Abstract
In this study, we describe the molecular characterization of a plasmid-located blaNDM-1 harbored by an Acinetobacter clinical isolate recovered from a patient in Turkey that putatively constitutes a novel Acinetobacter species, as shown by its distinct ARDRA (amplified 16S ribosomal DNA restriction analysis) profile and molecular sequencing techniques. blaNDM-1 was carried by a conjugative plasmid widespread among non-baumannii Acinetobacter isolates, suggesting its potential for dissemination before reaching more clinically relevant Acinetobacter species.
TEXT
Acinetobacter spp. belonging to the Acinetobacter baumannii group are a major cause of nosocomial intensive care unit (ICU) infections, with A. baumannii being the species with the highest clinical relevance, followed by Acinetobacter pittii and Acinetobacter nosocomialis (1, 2).
The implementation of molecular identification methods and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has led to increasing reports of infections caused by non-baumannii Acinetobacter isolates and to the identification of putative novel Acinetobacter species (3, 4). The increase of carbapenem resistance in Acinetobacter spp. is also of concern, and we are witnessing the worldwide dissemination of NDM enzymes among different Acinetobacter spp. (5). Here, we report the molecular characterization of blaNDM-1 from a putatively novel Acinetobacter species isolated in Turkey.
An Acinetobacter species isolate (JVAP01) was recovered from a six-year-old female patient admitted to the Adnan Menderes University Hospital, Turkey, in August 2009 with a urinary tract infection. Empirical treatment with cefuroxime axetil (75 mg/kg/day) and nitrofurantoin (1 mg/kg/day) was initiated and sustained upon availability of microbiological data, since remission of the patient's symptoms was observed. Therapy was continued for 1 week, and upon recovery, she was discharged from the hospital. The patient had no history of travel or admission to other hospitals in Turkey.
JVAP01 was initially reported as belonging to the Acinetobacter calcoaceticus-A. baumannii complex by the Phoenix system (Becton Dickinson, USA) but identified as A. pittii (97%) and as A. calcoaceticus (99%) by recA and 16S-23S rRNA gene internal transcribed spacer (ITS) sequencing, respectively (6). Multilocus sequence typing (MLST) performed according to the Pasteur MLST scheme for A. baumannii (http://pubmlst.org/abaumannii/) identified a novel recA allele (recA106), and JVAP01 was assigned a novel sequence type (ST606).
Amplified 16S ribosomal DNA restriction analysis (ARDRA) (7) showed a 2-5-1-1-3-17 ARDRA pattern for CfoI, AluI, MboI, RsaI, MspI, and BfaI, respectively, matching the band pattern of several strains from the Acinetobacter reference collection of Leiden University. This collection of strains apparently constituted a novel Acinetobacter species, preliminarily termed “NB14,” and the strains showed high similarity in amplified fragment length polymorphism (AFLP) fingerprint analysis (data not shown).
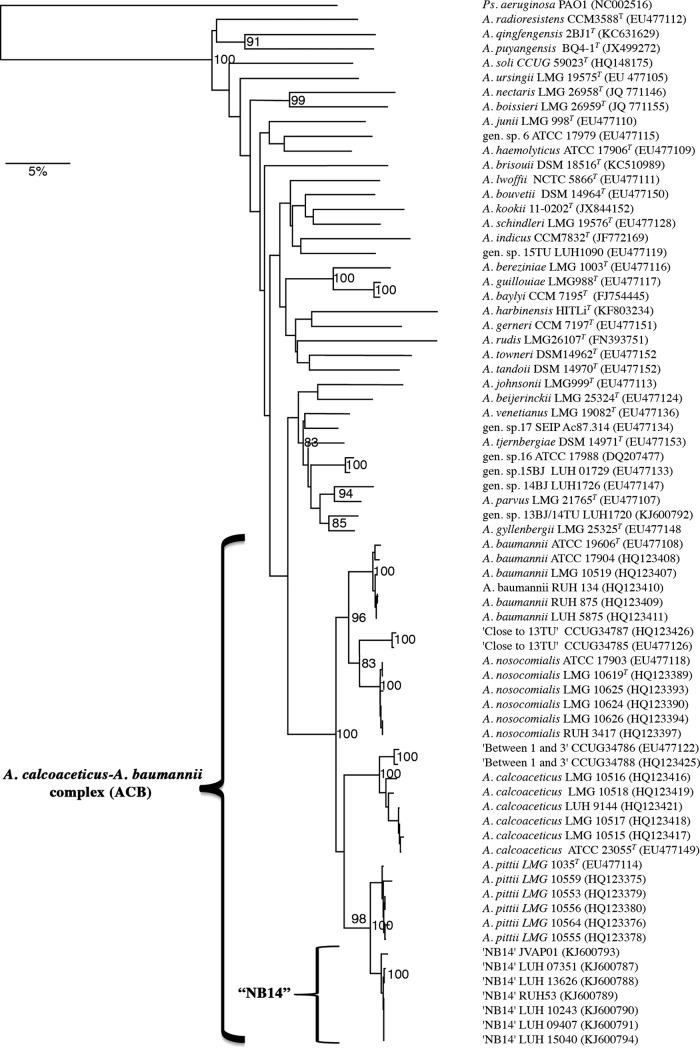
A comparative sequence analysis of the RNA polymerase β-subunit gene (rpoB) of JVAP01 was performed using the Bionumerics 5.1 software with default parameters as previously described (8, 9). The rpoB sequences of Acinetobacter spp. were used to create a neighbor-joining dendrogram for phylogenetic clustering with the Jukes-Cantor algorithm. As shown in Fig. 1, strains from each species of the A. calcoaceticus-A. baumannii complex formed respective clusters, supported by high bootstrap values, with strains belonging to NB14 forming a distinct branch that was closest to both A. pittii and A. calcoaceticus (Fig. 1). The bootstrap value for linkage of A. pittii to NB14 was 98%.
FIG 1.
Cluster analysis of the rpoB sequences from Acinetobacter spp. A rooted neighbor-joining tree based on the partial sequence of rpoB (nucleotide positions 2915 to 3775) of 70 strains of the A. calcoaceticus-A. baumannii complex, including those of NB14 and representatives of different species of the genus Acinetobacter, is shown. P. aeruginosa PAO1 was used as the outgroup. Bootstrap percentages based on 1,000 simulations are shown. GenBank accession numbers are given in parentheses. The bar indicates 5% sequence divergence.
The rpoB interspecies similarity values for the species of the A. calcoaceticus-A. baumannii complex ranged between 89.6 and 98% (data not shown), while the intraspecies similarity of NB14 strains ranged between 99.1 and 100%, and this group was closest to A. pittii (97 to 98%).
Antimicrobial susceptibility testing performed by Etest (AB-bioMérieux, Sweden) and interpreted according to EUCAST guidelines (version 4.0, 2014) showed that JVAP01 was resistant to β-lactams and kanamycin and susceptible to amikacin, gentamicin, ciprofloxacin, and colistin (Table 1). JVAP01 also yielded positive results in the imipenem-EDTA synergy test and the modified Hodge test (MHT), suggesting metallo-β-lactamase (MBL) production. PCR screening for class B and class D β-lactamase genes (10) was negative except for blaNDM-1, as confirmed by DNA sequencing. Of note, JVAP01 showed reduced susceptibility to amikacin despite carrying a full-length aphA6 gene, thus suggesting reduced expression of aphA6, as previously noted (11, 12).
TABLE 1.
Antimicrobial susceptibility testing by Etest for the indicated strains
| Antimicrobial(s) | MIC (mg/liter)a |
|||
|---|---|---|---|---|
| NB14 JVAP01 | A. baumannii ATCC 17978 | A. baumannii ATCC 17978-C | A. baumannii ATCC 17978-T | |
| Cefepime | >256 | 2 | >256 | >256 |
| Ceftazidime | >256 | 3 | >256 | >256 |
| Cefotaxime | >256 | 6 | >256 | >256 |
| Ciprofloxacin | 0.38 | 0.5 | 0.5 | 0.38 |
| Amikacin | 3 | 0.38 | 1 | 1.5 |
| Gentamicin | 0.125 | 0.19 | 0.25 | 0.5 |
| Kanamycin | 24 | 0.5 | 8 | 12 |
| Piperacillin-tazobactam | 128 | 0.32 | 96 | 32 |
| Imipenem | >32 | 0.25 | >32 | >32 |
| Meropenem | >32 | 0.38 | >32 | >32 |
| Tigecycline | 1 | 0.5 | 0.25 | 1 |
| Colistin | 0.38 | 0.125 | 0.38 | 0.38 |
| Rifampin | 2 | >32 | 32 | 32 |
C, transconjugant strain; T, transformed strain.
S1 nuclease pulse-field gel electrophoresis and Southern blot analysis (10) located both blaNDM-1 and aphA6 on a plasmid (pNDM-JVAP01) of approximately 50 kb that could not be assigned to any plasmid replicon type (13). pNDM-JVAP01 was successfully transferred to A. baumannii ATCC 17978 by both biparental mating and electroporation, but not to E. coli MC1061 (12). A. baumannii ATCC 17978 transconjugants and transformants acquired both blaNDM-1 and aphA6 as well as resistance to carbapenems and reduced susceptibility to kanamycin and amikacin, in agreement with MICs of the parental strain (Table 1). pNDM-JVAP01 was unstable in JVAP01, as it was spontaneously lost after 5 successive passages on blood agar plates in the absence of antibiotic pressure.
Genomic DNA from JVAP01 was used to sequence the pNDM-JVAP01 plasmid (paired-end 150-bp reads) in an Illumina MiSeq system, revealing the presence of a putative composite transposon Tn125 bracketing the blaNDM-1, bleMBL, trpF, dsbC, cutA1, groES, groEL, and ISCR21 array of genes (14). Overall, pNDM-JVAP01 is a molecule of 47,268 bp with a GC content of 40.8% also containing genes for a type IV secretion system presumably involved in plasmid conjugation and a Z toxin of unknown function. Interestingly, pNDM-JVAP01 displays 99.98% identity at the nucleotide level with pNDM-BJ01 (NC_019268) isolated from Acinetobacter lwoffii and more than 96% identity with NDM-harboring plasmids recovered from A. lwoffii, Acinetobacter bereziniae, and A. pittii (KJ547696, KF702385, and KJ003839, respectively), with only minor differences in the plasmid backbone sequences. The partial sequences of additional plasmids containing blaNDM-1 and recovered from A. haemolyticus (JQ080305) and A. junii (KJ018154) also show >99% identity with the nucleotide sequence of pNDM-JVAP01.
The appearance of several tentative novel species within the genus Acinetobacter makes accurate identification in routine diagnostic laboratories difficult, since phenotypic identification can only be considered presumptive. ITS and recA sequencing identification methods in this study provided inconclusive results, reflecting the limitation of methods that rely on the availability of curated databases (6). ARDRA analysis upon the inclusion of BfaI, however, highlighted the genetic relatedness between JVAP01 and Acinetobacter isolates tentatively belonging to a novel Acinetobacter species (NB14). Cluster analysis of rpoB sequences showed that JVAP01 formed a distinct monophyletic group that also included additional NB14 strains and that was closer to both A. pittii and A. calcoaceticus within the A. calcoaceticus-A. baumannii complex, providing further evidence that JVAP01 and NB14 strains represent a novel presumptive Acinetobacter species.
Genetic analysis of the JVAP01 strain revealed the common transposon Tn125 structure carrying blaNDM-1. Tn125 has been described as being inserted both in the chromosome and on plasmids of A. baumannii; however, in non-baumannii Acinetobacter species, Tn125 has been located exclusively on plasmids (5, 11, 12, 15–17). In JVAP01, Tn125 was integrated within a transferable plasmid of approximately 50 kb, highly similar to blaNDM-carrying plasmids recovered from non-baumannii Acinetobacter isolates and containing a type IV secretion system (5, 11, 15–17), suggesting the widespread nature of this plasmid and a role in blaNDM-1 transfer among different Acinetobacter species. This study reports the first identification of blaNDM-1 in a putatively novel Acinetobacter species which has been informally named NB14 and which is closely related to A. pittii, which has been recognized as the potential resistant reservoir for the dissemination of NDM-1, being found in food of animal origin and sewage and causing human infections and outbreaks in hospital units (18).
Nucleotide sequence accession numbers.
The rpoB sequences of NB14 strains and the pNDM-JVAP01 sequence were submitted to GenBank with accession numbers KJ600793 (JVAP01), KJ600787 (LUH 07351), KJ600788 (LUH 13626), KJ600789 (RUH 53), KJ600790 (LUH 10243), KJ600791 (LUH 09407), KJ600794 (LUH 15040), and KM923969.1 (pNDM-JVAP01).
ACKNOWLEDGMENTS
We thank the team of the curators of the Institute Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at http://pubmlst.org/abaumannii/.
We have no conflicts of interests to declare.
This study was supported by the Ministerio de Economía y Competitividad, Instituto de Salud Carlos III, cofinanced by European Regional Development Fund (ERDF) “A Way to Achieve Europe,” the Spanish Network for Research in Infectious Diseases (REIPI RD12/0015), and the Spanish Ministry of Health (grant number FIS 11/02024). This study was also supported by grant 2014 SGR 0653 from the Departament d'Universitats, Recerca i Societat de la Informació, of the Generalitat de Catalunya, by grant 10030 of the Adnan Menderes University Scientific Research Foundation, by funding from the European Community (SATURN, contract HEALTH-F3-2009241796), and by a research grant of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID, 2014) to P.E. D.R. was the recipient of a Sara Borrell postdoctoral contract (CD12/00482) from the Spanish Health Institute (Instituto Carlos III).
REFERENCES
- 1.Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front Microbiol 3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Espinal P, Roca I, Vila J. 2011. Clinical impact and molecular basis of antimicrobial resistance in non-baumannii Acinetobacter. Future Microbiol 6:495–511. doi: 10.2217/fmb.11.30. [DOI] [PubMed] [Google Scholar]
- 3.Nemec A, Krizova L, Maixnerova M, Sedo O, Brisse S, Higgins PG. 2015. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int J Syst Evol Microbiol 65:934–942. doi: 10.1099/ijs.0.000043. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Chen T, Yu R, Lu X, Zong Z. 2013. Acinetobacter pittii and Acinetobacter nosocomialis among clinical isolates of the Acinetobacter calcoaceticus-baumannii complex in Sichuan, China. Diagn Microbiol Infect Dis 76:392–395. doi: 10.1016/j.diagmicrobio.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y, Liu L, Li X, Chen Y, Jiang Y, Wang Y, Yu Y, Xie X. 2015. Spread of a common blaNDM-1-carrying plasmid among diverse Acinetobacter species. Infect Genet Evol 32:30–33. doi: 10.1016/j.meegid.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin Microbiol Infect 18:1097–1103. doi: 10.1111/j.1469-0691.2011.03696.x. [DOI] [PubMed] [Google Scholar]
- 7.Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, de Vos P, Claeys G, Verschraegen G. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol 33:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Scola B, Gundi VA, Khamis A, Raoult D. 2006. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemec A, Musilek M, Sedo O, De Baere T, Maixnerova M, van der Reijden TJ, Zdrahal Z, Vaneechoutte M, Dijkshoorn L. 2010. Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int J Syst Evol Microbiol 60:896–903. doi: 10.1099/ijs.0.013656-0. [DOI] [PubMed] [Google Scholar]
- 10.Espinal P, Fugazza G, Lopez Y, Kasma M, Lerman Y, Malhotra-Kumar S, Goossens H, Carmeli Y, Vila J. 2011. Dissemination of an NDM-2-producing Acinetobacter baumannii clone in an Israeli rehabilitation center. Antimicrob Agents Chemother 55:5396–5398. doi: 10.1128/AAC.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang X, Hao Q, Yang X, Xiao X, Luan C, Yang Y, Cui Y, Yang R, Gao GF, Song Y, Zhu B. 2012. Novel plasmid and its variant harboring both a bla(NDM-1) gene and type IV secretion system in clinical isolates of Acinetobacter lwoffii. Antimicrob Agents Chemother 56:1698–1702. doi: 10.1128/AAC.06199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roca I, Mosqueda N, Altun B, Espinal P, Akova M, Vila J. 2014. Molecular characterization of NDM-1-producing Acinetobacter pittii isolated from Turkey in 2006. J Antimicrob Chemother 69:3437–3438. doi: 10.1093/jac/dku306. [DOI] [PubMed] [Google Scholar]
- 13.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother 54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Wu C, Zhang Q, Qi J, Liu H, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-beta-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152. doi: 10.1371/journal.pone.0037152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Chen Y, Jia X, Luo Y, Song Q, Zhao W, Wang Y, Liu H, Zheng D, Xia Y, Yu R, Han X, Jiang G, Zhou Y, Zhou W, Hu X, Liang L, Han L. 2012. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect 18:E506–E513. doi: 10.1111/1469-0691.12035. [DOI] [PubMed] [Google Scholar]
- 17.Zong Z, Zhang X. 2013. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother 68:1007–1010. doi: 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 18.Bogaerts P, Huang TD, Rezende de Castro R, Bouchahrouf W, Glupczynski Y. 2013. Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J Antimicrob Chemother 68:2414–2415. doi: 10.1093/jac/dkt201. [DOI] [PubMed] [Google Scholar]