncytia was observed 3 to 4 days after initiation of infection in the no-drug culture. When extensive CPE was seen in the drug-containing wells, a new passage was generated by infecting fresh MT-2 cells with 400 μl of the harvested virus using a drug concentration 2-fold higher than in the previous passage. Two control cultures containing either no drug or the previous drug concentration were also set up as controls for viral growth. Successive viral passages were obtained by repeating this procedure. The duration of each culture at a given drug concentration was dependent upon the development of CPE in the drug wells and ranged from 3 to 74 days. Viral supernatants were prepared and sequenced (population sequencing) as described below.
Virus sample preparation and sequencing.
Viral supernatants were treated with 4 units of DNase I (New England BioLabs, Ipswich, MA, USA) for 45 min at room temperature, and viral RNA was extracted from 200 μl viral supernatant using the EZ1 Virus Minikit v2.0 with the BioRobot EZ1 workstation (Qiagen, Valencia, CA, USA) and eluted in 60 μl. Viral RNA was reverse transcribed into cDNA using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Buckinghamshire, United Kingdom) according to the manufacturer's instructions with the HIV-1-specific primer R4395 (5′-CAGTCTACTTGTCCATGCATGGCTTCTCC-3′; final concentration, 0.5 μM). The viral cDNA (2.5 μl of a 40-μl reaction mixture) was amplified through 35 cycles of PCR using the primers RT18 (5′-GGAAACCAAAAATGATAGGGGGAATTGGAGG-3′) and RT4361 (5′-GCTGACATTTATCACAGCTGGCTAC-3′) to generate a 1,985-bp viral-DNA fragment containing the entire RT portion of the pol gene (spanning nucleotide 200 of protease to nucleotide 105 of integrase). The PCR was performed using Phusion High Fidelity polymerase (New England BioLabs) in a C1000 Touch Thermal Cycler (Bio-Rad, Hercules, CA, USA), and the PCR product was purified using the QIAquick PCR purification kit (Qiagen) and sequenced through the integrase region on both DNA strands (ELIM Biopharmaceuticals, Inc., Hayward, CA, USA). Sequencing data were analyzed using SeqMan and MegAlign (DNAStar, Madison, WI, USA). For the clonal sequencing analysis, PCR products of interest were subcloned (Zero Blunt TOPO PCR Cloning Kit; Invitrogen, Carlsbad, CA, USA) and grown overnight in bacterial plates containing kanamycin. Twenty clones were picked and grown overnight (37°C with shaking at 225 rpm) in 3 ml culture medium containing kanamycin. Plasmid DNA was purified from the culture (QIAprep kit; Qiagen) and sequenced as indicated above.
Phenotyping analyses in MT-2 cells.
The phenotypes of selected viral pools of interest were determined in a 5-day multicycle antiviral assay in MT-2 cells using a luciferase-based viability readout (CellTiterGlo; Promega, Madison, WI, USA) as described previously (29). Briefly, MT-2 cells (2.4 million) were incubated with virus for 3 h at 37°C in 1-ml screw-cap tubes. The amount of virus used was normalized to yield a signal-to-noise (S/N) ratio in the range of 4 to 7, which was equivalent to an MOI of around 0.003 for wild-type HIV-1IIIB based on the provided titer. The S/N ratio was calculated from the 100 nM EFV control (maximum cell survival) and the no-drug control (minimum cell survival). Fivefold dilutions of the drugs of interest were prepared and transferred in triplicate to the inside wells of the 96-well assay plates. After the 3-hour incubation, the infected MT-2 cells were diluted 1:14 to a concentration of 0.17 million cells/ml with tissue culture medium, and 50 μl of cell suspension was transferred to all the wells in the assay plates. After 5 days of incubation at 37°C, 100 μl of CellTiterGlo reagent was added to each well, and the luminescence was measured using an Envision plate reader (PerkinElmer, Shelton, CT, USA). The percent inhibition in the drug-containing wells in comparison to the fully protected EFV control and the associated EC50 were plotted and calculated using Excel (Microsoft, Redmond, WA, USA) and XL Fit (IDBS, Alameda, CA, USA). The statistical significance of the fold changes (FCs) for the mutants compared to the wild-type control was calculated using Excel (two-tailed Student's t test).
Comparative resistance profiling of TAF and TFV in recombinant isolates.
The in vitro antiviral resistance profiling was performed by Monogram Biosciences (South San Francisco, CA, USA) using the single-cycle PhenoSense assay (30). The potencies of TAF and TFV were determined for 24 patient-derived recombinant HIV-1 variants from the Monogram collection. These recombinants were selected based on their genotypic resistance to multiple NRTIs so as to span a wide range of NRTI resistances. The activities of TAF and TFV against the tested viruses were expressed as the FC in the calculated EC50 relative to that of the wild-type reference strain, HIV-1NL4-3.
Efficacy and resistance profiling of HIV-1 primary isolates in fresh human PBMCs.
Resistance profiling of 7 HIV-1 primary isolates (isolate IDs, 1064-52, 52-52, 8070_1, 4736_4, A-17, 5705-72, and MDR-769) was conducted at Southern Research Institute (SRI) (Frederick, MD, USA) using human PBMCs seronegative for HIV and hepatitis B virus (HBV) freshly isolated from screened donors as described previously (31). Briefly, phytohemagglutinin-activated PBMCs cultured in the presence of interleukin-2 were plated in 96-well plates, infected with HIV-1 primary isolates and incubated for 7 days in the presence of drug. Drug inhibition was determined based on reverse transcriptase activity. Seven HIV-1 primary isolates with resistance mutations against one or more drug classes (protease inhibitor [PI], NRTI, non-NRTI [NNRTI], and integrase strand transfer inhibitor [INSTI]) were tested for susceptibility against TAF; AZT, nevirapine (NVP), indinavir (IDV), enfuvirtide (T20), raltegravir (RAL), and EVG were used as controls.
RESULTS
Resistance selections with TAF and TFV.
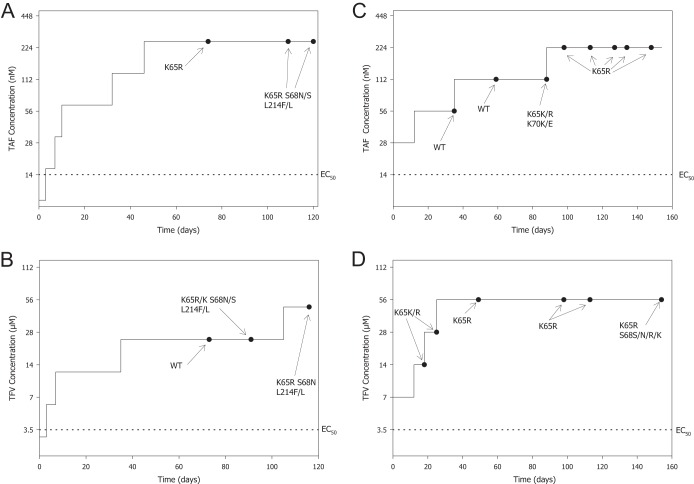
We conducted resistance selection experiments with TAF and TFV in parallel in order to compare the resistance profiles of the two compounds. TFV is usually used in vitro instead of TDF due to the technical limitations of the low stability of TDF in culture medium. The dose escalation method was used in these experiments. We assessed the effect of the increasing selective pressure on the wild-type HIV-1IIIB from TAF and TFV by analyzing the genotypic and phenotypic changes in the virus over time. Two selection experiments were conducted. The first experiment started at concentrations of TAF and TFV below the EC50 for each drug (the time course and genotypic changes are shown in Fig. 2A and B, respectively), and the second experiment started at concentrations of TAF and TFV corresponding to twice the EC50 for each drug (the time course and genotypic changes are shown in Fig. 2C and D, respectively). The durations of the experiments were >115 days and >147 days for the first and second experiments, respectively. The outcomes of the 4 selection experiments were nearly identical, resulting in the development of the K65R amino acid substitution in RT, which was accompanied by an S68N substitution (full mutation or a mixture with the wild type) in 3 of 4 cases. The development of the L214F polymorphism that was observed in the first experiment for both drugs was also observed in the no-drug control (data not shown) and may be due to the presence of the amino acid substitution at a low level in the starting material. A transient K70K/E substitution was observed, along with a K65K/R substitution in the second experiment with TAF, but was not detected at subsequent time points (Fig. 2C). Clonal sequencing analyses showed that the K70E and K65R substitutions arose on different genomes, with the K65R-containing virus outgrowing the K70E-containing virus with increased time in culture and drug concentration (Fig. 3). The clonal sequencing analysis also evidenced the presence of an S68N or S68I substitution in 40% of the clones at the last time point analyzed (day 148) that was not detected by population sequencing. The drug concentrations at which the K65R substitution first appeared varied between experiments from about 4 times the EC50 to 15 times the EC50, independent of the drug used, likely a reflection of the stochastic nature associated with this experimental model. Importantly, attempts to increase the drug concentrations beyond 16 times the EC50 of each drug (to 24 times the EC50, or 336 nM, for TAF and 24 times the EC50, or 84 μM, for TFV) over >5 weeks in the second experiment did not yield viable virus.
FIG 2.
HIV-1 selection time courses using either TAF or TFV. Selection experiments were started either at a concentration below the EC50 (TAF [A] and TFV [B]) or at a concentration of twice the EC50 (TAF [C] and TFV [D]). Amino acid changes compared to the HIV-1IIIB starting material (time zero) are shown. WT (wild type) indicates that no changes from the HIV-1IIIB starting material were found.
FIG 3.

Clonal sequencing analysis of TAF-selected viruses from Fig. 2C. Twenty clones were analyzed at each time point.
Phenotypic analyses of the 2 final viruses obtained in the second experiment were carried out using a 5-day cytopathic assay in MT-2 cells. The fold changes from the wild-type control observed for the viruses selected by either TAF or TFV were very similar. Susceptibility to TAF was 6.5-fold above that of the wild type for both selected viruses, and TFV susceptibilities were 5.5- and 5.1-fold above that of the wild type for the TAF- and TFV-selected viruses, respectively (Table 1). Differences in the fold changes from the wild type between TAF and TFV for each of the selected viruses were not statistically significant (Student's t test; P = 0.17 and 0.25 for TAF- and TFV-selected viruses, respectively). Consistent with the presence of the K65R substitution, reduced FTC susceptibility was also observed at similar levels in the 2 selected viruses (8.5- and 6.7-fold from that of the wild type), while susceptibility to the control drugs EFV and EVG was near wild-type levels. These data provide evidence that under these selection conditions (low doses of drug near the EC50), TAF and TFV have the same resistance profile, resulting in a mutant virus with the K65R amino acid substitution and similar phenotypic profiles for both TAF and TFV.
TABLE 1.
Characteristics and drug susceptibilities of selected viruses
| Selecting drug | Time point (day) | Concn (FC over EC50) | Selected virus |
|||||
|---|---|---|---|---|---|---|---|---|
| Amino acid substitution(s) | Susceptibility (FC from wild-type control [HIV-1IIIB])a |
|||||||
| TAF | TFV | FTC | EFV | EVG | ||||
| TAF | 148 | 224 nM (16×) | K65R | 6.5b | 5.5b | 8.5b | 1.4 | 1.7 |
| TFV | 154 | 56 μM (16×) | K65R S68(S/N/R/K) | 6.5b | 5.1b | 6.7b | 1.5 | 1.4 |
EC50 against HIV-1IIIB in MT-2 standard assay was 10 nM, 2.9 μM, 1.2 nM, 0.77 μM, 0.26 μM, and 1.5 nM for TAF, TFV, EVG, FTC, ZDV, and EFV respectively. TAF, tenofovir alafenamide; TFV, tenofovir; FTC, emtricitabine; AZT, zidovudine; EFV, efavirenz; EVG, elvitegravir. Fold changes of the average EC50 were obtained from 5 independent experiments.
P < 0.05 (t test).
Comparative resistance profiling of TAF and TFV.
The antiviral phenotypic susceptibilities of TAF and TFV were analyzed against a panel of 24 patient-derived HIV-1 recombinant isolates in the Monogram Biosciences PhenoSense assay (Table 2). The mutants in the panel were chosen to represent a wide array of NRTI resistance-associated amino acid substitutions and were expected to display a wide spectrum of TFV susceptibilities, from hypersusceptible to highly resistant. The choice of the mutants present in the panel, however, is not representative of their frequencies in the HIV-1-infected population and was meant to capture the diversity of EC50 FCs from the wild type observed for TFV. In the PhenoSense assay, clinical susceptibility cutoffs for TDF have been established at 1.4 (lower cutoff) and 4 (higher cutoff) (14). A TFV FC from the wild type of <1.4 indicates full sensitivity to TDF, a TFV FC of >4 indicates resistance to TDF, and a TFV FC between 1.4 and 4 indicates reduced sensitivity to TDF.
TABLE 2.
Phenotypic susceptibilities of 24 recombinant HIV-1 isolates with NRTI amino acid substitutions against TAF and TFV
| Virus ID | EC50 FCa |
Resistance category | NRTI amino acid substitution(s) | |
|---|---|---|---|---|
| TAF | TFV | |||
| 13 | 0.34 | 0.41 | NRTI | L74V |
| 16 | 0.40 | 0.47 | NRTI + M184V | L74V Y115F M184Vb |
| 5 | 0.50 | 0.48 | M184V | M184V |
| 14 | 0.43 | 0.50 | NRTI | L74V |
| 22 | 0.56 | 0.53 | Q151M + M184V | Q151M M184Vb |
| 15 | 0.50 | 0.59 | NRTI + M184V | L74V Y115F M184Vb |
| 6 | 0.67 | 0.65 | M184V | M184Vb |
| 21 | 0.82 | 0.82 | Q151M + M184V | A62V V75V/I F116Y Q151M M184Vb |
| 20 | 0.91 | 0.93 | Q151M | F116Y Q151Mb |
| 11 | 0.78 | 0.98 | K65R + M184V | A62A/V K65R M184Vb |
| 12 | 1.09 | 1.18 | K65R + M184V | K65R M184Vb |
| 9 | 1.68 | 1.48 | K65R | K65Rb |
| 10 | 1.91 | 1.71 | K65R | K65Rb |
| 17 | 1.62 | 1.81 | 3 TAMs | M41L L74V L210W T215Y |
| 3 | 2.11 | 2.27 | 6 TAMs + M184V | M41L D67N K70R M184V L210W T215Y K219E |
| 19 | 3.43 | 2.82 | Q151M complex | A62V V75I F77L Y115F F116Y Q151Mb |
| 1 | 3.62 | 3.48 | 6 TAMs | M41L D67N K70R L210W T215F K219Qb |
| 18 | 8.80 | 3.80 | 5 TAMs | M41L D67N T69D L74V L210W T215Y K219Rb |
| 2 | 4.77 | 4.01 | 6 TAMs | M41L D67N K70R L210W T215Y K219Eb |
| 4 | 9.16 | 6.11 | 6 TAMs + M184V | M41L D67N K70R M184V L210W T215Y K219Eb |
| 24 | 9.07 | 9.60 | Q151M complex + K65R | A62V K65R K70K/R V75I F77L F116Y Q151Mb |
| 8 | 20.0 | 18.0 | T69 insertion | M41L T69insc L74V L210W T215Yb |
| 23 | 22.0 | 19.0 | T69 insertion | M41L A62V T69ins L210W T215Yb |
| 7 | 23.0 | 20.0 | T69 insertion | A62V T69ins V75Ib |
Susceptibilities are expressed as the FC in the EC50 from that of the wild-type control. The wild-type EC50s for TAF and TFV were 10 nM and 0.6 μM, respectively. The Monogram Biosciences assay resistance cutoffs for TFV/TAF are 1.4-fold and 4.0-fold above that of the wild type for reduced susceptibility and resistance, respectively.
Presence of NNRTI resistance-associated amino acid substitutions (not shown).
T69ins, T69 insertion.
The panel of recombinant mutants showed TFV FCs from the wild type ranging from 0.41 to 20. Eleven isolates were susceptible to TFV, 7 isolates showed reduced susceptibility to TFV, and 6 isolates showed resistance to TFV. Most isolates that were susceptible to TFV (8 of 11) had the M184V amino acid substitution plus a few additional NRTI resistance-associated substitutions, including two isolates with K65R plus M184V. None of the isolates that were susceptible to TFV had TAMs (M41L, D67N, K70R, L210W, T215Y/F, and K219Q/E/N/R). Isolates with reduced susceptibility to TFV rarely had the M184V substitution (only 1 of 7) and had an increasingly complex resistance pattern with either 3 or more TAMs, the K65R substitution alone, or the Q151M multidrug resistance (MDR) complex (see below). Isolates resistant to TFV had either 6 TAMs, a Q151M complex plus K65R, or double insertions at T69 plus other NRTI resistance-associated substitutions and/or TAMs. The highest level of resistance to TFV was observed for isolates with T69 double insertions.
Although TAF is more potent than TFV against wild-type HIV-1 in vitro (10 nM versus 600 nM), susceptibility to TAF measured as the fold change from the wild type for this panel of mutants was almost identical to that to TFV, ranging from 0.34- to 23-fold that of the wild type. This was expected, as TFV-DP is the common active moiety for both compounds. Some of the variability observed between the 2 compounds was most likely stochastic, and only virus number 18 was categorized differently for TAF than for TFV based on the TFV cutoffs. (Susceptibility cutoffs for TAF have not been established.) As a result, there was a strong correlation between the fold changes from the wild type for TAF and TFV (R2 = 0.97 [data not shown]), indicating that TAF and TFV have virtually the same resistance profile against the panel of NRTI-resistant mutants tested in the PhenoSense assay.
Susceptibilities to TAF of HIV-1 primary isolates.
The antiviral activity of TAF in PBMCs was tested against a panel of 7 primary HIV-1 isolates with resistance-associated amino acid substitutions across multiple drug classes (Table 3). Each isolate was tested in a single experiment against TAF and internal control compounds using the commercially available PBMC assay from SRI (Frederick, MD, USA). Five of the 7 isolates were single-class-resistant mutants, including one NNRTI-resistant mutant (NNRTI-R), 2 protease inhibitor-resistant mutants (PI-R), and 2 integrase strand transfer inhibitor-resistant mutants (INSTI-R). The 2 remaining isolates had either NRTI resistance (NRTI-R) plus NNRTI-R or NRTI-R plus PI-R. TAF demonstrated antiviral activity against the NNRTI-R single-class-resistant mutant (<2-fold that of the wild type) and showed hypersusceptibility to the PI-R and INSTI-R single-class-resistant mutants (the FC was ≤0.5-fold that of the wild type). A comparable activity profile was observed for the NRTI internal control AZT against these mutants. For the 2 viruses that contained NRTI resistance-associated amino acid substitutions, TAF showed 2.1- and 5.4-fold-reduced susceptibilities, respectively, that were associated with the presence of a number of resistance substitutions, namely, 3 TAMs plus M184V (D67N, K70R, M184V, and K219E) in the first isolate (5705-72) and a multidrug resistance Q151M substitution complex (A62V, V75I, F116Y, and Q151M) plus K65R plus TAMs (M41L, D67N, L210W, and T215Y) in the second isolate (MDR-769). As these viruses harbored AZT resistance substitutions, much higher resistance was observed for AZT in those isolates (33.1- and >89-fold above those of the wild-type isolates, respectively). Control compounds for the resistance classes studied, NVP for NNRTI-R, IDV for PI-R, and RAL and EVG for INSTI-R, showed the expected reductions in susceptibility of their respective resistant isolates, while the control compound, T-20, showed hypersusceptibility of all mutants when tested.
TABLE 3.
TAF activities against drug-resistant primary HIV-1 isolates in PBMCs
| Isolate ID | Resistance class(es) | Resistance-associated amino acid substitution(s)b | EC50 fold changea |
||||||
|---|---|---|---|---|---|---|---|---|---|
| TAF | AZT | NVP | IDV | T20 | RAL | EVG | |||
| A-17 | NNRTI-R | RT: K103N Y181C | 1.7 | 0.7 | >380 | − | 0.2 | − | − |
| 1064-52 | PI-R | RT: D67N | 0.5 | 1.0 | − | 39.3 | 0.4 | − | − |
| PR: I54V V82F L90M | |||||||||
| 52-52 | PI-R | PR: M46I I54V V82T | 0.4 | 0.4 | − | 15.2 | 0.2 | − | − |
| 8070_1 | INSTI-R | IN: G140S Y143H Q148H | 0.2 | 0.2 | − | − | − | 250 | 222 |
| 4736_4 | INSTI-R | IN: E92Q N155H | 0.1 | 0.2 | − | − | − | 18.9 | 101 |
| 5705-72 | NRTI-R, NNRTI-R | RT: D67N K70R K103N M184V K219E | 2.1 | 33.1 | 279 | − | 0.6 | − | − |
| MDR-769 | NRTI-R, PI-R | RT: M41L A62V K65R D67N V75I F116Y Q151M L210W T215Y | 5.4 | >89 | − | 210 | 0.7 | − | − |
| PR: M46L I54V V82A I84V L90M | |||||||||
The fold changes calculated from the average EC50 across wild-type isolates were as follows: 3.4 nM (TAF), 11.2 nM (AZT), 25.1 nM (NVP), 12.0 nM (IDV), 39.4 nM (T20), 3.1 nM (RAL), and 1.0 nM (EVG). −, not tested.
IN, integrase; PR, protease.
DISCUSSION
TAF is the new prodrug of the HIV-1 NtRTI tenofovir. TAF has shown improved safety and pharmacologic profiles, as well as a more potent antiviral effect, compared to the current prodrug of tenofovir (TDF) in early clinical studies (2, 3). Treatment of HIV-infected subjects with the 25-mg therapeutic dose of TAF is associated with a 5-fold increase in the quantity of the active moiety, TFV-diphosphate, that is delivered to target cells in vivo compared to treatment with 300 mg TDF. In this report, we have described experiments aimed at characterizing the in vitro resistance profile of TAF, and we discuss a model linking our in vitro results with the increased delivery of TFV-DP observed in vivo upon dosing with TAF.
Resistance selection experiments using TAF and TFV in parallel gave rise to mutants harboring the K65R amino acid substitution in RT in all cases. Phenotypic analyses of the selected mutants showed virtually the same fold changes compared to the wild type regardless of the selecting drug. The resistance selection experiments were performed because of the differences in the chemical compositions of TAF and TFV, but the results were expected, since the active moiety for both TAF and TFV is the same: TFV-DP (as shown in Fig. 1). The S68N substitution observed in some of the selections is not novel (9) and may play a role similar to that previously seen with the S68G substitution in restoring the replication capacity associated with K65R (18). The presence of wild-type virus at a low drug concentration during the course of the selections is a reflection of the high MOI that is used in drug resistance selection experiments, where the quantity of virus from passage to passage is not controlled, unlike the conditions used for EC50 determination. The presence of wild-type virus does not indicate that the transient wild-type virus was resistant to the drug, but rather, that the wild-type virus was not yet fully depleted at that time and was still the major species detected by population sequencing. This is also substantiated by the data in Table 1, where the EC50s for TAF and TFV for the selected viruses are about 3 times lower than the concentrations at which the viruses can grow. Importantly, the selected resistant viruses harboring the K65R amino acid substitution were not able to grow at the highest concentrations tested, suggesting that TAF and TFV retain antiviral activity at higher concentrations against K65R mutants. This further suggests that the 5-fold-higher loading of TFV-DP to target cells obtained upon dosing with TAF compared to TDF has the possibility to overcome some level of resistance conferred by the K65R mutation in vivo.
As in the resistance selection experiments, TAF and TFV showed the same resistance profile when tested against a panel of patient-derived recombinant HIV-1 isolates with NRTI resistance-associated amino acid substitutions. As TFV-DP is the active moiety in target cells for both TAF and TFV, the high degree of correlation between the 2 compounds confirmed in the PhenoSense assay was expected. Notably, though, the absolute activity of TAF is much greater than that of TFV due to its higher cellular permeability. Eight of the 11 patient-derived isolates with sensitivity to TAF and TFV (FCs of <1.4, based on the TDF cutoffs) also harbored the M184V substitution. These results are in agreement with previous reports (8, 14, 23) that showed enhanced sensitivity of TFV in the presence of the M184V substitution. Importantly, the M184V-enhanced sensitivity to TAF and TFV was also observed for 2 isolates that also harbored the K65R substitution. Sensitivity to TAF and TFV was observed in 3 of the 5 isolates harboring the Q151M amino acid substitution, while isolates harboring the infrequent T69 double insertion showed the highest level of resistance to TAF and TFV, with rarely observed high FCs between 18 and 23. In a recent survey of 1,303 treatment-experienced subjects with phenotypic data, only 6 samples (<0.5%) had TFV FCs of >10 in the Monogram assay (unpublished data). Other samples that showed resistance to TAF and TFV (FCs of >4, based on TDF cutoffs) had 5 or 6 TAMs or the Q151M MDR complex plus the K65R substitution. Samples with reduced sensitivity to TAF and TFV (FCs between 1.4 and 4, based on TDF cutoffs) included 2 samples with the K65R substitution alone and samples with 3 and 6 TAMs or the Q151M MDR complex, indicating an overlap in the genetic makeup of viruses showing either reduced sensitivity or resistance to TFV.
As expected, the results reported in this paper indicate that TAF is active against PI-R, NNRTI-R, and INSTI-R single-class-resistant primary HIV-1 isolates in PBMCs. The two isolates with NRTI-R amino acid substitutions showed only moderately reduced susceptibility to TAF (2.1- and 5.4-fold above that of the wild type) that could possibly be overcome in vivo as a result of the increased TFV-DP level observed in PBMCs upon TAF dosing (2, 3). Hypersusceptibility to TAF and/or AZT (2.5- to 10-fold) was noted in INSTI-R and PI-R single-class-resistant mutants in this small data set of HIV-1 primary isolates. Further studies are required to assess the significance of these findings, as prior studies with similar mutants did not indicate such hypersusceptibility (32–34).
The data presented here have shown that qualitatively, the in vitro resistance profiles of TAF and TFV are identical. This finding was expected, as both TAF and TFV produce the same RT-inhibitory moiety in target cells, TFV-DP. However, as TAF loads the active inhibitor (TFV-DP) in vivo at concentrations 5 times higher than those with TDF (2, 3), it is conceivable that viruses able to escape the inhibitor (TFV-DP) when dosed with TDF because of mild to moderate resistance may not escape inhibition in the presence of 5 times more TFV-DP upon dosing with TAF. The 5-fold increase in the TFV-DP concentration would have the effect of increasing the resistance cutoffs by up to 5-fold, as modeled in Fig. 4. The projected TAF clinical cutoffs would theoretically be 5-fold higher than the TDF cutoffs, resulting in a lower cutoff of 7 (5 × 1.4) and a higher cutoff of 20 (5 × 4). Consequently, 18 of 24 (75%) viruses from the panel tested here would be considered sensitive to TAF (FC < 7) (up from 11 of 24 [46%]), 3 of 24 (12.5%) would be in the reduced sensitivity to TAF category (FCs between 7 and 20) (down from 6 of 24 [25%]), and 3 of 24 would be resistant to TAF (FC > 20) (down from 7 of 24 [29%]). These predictions were supported by in vitro experiments in MT-2 cells that mimicked the 5-fold-increased in vivo concentration of TFV-DP associated with TAF treatment compared to TDF (35). The results from these in vitro experiments showed that TAF, but not TFV, could prevent the breakthrough of viruses with multiple NRTI resistance mutations. Preliminary clinical observations (phase 2 and phase 3 studies) have shown a very low incidence of TFV genotypic resistance in treatment-naive patients treated either with TAF (1 in 978) or TDF (3 in 925) (unpublished data), and longer follow-up is needed to show a potential difference between TAF and TDF.
FIG 4.

TAF susceptibility in the Monogram panel (data from Table 3) and number of samples in each sensitivity category based on current cutoffs (TDF dosing) and projected cutoffs (5 times increased upon TAF dosing). The clinical cutoffs for TDF susceptibility in the Monogram assay are shown (an FC of <1.4 indicates sensitivity to TDF, an FC between 1.4 and 4 indicates reduced sensitivity to TDF, and an FC of >4 indicates resistance to TDF). Clinical cutoffs for TAF have not been established.
ACKNOWLEDGMENTS
We thank Michael Abram and Kirsten White for their critical review of the manuscript, Mike Tran for his help with the figures, and Heidi Fisher for her editorial assistance.
REFERENCES
- 1.Anonymous. 2003. VIREAD (tenofovir disoproxil fumarate) tablets. U.S. prescribing information. Gilead Sciences, Inc., Foster City, CA. [Google Scholar]
- 2.Ruane PJ, Dejesus E, Berger D, Markowitz M, Bredeek UF, Callebaut C, Zhong L, Ramanathan S, Rhee MS, Fordyce MW, Yale K. 2013. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of tenofovir alafenamide as 10-day monotherapy in HIV-1-positive adults. J Acquir Immune Defic Syndr 63:449–455. doi: 10.1097/QAI.0b013e3182965d45. [DOI] [PubMed] [Google Scholar]
- 3.Sax PE, Zolopa A, Brar I, Elion R, Ortiz R, Post F, Wang H, Callebaut C, Martin H, Fordyce MW, McCallister S. 2014. Tenofovir alafenamide vs. tenofovir disoproxil fumarate in single tablet regimens for initial HIV-1 therapy: a randomized phase 2 study. J Acquir Immune Defic Syndr 67:52–58. doi: 10.1097/QAI.0000000000000225. [DOI] [PubMed] [Google Scholar]
- 4.Lee WA, He G-X, Eisenberg E, Cihlar T, Swaminathan S, Mulato A, Cundy KC. 2005. Selective intracellular activation of a novel prodrug of the human immunodeficiency virus reverse transcriptase inhibitor tenofovir leads to preferential distribution and accumulation in lymphatic tissue. Antimicrob Agents Chemother 49:1898–1906. doi: 10.1128/AAC.49.5.1898-1906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bam RA, Birkus G, Babusis D, Cihlar T, Yant SR. 2014. Metabolism and antiretroviral activity of tenofovir alafenamide in CD4 T-cells and macrophages from demographically diverse donors. Antivir Ther 19:669–677. doi: 10.3851/IMP2767. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut C, Stepan G, Tian Y, Miller MD. 2015. In vitro virology profile of tenofovir alafenamide, a novel oral prodrug of tenofovir with improved antiviral activity compared to that of tenofovir disoproxil fumarate. Antimicrob Agents Chemother 59:5909–5916. doi: 10.1128/AAC.01152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller MD. 2004. K65R, TAMS and tenofovir. AIDS Rev 6:22–33. [PubMed] [Google Scholar]
- 8.Wainberg MA, Miller MD, Quan Y, Salomon H, Mulato AS, Lamy PD, Margot NA, Anton KE, Cherrington JM. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antivir Ther 4:87–94. [DOI] [PubMed] [Google Scholar]
- 9.Margot NA, Waters JM, Miller MD. 2006. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob Agents Chemother 50:4087–4095. doi: 10.1128/AAC.00816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laflamme G, Grant D, White K, Stray K, Boojamra C, Zhang L, Mackman R, Ray AS, Miller M, Cihlar T. 2007. Novel nucleotide inhibitor GS-9148 selects for a K70E mutation in HIV-1 reverse transcriptase and low-level resistance in vitro, poster H-1037. Abstr 47th Intersci Conf Antimicrob Agents Chemother. [Google Scholar]
- 11.Margot NA, Lu B, Cheng A, Miller MD. 2006. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in Study 903. HIV Med 7:442–450. doi: 10.1111/j.1468-1293.2006.00404.x. [DOI] [PubMed] [Google Scholar]
- 12.Kagan RM, Lee TS, Ross L, Lloyd RM Jr, Lewinski MA, Potts SJ. 2007. Molecular basis of antagonism between K70E and K65R tenofovir-associated mutations in HIV-1 reverse transcriptase. Antiviral Res 75:210–218. doi: 10.1016/j.antiviral.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 13.McColl DJ, Margot NA, Wulfsohn M, Coakley DF, Cheng AK, Miller MD. 2004. Patterns of resistance emerging in HIV-1 from antiretroviral-experienced patients undergoing intensification therapy with tenofovir disoproxil fumarate. J Acquir Immune Defic Syndr 37:1340–1350. doi: 10.1097/00126334-200411010-00002. [DOI] [PubMed] [Google Scholar]
- 14.Miller MD, Margot N, Lu B, Zhong L, Chen S-S, Cheng A, Wulfsohn M. 2004. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis 189:837–846. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- 15.Wolf K, Walter H, Beerenwinkel N, Keulen W, Kaiser R, Hoffmann D, Lengauer T, Selbig J, Vandamme A-M, Korn K, Schmidt B. 2003. Tenofovir resistance and resensitization. Antimicrob Agents Chemother 47:3478–3484. doi: 10.1128/AAC.47.11.3478-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White KL, Margot NA, Wrin T, Petropoulos CJ, Miller MD, Naeger LK. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob Agents Chemother 46:3437–3446. doi: 10.1128/AAC.46.11.3437-3446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das K, Bandwar RP, White KL, Feng JY, Sarafianos SG, Tuske S, Tu X, Clark AD Jr, Boyer PL, Hou X, Gaffney BL, Jones RA, Miller MD, Hughes SH, Arnold E. 2009. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J Biol Chem 284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svarovskaia ES, Feng JY, Margot NA, Myrick F, Goodman D, Ly JK, White KL, Kutty N, Wang R, Borroto-Esoda K, Miller MD. 2008. The A62V and S68G mutations in HIV-1 reverse transcriptase partially restore the replication defect associated with the K65R mutation. J Acquir Immune Defic Syndr 48:428–436. doi: 10.1097/QAI.0b013e31817bbe93. [DOI] [PubMed] [Google Scholar]
- 19.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B. 2004. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem 279:509–516. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 20.Quan Y, Gu Z, Li X, Li Z, Morrow CD, Wainberg MA. 1996. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (-)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J Virol 70:5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ly JK, Margot NA, MacArthur H, Hung M, Miller MD, White KL. 2007. The balance between NRTI discrimination and excision drives the susceptibility of HIV-1 RT mutants K65R, M184V and K65R+M184V. Antivir Chem Chemother 18:307–316. doi: 10.1177/095632020701800603. [DOI] [PubMed] [Google Scholar]
- 22.Xu HT, Asahchop EL, Oliveira M, Quashie PK, Quan Y, Brenner BG, Wainberg MA. 2011. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J Virol 85:11300–11308. doi: 10.1128/JVI.05584-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naeger LK, Margot NA, Miller MD. 2001. Increased drug susceptibility of HIV-1 reverse transcriptase mutants containing M184V and zidovudine-associated mutations: analysis of enzyme processivity, chain-terminator removal and viral replication. Antivir Ther 6:115–126. [PubMed] [Google Scholar]
- 24.Palmer S, Margot N, Gilbert H, Shaw N, Buckheit R Jr, Miller M. 2001. Tenofovir, adefovir, and zidovudine susceptibilities of primary human immunodeficiency virus type 1 isolates with non-B subtypes or nucleoside resistance. AIDS Res Hum Retrovir 17:1167–1173. doi: 10.1089/088922201316912772. [DOI] [PubMed] [Google Scholar]
- 25.Johnston E, Dupnik KM, Gonzales MJ, Winters MA, Rhee SY, Imamichi T, Shafer RW. 2005. Panel of prototypical infectious molecular HIV-1 clones containing multiple nucleoside reverse transcriptase inhibitor resistance mutations. AIDS 19:731–733. doi: 10.1097/01.aids.0000166098.54564.0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 27.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol 75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naeger LK, Margot NA, Miller MD. 2002. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 46:2179–2184. doi: 10.1128/AAC.46.7.2179-2184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margot NA, Hluhanich RM, Jones GS, Andreatta KN, Tsiang M, McColl DJ, White KL, Miller MD. 2012. In vitro resistance selections using elvitegravir, raltegravir, and two metabolites of elvitegravir M1 and M4. Antiviral Res 93:288–296. doi: 10.1016/j.antiviral.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Petropoulos CJ, Parkin NT, Limoli KL, Lie YS, Wrin T, Huang W, Tian H, Smith D, Winslow GA, Capon DJ, Whitcomb JM. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother 44:920–928. doi: 10.1128/AAC.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ptak RG, Gallay PA, Jochmans D, Halestrap AP, Ruegg UT, Pallansch LA, Bobardt MD, de Bethune MP, Neyts J, De Clercq E, Dumont JM, Scalfaro P, Besseghir K, Wenger RM, Rosenwirth B. 2008. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob Agents Chemother 52:1302–1317. doi: 10.1128/AAC.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margot NA, Gibbs CS, Miller MD. 2010. Phenotypic susceptibility to bevirimat in isolates from HIV-1-infected patients without prior exposure to bevirimat. Antimicrob Agents Chemother 54:2345–2353. doi: 10.1128/AAC.01784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McColl DJ, Chen X. 2010. Strand transfer inhibitors of HIV-1 integrase: bringing IN a new era of antiretroviral therapy. Antiviral Res 85:101–118. doi: 10.1016/j.antiviral.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Shimura K, Kodama E, Sakagami Y, Matsuzaki Y, Watanabe W, Yamataka K, Watanabe Y, Ohata Y, Doi S, Sato M, Kano M, Ikeda S, Matsuoka M. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J Virol 82:764–774. doi: 10.1128/JVI.01534-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margot N, Liu Y, Babusis D, Miller MD, Callebaut C. 2013. Antiviral activity of tenofovir alafenamide (TAF) against major NRTI-resistant viruses: improvement over TDF/TFV is driven by higher TFV-DP loading in target cells, poster 23. International Workshop on HIV and Hepatitis Virus Drug Resistance and Curative Strategies. [Google Scholar]