Abstract
Fungal infections are a leading cause of morbidity and death for hospitalized patients, mainly because they remain difficult to diagnose and to treat. Diseases range from widespread superficial infections such as vulvovaginal infections to life-threatening systemic candidiasis. For systemic mycoses, only a restricted arsenal of antifungal agents is available. Commonly used classes of antifungal compounds include azoles, polyenes, and echinocandins. Due to emerging resistance to standard therapies, significant side effects, and high costs for several antifungals, there is a need for new antifungals in the clinic. In order to expand the arsenal of compounds with antifungal activity, we previously screened a compound library using a cell-based screening assay. A set of novel benzimidazole derivatives, including (S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl)benzimidazole (EMC120B12), showed high antifungal activity against several species of pathogenic yeasts, including Candida glabrata and Candida krusei (species that are highly resistant to antifungals). In this study, comparative analysis of EMC120B12 versus fluconazole and nocodazole, using transcriptional profiling and sterol analysis, strongly suggested that EMC120B12 targets Erg11p in the ergosterol biosynthesis pathway and not microtubules, like other benzimidazoles. In addition to the marker sterol 14-methylergosta-8,24(28)-dien-3β,6α-diol, indicating Erg11p inhibition, related sterols that were hitherto unknown accumulated in the cells during EMC120B12 treatment. The novel sterols have a 3β,6α-diol structure. In addition to the identification of novel sterols, this is the first time that a benzimidazole structure has been shown to result in a block of the ergosterol pathway.
INTRODUCTION
Fungal infections represent a serious and currently unresolved health problem, especially in industrialized countries. Fungal infections account for ∼17% of intensive care unit infections in Europe, and similar numbers are reported from the United States (1–3). In addition, emerging resistance to commercially available antifungals has been reported (4, 5). Treatment, especially of systemic infections, is accompanied not only by intermediate success rates but also by high costs (6–9). In addition, common non-life-threatening superficial infections, such as recurrent vulvovaginal candidiasis, impose significant restrictions on patients, resulting in reduced quality of life. Due to the eukaryotic nature of fungal pathogens, well-tolerated antifungals are identified much less frequently than antibiotics targeting bacteria. Using an assay that mimics the smallest unit of a natural infection through incubation of the pathogen in the presence of host cells, we previously identified a benzimidazole, EMC120B12 [(S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl)benzimidazole] (10, 11), with large antifungal effects even against resistant yeasts such as Candida krusei (12). Our initial studies using transcriptional profiling indicated that the ergosterol pathway might be the target of the new compound. This was somewhat surprising, since benzimidazoles such as benomyl and nocodazole are known to target tubulin structures in eukaryotes, thereby blocking cell division (13, 14). Clinically relevant ergosterol biosynthesis inhibitors include the allylamines (naftifine and terbinafine) inhibiting squalene epoxidase (Erg1p) for topical (e.g., tinea pedis) and systemic treatment, the azoles (fluconazole, posaconazole, and voriconazole, for example) inhibiting C14-demethylase (CYP51 or Erg11p) for local and systemic treatment, and one morpholine (amorolfine) inhibiting both C14-reductase (Erg24p) and Δ8,7-isomerase (Erg2p) for treatment of onychomycosis (15–17).
To further analyze the mode of action of EMC120B12 and to identify its target, we performed a detailed comparative analysis on a genome-wide level, using transcriptional profiling in the presence and absence of the benzimidazole nocodazole, the azole fluconazole, and the benzimidazole EMC120B12. In addition, we analyzed the contents of individual sterols in Candida albicans and C. krusei strains after incubation with the drugs, in order to identify the precise target. Changes in sterol patterns, namely, accumulation of precursors and/or abnormal sterols, have been shown to allow the identification of the target enzyme(s) of ergosterol biosynthesis inhibitors. For this purpose, our recently developed whole-cell assay, employing liquid-liquid extraction and gas chromatography-ion trap mass spectrometry (GC-IT-MS), was used (16). Interestingly, under treatment with EMC120B12, not only the marker sterol for C14-demethylase inhibition, i.e., 14-methylergosta-8,24(28)-dien-3β,6α-diol (compound 4), but also hitherto unknown related sterols accumulated in C. krusei and C. albicans. Fluconazole showed the same effect but only at a much higher concentration. The highly similar sterol patterns after incubation with EMC120B12 and fluconazole and the highly similar transcriptional profiles strongly suggest that the compounds act on the same target, namely, Erg11p.
MATERIALS AND METHODS
Strains and media.
The Candida strains used in this study included clinical isolates of C. albicans SC5314 (18), with low MICs of 1 μg/ml for fluconazole and 0.125 μg/ml for EMC120B12, and the previously described strain Can741 (C. albicans 971), with MICs of 32 μg/ml for fluconazole and 8 μg/ml for EMC120B12, and clinical isolates of C. krusei (CK201 and CK242), both showing intrinsic resistance against fluconazole (MICs of 32 μg/ml) but exhibiting low MICs for EMC120B12 (MICs of 1 μg/ml). All MICs for each strain used in this work were determined as reported previously by us (11), according to EUCAST guidelines (19). Yeast strains were cultured overnight in yeast extract-peptone-dextrose (YPD) medium containing 2% glucose (Difco), from glycerol stock cultures, or were plated onto YPD agar plates (2% Bacto agar; Difco) for 48 h at 30°C and then transferred into the respective medium, if required.
Tubulin staining.
The mitotic spindles in proliferating Candida cells were analyzed by immunofluorescence according to the method described by Kilmartin and Adams (20), with some modifications. Cells were inoculated (optical density at 600 nm [OD600] of 0.2) and grown to an OD600 of 0.5, and then a test compound (nocodazole at 7.5 μg/ml, fluconazole at 0.5 μg/ml, or EMC120B12 at 0.1 μg/ml) or the control (dimethyl sulfoxide [DMSO]) was added for another 2.5 h. Antitubulin antibody (YOL1/34; Abcam) and DyLight 488-labeled secondary antibodies (Abcam) were incubated with cells for 1 h each, at room temperature, and the cells were washed at least three times after antibody incubation. Staining with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) was performed according to the manufacturer's instructions, before microscopic images were obtained.
Cultivation of C. albicans SC5314 for DNA microarray experiments.
To compare the effects of EMC120B12, fluconazole, and nocodazole on the transcriptome of C. albicans, 100 ml RPMI 1640 medium (Invitrogen, Karlsruhe, Germany) with 10% fetal calf serum (FCS) (Gibco Life Technologies GmbH, Karlsruhe, Germany) was inoculated with C. albicans SC5314 from an overnight culture (in 10 ml YPD medium [containing 1% yeast extract, 2% Bacto peptone, and 2% glucose] at 30°C), to an OD600 of 0.4, and cells were grown for 3 h at 37°C, with or without the amount of test compound corresponding to the 50% inhibitory concentration (IC50). Cells were centrifuged (1,200 × g for 3 min), and the supernatant was discarded. Subsequently, cells were resuspended in the remaining 500 μl of YPD medium and dropped into liquid N2, to generate cell beads for RNA isolation. The experiments were performed in triplicate.
Experimental design.
In total, three biological replicates were performed with each drug. All experiments were managed as dye swaps. Hybridization experiments included an untreated reference sample and a sample of cells treated with EMC120B12, fluconazole, or nocodazole.
Printing and processing of whole-genome DNA microarrays.
Whole-genome C. albicans SC5314 DNA microarrays were printed as described previously (11). For data analysis, a “gal file” was produced using TAS Application Suite software (version 2.7.1.18; Genomic Solutions, Huntingdon, United Kingdom) (11). To avoid nonspecific binding, whole-genome C. albicans SC5314 DNA microarrays were blocked as described previously (11).
RNA handling and hybridization.
RNA isolation, preparation for microarray analysis, and hybridization were performed exactly as described previously (11).
Scanning process.
Completely dried DNA microarrays were scanned using a GenePix 4300A microarray scanner (Molecular Devices, Sunnyvale, CA) (resolution, 10 μm; photomultiplier [PMT] setting, auto-PMT). Pictures of both channels were saved as 16-bit TIFF files.
Data analysis.
Raw data were created using Molecular Devices GenePix Pro 7.0 imaging software (Molecular Devices, Sunnyvale, CA). The data were statistically analyzed using R software (version 2.10.1), the Limma package (version 3.2.1), and the StatMod package (version 1.4.2) (21, 22), as described previously (11, 22–25). Background correction was performed using the “normexp” method, with an offset of 50 (26). The data were normalized with “print-tip-loess” normalization (27). Spots that were classified as not found by the image analysis software were ignored for fitting of the linear model. Multiple-hypothesis testing was performed (28, 29). Genes were regarded as differentially expressed and were investigated when the adjusted P values were 0.05 (therefore nominally controlling the expected false discovery rate [FDR] to less than 0.05) and the genes showed at least 2-fold upregulation or downregulation. More than 80% of all genes were detectable in the arrays with very high reproducibility, reflecting the high quality of the arrays.
Sterol analysis. (i) Cultivation and extraction of cells.
The Candida spp. selected were treated with several different concentrations of the test compounds EMC120B12 and fluconazole, as indicated below, and with nocodazole at 7.5 μg/ml. The concentrations of the compounds were chosen on the basis of the resistance characteristics of the isolates selected, i.e., the MICs determined with the EUCAST test (11). All compounds were dissolved in DMSO at concentrations that required a maximum of 10 μl to be added to the medium. The strains were grown in 24-well plates, with a final volume of 2 ml. The cell culture plates were inoculated with 107 cells and then incubated with the compounds for 48 ± 2 h at 30°C, without shaking. The analysis was performed with at least two biological replicates. After cultivation in the presence or absence of antifungal compounds, the cells were pelleted at 10,000 × g for 5 min, the supernatant was discarded, and the pellet was washed twice using 1 ml of phosphate-buffered saline (PBS). To the pellet, 1 ml of 2 M NaOH was added. The suspension was sonicated for 5 min, to separate cells adhering to each other. The suspensions were transferred into 4-ml glass vials with Teflon septa, and the vials were flooded with N2 and closed tightly. The sealed vials were incubated for 1 h at 70°C, in order to break the cells and to saponify the sterol esters, and the suspensions were transferred into 2-ml micro-test tubes. The glass vials were rinsed with 700 μl tert-butyl methyl ether (TBME) (Sigma-Aldrich, Steinheim, Germany) and combined with the lysed cell suspensions. Each suspension received 50 μl internal standard (cholestane, 10 mg/liter; Sigma-Aldrich, Steinheim, Germany) and was shaken vigorously for 1 min. The mixture was then centrifuged at 10,000 × g for 5 min, resulting in two separated phases. The upper organic phase was carefully removed and transferred to a new microcentrifuge glass vial, which contained 40 ± 5 mg of a 7:1 mixture of anhydrous Na2SO4 and primary secondary amine (PSA) for dispersive solid-phase extraction (dSPE) (Agilent Technologies, Waldbronn, Germany). The extraction was repeated with 750 μl of TBME, and the combined organic phases were shaken for at least 1 min with the PSA mixture, which removed interfering sugar and fatty acid residues. After centrifugation for 5 min at 10,000 × g, approximately 1 ml of the supernatant from each sample was transferred to a 1.5-ml brown glass autosampler vial. The samples were dried in a SpeedVac vacuum concentrator for 20 min. Each residue was dissolved in 950 μl of TBME, and 50 μl of the N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA)/N-trimethylsilylimidazole (TSIM) (9:1) silylation reagent mixture (Macherey-Nagel, Düren, Germany) was added. The samples were gently shaken and maintained at room temperature for at least 30 min, for completion of the silylation reaction, before being subjected to GC-IT-MS analysis.
(ii) GC-IT-MS.
The gas chromatograph (CP3800)-ion trap mass spectrometer (2100; Varian, Darmstadt, Germany) had the following settings: injector temperature, 250°C; injection volume, 1 μl (splitless time, 1 min). The stationary phase was a 30-m VF-5ms capillary column with a 10-m EZ-Guard column (Agilent, Waldbronn, Germany), with an inner diameter of 0.25 mm and a film thickness of 0.25 μm. The mobile phase was helium (99.999%) delivered at a constant flow rate of 1.4 ml/min. The transfer line temperature was 270°C, and the ion trap temperature was 200°C. The GC oven started at 50°C and ramped up to 260°C (heating rate, 50°C/min), followed by a gradient of 4°C/min up to 310°C, with a hold time of 1 min. The total run time was 18.7 min. The mass spectrometer operated in 3 segments. In the first segment (solvent cutoff), the mass spectrometer was switched on after 9 min. In the second segment, the mass spectrometer scanned from m/z 50 to m/z 450. In the last segment, the mass spectrometer scanned from m/z 100 to m/z 650 (with electron ionization at 70 eV). Data analysis and instrument control were performed with Varian Workstation 6.9 SP 1 software (Varian, Darmstadt, Germany). The sterols were analyzed as trimethylsilyl (TMS) ethers. The sterol TMS ethers were identified by their mass spectra and relative retention times (RRTs), according to the method described by Müller et al. (16).
Microarray data accession number.
The microarray data were deposited in GEO under accession number GSE64976.
RESULTS
Antifungal properties of EMC120B12.
EMC120B12 is a novel benzimidazole derivative that was identified by us previously (11) and showed high antifungal activities against the major Candida species, including C. albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, and C. krusei, of which C. krusei is especially notorious for showing high-level resistance to azoles. In our previous publication, we demonstrated that EMC12B12 showed only limited toxicity for three mammalian cell lines tested. HeLa cells showed the greatest sensitivity to EMC120B12 (IC50, 62.5 μM), whereas A549 and CHO-K1 cells showed lower sensitivity (IC50, 125 μM). With an average EMC120B12 MIC for C. albicans (61 C. albicans isolates tested according to EUCAST guidelines [19]) of 1.1 μg/ml, the selectivity/toxicity index (IC50 in mammalian cells/MIC) was determined to be between 57 and 114. In order to identify the mode of action of EMC120B12, we performed detailed comparative analyses on both a genome-wide level and a biochemical level, using transcriptional profiling and sterol analysis, respectively.
Transcriptional analysis.
As shown in Table 1, the strain incubated with fluconazole showed a profile highly similar to the profile identified in the presence of EMC120B12. In both cases, genes encoding enzymes required for ergosterol biosynthesis were most prominently induced. In total, 13 genes directly involved in ergosterol biosynthesis, plus the regulatory factor UPC2, were induced to almost identical levels in SC5314 cells incubated with EMC120B12 or with fluconazole. For these genes, no changes in expression levels were observed in the presence of nocodazole. Gene ontology (GO) term analysis (http://www.candidagenome.org/cgi-bin/GO/goTermFinder) confirmed this finding, as it revealed a clustering of differentially regulated genes associated with the ergosterol biosynthetic process for EMC120B12 (cluster frequency, 28.9%; background frequency, 0.4%), as well as for fluconazole (cluster frequency, 32.3%; background frequency, 0.4%). In contrast, comparing the profiles of EMC120B12 and fluconazole with that of the benzimidazole nocodazole showed no significant similarity. In the presence of nocodazole, genes related to amino acid metabolism were slightly affected (arginine metabolic process; cluster frequency, 5.7%; background frequency, 0.2%); however, genes required for ergosterol biosynthesis were not induced significantly (Table 1). Thus, comparison of the responses of C. albicans to EMC120B12 and fluconazole versus the response to the benzimidazole nocodazole indicates distinct targets. These data can be found in Table 1.
TABLE 1.
Comparison of genes differentially expressed by C. albicans SC5314 in the presence of EMC120B12, fluconazole, and nocodazole, based on transcriptional profiling
| Gene name and function | EMC120B12 |
Fluconazole |
Nocodazole |
|||
|---|---|---|---|---|---|---|
| Fold expression | Adjusted P | Fold expression | Adjusted P | Fold expression | Adjusted P | |
| Ergosterol biosynthesis | ||||||
| ERG1 | 4.34 | 0.0001 | 4.76 | 0.0007 | −1.07 | 0.4709 |
| ERG2 | 2.96 | 0.0068 | 2.82 | 0.1655 | 1.10 | 0.3781 |
| ERG3 | 6.95 | <0.0001 | 4.52 | 0.0070 | −1.66 | 0.0131 |
| ERG4 | 3.08 | 0.0014 | 2.16 | 0.0181 | −1.01 | 0.9403 |
| ERG5 | 3.05 | 0.0001 | 2.21 | 0.0263 | −1.04 | 0.6648 |
| ERG6 | 11.36 | <0.0001 | 8.34 | 0.0007 | −1.19 | 0.2257 |
| ERG10 | 2.60 | 0.0009 | 2.05 | 0.0362 | −1.20 | 0.0145 |
| ERG11 | 8.41 | <0.0001 | 5.02 | 0.0024 | −1.12 | 0.1592 |
| ERG13 | 4.91 | <0.0001 | 3.32 | 0.0074 | −1.11 | 0.1258 |
| ERG24 | 3.32 | 0.0016 | 2.83 | 0.0033 | 1.01 | 0.8406 |
| ERG25 | 2.64 | 0.0002 | 2.82 | 0.0070 | 1.02 | 0.7591 |
| ERG251 | 4.66 | <0.0001 | 4.90 | 0.0025 | −1.37 | 0.0008 |
| ERG26 | 2.09 | 0.0002 | 2.25 | 0.0061 | −1.03 | 0.7119 |
| UPC2 | 3.47 | 0.0002 | 4.31 | 0.0024 | 1.14 | 0.1577 |
| Response to antimycotics | ||||||
| CHT2 | −3.90 | <0.0001 | −5.34 | 0.0007 | −11.51 | <0.0001 |
| CRH11 | 2.02 | 0.0069 | 1.74 | 0.2978 | −1.13 | 0.2070 |
| CSH1 | 2.12 | 0.0055 | 1.75 | 0.5433 | 2.24 | 0.0002 |
| DDR48 | 13.84 | <0.0001 | 12.31 | 0.0070 | 1.64 | 0.0025 |
| FRP1 | 2.01 | 0.0008 | 1.57 | 0.3109 | 1.55 | 0.0004 |
| FTH1 | 2.03 | 0.0005 | 2.21 | 0.0235 | 1.66 | 0.0008 |
| HYR1 | 2.72 | 0.0038 | 1.92 | 0.2725 | −2.32 | 0.0001 |
| NHP6A | −2.04 | 0.0014 | −1.34 | 0.9995 | −2.32 | <0.0001 |
| orf19.6688 | −3.39 | 0.0001 | −3.00 | 0.1915 | −4.24 | <0.0001 |
| orf19.7504 | 2.46 | 0.0077 | 4.11 | 0.0024 | 1.07 | 0.3812 |
| PGA7 | 2.46 | 0.0005 | 1.53 | 0.6487 | 1.49 | 0.0045 |
| PHR2 | 4.38 | 0.0001 | 5.22 | 0.0362 | 1.21 | 0.1683 |
| POL30 | −2.00 | 0.0069 | −2.68 | 0.0007 | −1.20 | 0.0336 |
| RTA2 | 4.18 | 0.0003 | 5.31 | 0.0024 | 2.23 | 0.0009 |
| Other functions | ||||||
| HTA2 | −2.32 | 0.0038 | −3.16 | 0.0021 | −2.90 | <0.0001 |
| MAL2 | −2.96 | 0.0002 | −2.62 | 0.2452 | −1.72 | 0.0013 |
| MAL31 | −2.75 | 0.0038 | −1.96 | 0.3062 | −1.61 | 0.0130 |
| orf19.2125 | 3.12 | 0.0002 | 3.60 | 0.0048 | −1.15 | 0.2970 |
| orf19.4476 | 2.34 | 0.0077 | 2.42 | 0.0303 | 3.51 | 0.0001 |
| orf19.5799 | 2.59 | <0.0001 | 3.47 | 0.0070 | 1.54 | 0.0007 |
| orf19.6840 | 2.43 | 0.0003 | 3.29 | 0.0172 | −1.01 | 0.9456 |
| orf19.7455 | 3.07 | 0.0002 | 4.80 | 0.0389 | −1.03 | 0.8849 |
| orf19.90 | 2.93 | 0.0006 | 2.79 | 0.0953 | −1.37 | 0.0081 |
| PGA23 | 5.50 | <0.0001 | 2.94 | 0.0070 | −4.36 | <0.0001 |
| PGA26 | −3.35 | 0.0003 | −3.11 | 0.6235 | −3.80 | <0.0001 |
| PST3 | 2.10 | <0.0001 | 1.84 | 0.1448 | 1.30 | 0.0254 |
| RBT1 | −2.63 | 0.0002 | −2.70 | 0.1294 | −1.37 | 0.0026 |
| SET3 | 3.19 | <0.0001 | 3.25 | 0.0151 | −1.1 | 0.2382 |
| ARO10 | −5.25 | 0.0007 | −3.66 | 0.0626 | −6.96 | <0.0001 |
| CDG1 | −2.04 | 0.0003 | −1.78 | 0.3509 | −3.09 | <0.0001 |
| CHA1 | −2.46 | 0.0007 | −1.87 | 0.3062 | −3.00 | <0.0001 |
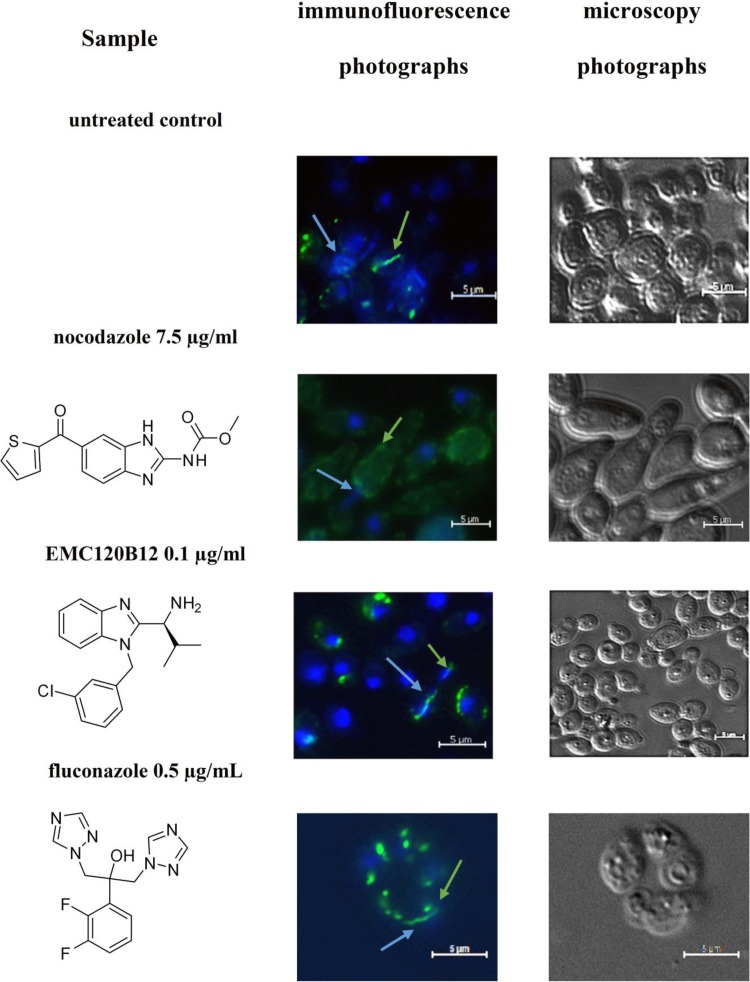
The differences in the actions of nocodazole and EMC120B12 on C. albicans could be confirmed by analyzing the tubulin structures of mitotic spindles in vivo in the presence and absence of all three compounds. As shown in Fig. 1, EMC120B12 and fluconazole had no effect on the formation of mitotic spindles in the cells. As expected, however, nocodazole completely blocked formation of the tubulin spindle. These results confirmed, both on a transcriptional level and by cell biological methods, that the benzimidazole EMC120B12, in contrast to other benzimidazoles such as nocodazole, had no detectable effect on tubulin structures. Instead, the similarity of the transcriptional profiles for fluconazole and EMC120B12 strongly suggested the ergosterol pathway as a common target.
FIG 1.
Tubulin staining of C. albicans. C. albicans was incubated with three antifungal compounds, as indicated, or DMSO as the control treatment. The presence or absence of mitotic spindles was monitored using antitubulin antibodies coupled to Alexa 488. Green strings and dots indicate tubulin structures, such as mitotic spindles. Nuclei were stained using DAPI. Blue arrows, nuclei; green arrows, Candida tubulin.
Analysis of sterol patterns to determine the target enzyme of EMC120B12.
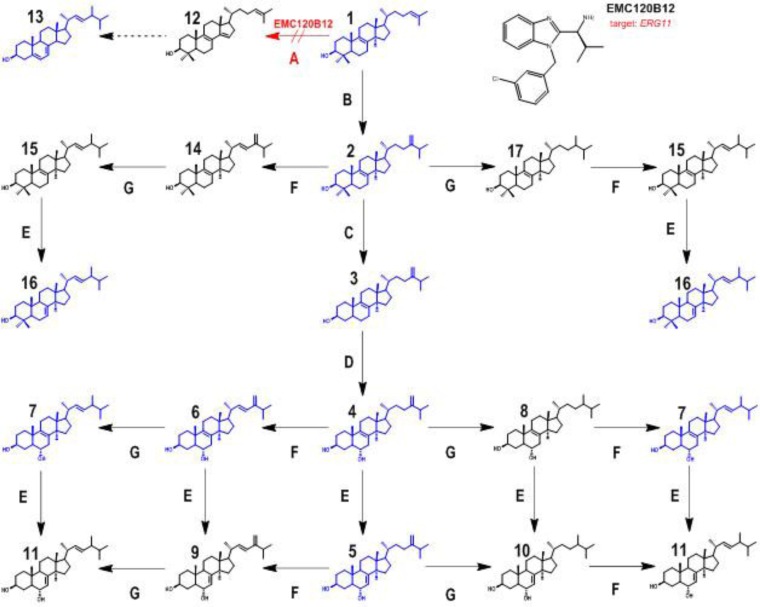
In a biochemical approach, we analyzed the sterol patterns for more susceptible and resistant isolates of C. albicans and C. krusei isolates in the presence and absence of the three compounds EMC120B12, fluconazole, and nocodazole. Analysis of the changes in the sterol patterns upon incubation with potential antifungal compounds is a highly efficient method for identification of a defined target enzyme. To identify the target enzyme of EMC120B12, we analyzed the inhibitory effects of EMC120B12 on the distal ergosterol biosynthesis pathway, in which eight distinct enzymes are involved in the conversion of lanosterol to ergosterol (16, 30–32). Treatment with ergosterol biosynthesis inhibitors leads, depending on the target enzyme, to accumulation of typical ergosterol precursors, e.g., lanosterol (compound 1) (inhibition of C14-demethylase) and/or aberrantly formed sterols such as lichesterol (inhibition of sterol Δ8/7-isomerase) or 14-methylergosta-8,24(28)-dien-3β,6α-diol (compound 4; inhibition of C14-demethylase) (Fig. 2) (16, 33–38). We used the GC-IT-MS technique to analyze the sterol patterns of selected C. albicans and C. krusei strains after incubation with EMC120B12, and we compared the results with those obtained with fluconazole and nocodazole (Fig. 3 and 4). Sterol patterns were analyzed for clinical isolates of C. albicans with low MICs (SC5314) (Fig. 4A1 to A6) and resistance against fluconazole (Can741) (Fig. 4B1 to B6), as well as in clinical isolates of C. krusei (CK201 and CK242) that showed intrinsic resistance to fluconazole but low MICs for EMC120B12 (CK201 is shown in Fig. 4C1 to C6) (11). Since azoles such as fluconazole are known to be fungistatic, rather than fungicidal, concentrations higher than the MIC were used. In the following paragraphs, we first describe the identification of the isolated sterols by GC-MS. Both MS data and chromatographic data (retention times) were needed to assign structures to the sterols. After assigning the structures, we describe the observed effects of the three compounds on the sterol patterns of the Candida isolates mentioned above.
FIG 2.
Sterol biosynthesis pathway after inhibition of sterol C14-demethylase (encoded by ERG11). Enzyme A, sterol C14-demethylase (ERG11); enzyme B, sterol C24-methyltransferase (ERG6); enzyme C, sterol C4-demethylase (ERG25, ERG26, and ERG27); enzyme D, sterol C5-desaturase (ERG3); enzyme E, sterol Δ8(7)-isomerase (ERG2); enzyme F, sterol C22-desaturase (ERG5); enzyme G, Δ24(28)-reductase (ERG4). Compound 1, lanosta-8,24-dien-3β-ol (lanosterol); compound 2, 4,4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol); compound 3, 14-methylergosta-8,24(28)-dien-3β-ol (14-methylfecosterol); compound 4, 14-methylergosta-8,24(28)-dien-3β,6α-diol; compound 5, 14-methylergosta-7,24(28)-dien-3β,6α-diol; compound 6, 14-methylergosta-8,22,24(28)-trien-3β,6α-diol; compound 7, 14-methylergosta-8,22-dien-3β,6α-diol; compound 8, 14-methylergosta-8-en-3β,6α-diol; compound 9, 14-methylergosta-7,22,24(28)-trien-3β,6α-diol; compound 10, 14-methylergosta-7-en-3β,6α-diol; compound 11, 14-methylergosta-7,22-dien-3β,6α-diol; compound 12, 4,4-dimethylergosta-8,14,24-trien-3β-ol (follicular fluid-meiosis-activating sterol [FF-MAS]); compound 13, ergosta-5,7,22-trien-3β-ol (ergosterol); compound 14, 4,4,14-trimethylergosta-8,22,24(28)-trien-3β-ol; compound 15, 4,4,14-trimethylergosta-8,22-dien-3β-ol; compound 16, 4,4,14-trimethylergosta-7,22-dien-3β-ol; compound 17, 4,4,14-trimethylergosta-8-en-3β-ol. Sterol structures shown in blue, structures detected in this study; sterol structures shown in black, structures postulated but not detected. Red arrow, inhibition of ERG11 caused by EMC120B.
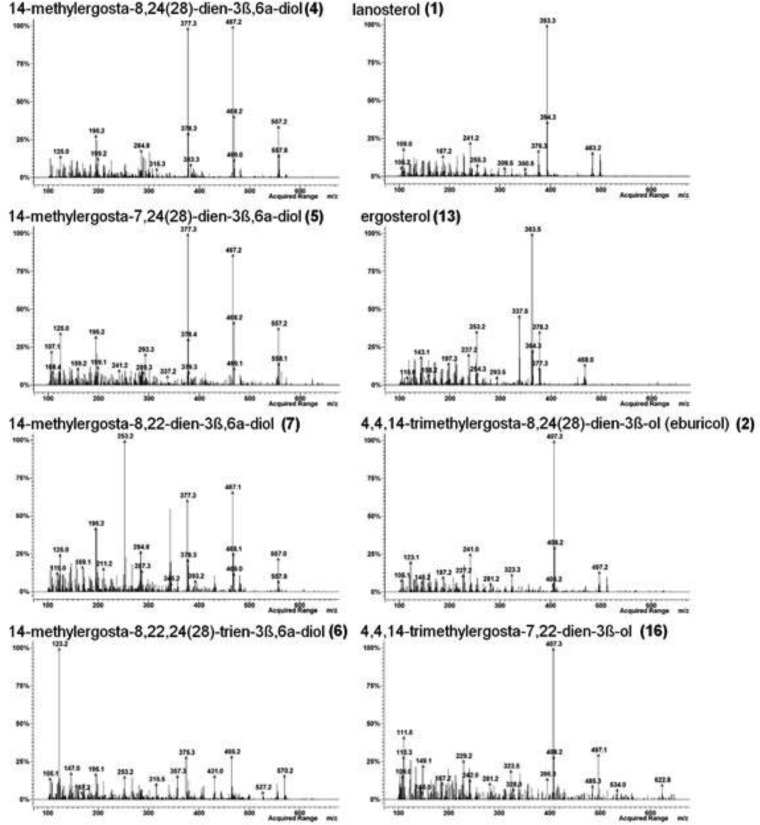
FIG 3.
Mass spectra of sterol TMS ethers detected in WT Candida krusei (CK242) after treatment with EMC120B12 (4 μg/ml) (see also Fig. 4). Left, sterols with a 3β,6α-diol structure; right, remaining sterols from Fig. 4.
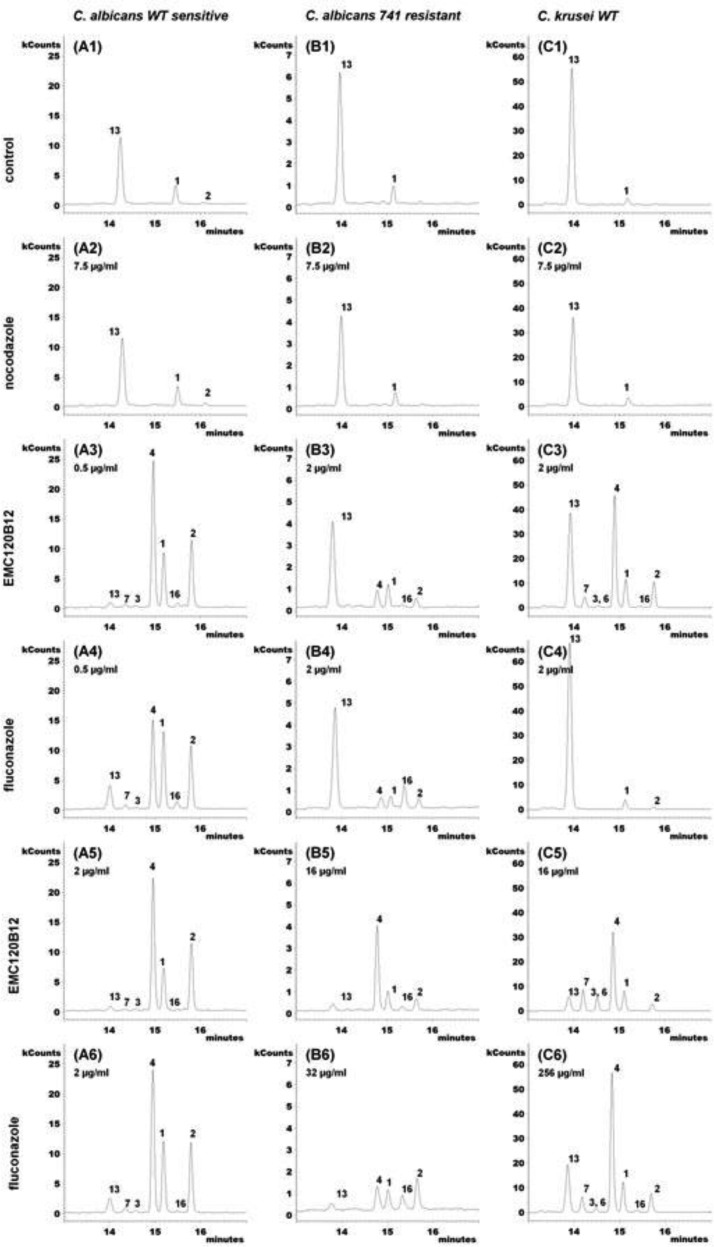
FIG 4.
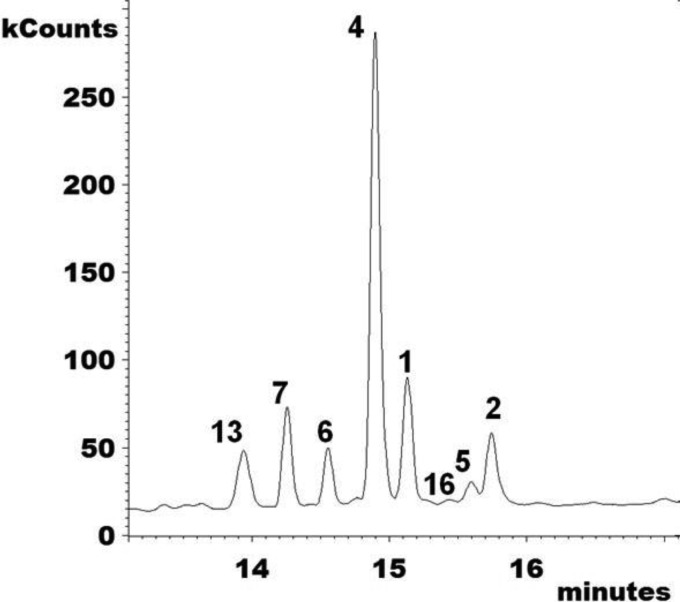
Extracted ion chromatograms (m/z 123, 253, 363, 377, 379, 393, 407, and 467) of accumulating sterols after treatment with EMC120B12, fluconazole, or nocodazole. (A1 to A6) WT Candida albicans (SC5314, sensitive); (A1) control; (A2) nocodazole at 7.5 μg/ml; (A3) EMC120B12 at 0.5 μg/ml; (A4) fluconazole at 0.5 μg/ml; (A5) EMC120B12 at 2 μg/ml; (A6) fluconazole at 2 μg/ml. (B1 to B6) Candida albicans Can741 (resistant); (B1) control; (B2) nocodazole at 7.5 μg/ml; (B3) EMC120B12 at 2 μg/ml; (B4) fluconazole at 2 μg/ml; (B5) EMC120B12 at 16 μg/ml; (B6) fluconazole at 32 μg/ml. (C1 to C6) WT Candida krusei (CK201); (C1) control; (C2) nocodazole at 7.5 μg/ml; (C3) EMC120B12 at 2 μg/ml; (C4) fluconazole at 2 μg/ml; (C5) EMC120B12 at 16 μg/ml; (C6) fluconazole at 256 μg/ml. Sterol 1, lanosterol (m/z 393); sterol 2, 4,4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol; m/z 407); sterol 3, 14-methylergosta-8,24(28)-dien-3β-ol (14-methylfecosterol; m/z 379); sterol 4, 14-methylergosta-8,24(28)-dien-3β,6α-diol (m/z 377); sterol 6, 14-methylergosta-8,22,24(28)-trien-3β,6α-diol (m/z 123); sterol 7, 14-methylergosta-8,22-dien-3β,6α-diol (m/z 253); sterol 13, ergosta-5,7,22-trien-3β-ol (ergosterol; m/z 363); sterol 16, 4,4,14-trimethylergosta-7,22-dien-3β-ol (m/z 407).
Identification of accumulating sterols by mass spectrometry.
For correlation of the MS data and the structures of the individual sterols isolated, literature data were extensively used. Furthermore, as outlined below, chromatographic data were included in the identification of the structures of the isolated sterols. On the basis of comprehensive analytical data in the literature, the sterols 4 (39, 40) and 2 (41–43) were identified unambiguously (Fig. 2). The mass spectrum of the bis-TMS ether of the known diol 4, the marker sterol for inhibition of Erg11p, showed a marginal molecular peak ([M]+) at m/z 572 but a strong fragmentation sequence of m/z 557 ([M−CH3]+), m/z 467 ([M−CH3−TMS]+), and m/z 377 ([M−CH3−2TMS]+) (Fig. 3). The relative retention time (RRT) was 1.43 (Table 2; see also Fig. 4).
TABLE 2.
Calculated and detected RRTs for sterol TMS ethers
| Sterol | Calculated RRT | Detected RRT |
|---|---|---|
| 3β,6α-Diol sterols | ||
| 14-Methylergosta-8,24(28)-dien-3β,6α-diol (compound 4) | 1.44 | 1.43 |
| 14-Methylergosta-7,24(28)-dien-3β,6α-diol (compound 5) | 1.47 | 1.49 |
| 14-Methylergosta-8,22,24(28)-trien-3β,6α-diol (compound 6) | 1.41 | 1.39 |
| 14-Methylergosta-8,22-dien-3β,6α-diol (compound 7) | 1.36 | 1.36 |
| 14-Methylergosta-8-en-3β,6α-diol (compound 8)a | 1.40 | |
| 14-Methylergosta-7,22,24(28)-trien-3β,6α-diol (compound 9)a | 1.44 | |
| 14-Methylergosta-7-en-3β,6α-diol (compound 10)a | 1.44 | |
| 14-Methylergosta-7,22-dien-3β,6α-diol (compound 11)a | 1.40 | |
| 4,4,14-Trimethyl sterols | ||
| 4,4,14-Trimethylergosta-8,24(28)-dien-3β-ol (eburicol) (compound 2) | 1.51 | 1.51 |
| 4,4,14-Trimethylergosta-8,22,24(28)-trien-3β-ol (compound 14)a | 1.48 | |
| 4,4,14-Trimethylergosta-8,22-dien-3β-ol (compound 15)a | 1.45 | |
| 4,4,14-Trimethylergosta-7,22-dien-3β-ol (compound 16) | 1.49 | 1.48 |
| 4,4,14-Trimethylergosta-8-en-3β-ol (compound 17)a | 1.48 | |
| 4,4,14-Trimethylergosta-7,24(28)-dien-3β-ol (compound 18)a | 1.55 |
This sterol has not been detected in our investigation but is postulated on the basis of expected biochemical conversions.
The mass spectrum of sterol 5 was very similar to that of sterol 4. This is typical for sterols differing only in the double bond positions of C7(8) and C8(9) (16, 44). Therefore, sterol 5 was identified as 14-methylergosta-7,24(28)-dien-3β,6α-diol. This assignment was confirmed by RRT considerations (see below). An additional double bond could be located in the side chain, giving a C22(23)-C24(28) diene system but with the consequence of having a fully saturated steroid backbone. However, there is no suitable enzymatic reaction in the ergosterol biosynthesis pathway that could manage the saturation of the sterol backbone. Furthermore, the fragmentation of the sterol 14-methylergosta-22,24(28)-dien-3β,6α-diol should differ strongly from the observed fragmentation of sterol 4, due to the doubly unsaturated side chain (compare this with compound 6, with a strong fragment of m/z 123). The only meaningful alternatives to the postulated structure of 14-methylergosta-7,24(28)-dien-3β,6α-diol (compound 5) are 14-methylergosta-8,22-dien-3β,6α-diol (compound 7) and 14-methylergosta-7,22-dien-3β,6α-diol (compound 11). Both of these options fail on the basis of the mass spectrum (see below) (Fig. 3) and RRT considerations (Table 2).
Structure 7 was already assigned to another 3β,6α-diol-type steroid found in this work. The mass spectrum of compound 7 (RRT, 1.36) showed the same fragmentation sequence as did those of compounds 4 and 5, i.e., m/z 557 ([M−CH3]+), m/z 467 ([M−CH3−TMS]+), and m/z 377 ([M−CH3−2TMS]+), but with different abundances of typical fragments. The base peak was not m/z 467 (compound 4) or m/z 377 (compound 5) but m/z 253 ([M−CH3−2TMS−SC]+). Observation of the base peak plus another fragment with an abundance of >50% at m/z 343 ([M−CH3−TMS−SC]+) is typical for Δ22-sterols but not for Δ24(28)-sterols (36). The only remaining alternative structure for compound 7 was 14-methylergosta-7,22-dien-3β,6α-diol (compound 11), which was excluded by RRT analysis (see below).
The [M]+ ion at m/z 570 of the bis-TMS ether of compound 6 indicated an additional double bond, compared to compounds 4, 5, and 7. The fragmentation sequence of this sterol was less abundant, because there was a very strong base peak at m/z 123, equivalent to a doubly unsaturated side chain. As mentioned above, Δ7- and Δ8-sterols show virtually the same fragmentation; therefore, the observed sterol could be either 14-methylergosta-7,22,24(28)-trien-3β,6α-diol (compound 9) or 14-methylergosta-8,22,24(28)-trien-3β,6α-diol (compound 6). RRT analysis clearly indicated the structure 14-methylergosta-8,22,24(28)-trien-3β,6α-diol (compound 6; see below).
According to the identification of 3β,6α-diol sterols, the structure of compound 16 was confirmed by comparing its mass spectrum and RRT with those of the known sterol 4,4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol, compound 2). The proposed structure of compound 16 is 4,4,14-trimethylergosta-7,22-dien-3β-ol, but there are two more theoretical structures, namely, 4,4,14-trimethylergosta-7,24(28)-dien-3β-ol (compound 18, not shown in Fig. 2) and 4,4,14-trimethylergosta-8,22-dien-3β-ol (compound 15). Sterol 18, containing a C7(8) double bond in ring B, was not considered in Fig. 2 because the enzyme sterol Δ8/7-isomerase does not accept 4,4-dimethyl sterols such as eburicol (compound 2) or lanosterol (compound 1) as substrates (16, 40, 45–47). Structure 15 was excluded on the basis of chromatographic data (see below).
Analysis of chromatographic data.
A well-established tool in GC analysis is the use of retention indices and the application of functional group increment rules (48, 49). This allows the precise prediction of chromatographic behavior for clusters of closely related compounds. In our investigation, all analyses of sterols were carried out under identical conditions; therefore, the relative retention times (RRTs) determined relative to the internal standard cholestane (RRT, 1.00) were comparable. Analysis of RRTs of relevant known sterol TMS ether pairs (Table 3) allowed conclusions to be drawn regarding the structures of unknown sterol TMS ethers. The absolute retention times for specific sterols were reproducible, varying by less than 1% in one batch. Shifts of RRTs (always in the same direction for all sterols in the batch) were not greater than ±0.02 over a period of >1 year. In our case, the novel sterols differ only in the number of double bonds (two or three) and/or the position of a double bond in the sterol backbone [C7(8), C8(9), C22(23), or C24(28)]. The following ΔRRT rules were observed: (i) the shift of a double bond from C7(8) to C8(9) leads to an increase in RRT of +0.04, (ii) the shift of a double bond from C24(28) to C22(23) leads to an increase in RRT of +0.07, (iii) the introduction of a second double bond in the side chain at C24(28), next to an existing double bond at C22(23), leads to an increase in RRT of +0.04, and (iv) saturation of the double bond at C22(23) next to an existing double bond at C24(28) leads to an increase in RRT of +0.03. This chromatographic behavior of the sterol TMS ether pairs is in line with literature findings (36, 44, 50). On the basis of the RRTs of the known compounds 14-methylergosta-8,24(28)-dien-3β,6α-diol (compound 4) and 4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol, compound 2), the RRTs of all other detected sterols in Fig. 2 and Table 2 could be calculated with our ΔRRT rules. The detected RRTs of known sterols 2 and 4 were also calculated, in order to confirm the reliability of our approach. The differences between the calculated RRT and the detected RRT were marginal in all cases. With this additional information, the novel sterols were identified using mass spectra and RRT data.
TABLE 3.
ΔRRTs for relevant sterol TMS ether pairs
| Sterol 1 | Sterol 2 | ΔRRTa |
|---|---|---|
| Ergosta-7-en-3β-ol | Ergosta-8-en-3β-ol | +0.04 |
| Ergosta-7,22-dien-3β-ol | Ergosta-8,22-dien-3β-ol | +0.04 |
| Ergosta-7,24(28)-dien-3β-ol | Ergosta-8,24-dien-3β-ol | +0.04 |
| Ergosta-7,24(28)-dien-3β-ol | Ergosta-7,22-dien-3β-ol | +0.07 |
| Ergosta-8,24(28)-dien-3β-ol | Ergosta-8,22-dien-3β-ol | +0.07 |
| Ergosta-5,7,24(28)-trien-3β-ol | Ergosta-5,7,22-trien-3β-ol | +0.06 |
| Ergosta-7,22,24(28)-trien-3β-ol | Ergosta-7,22-dien-3β-ol | +0.04 |
| Ergosta-8,22,24(28)-trien3β-ol | Ergosta-8,22-dien-3β-ol | +0.04 |
| Ergosta-7,24(28)-dien-3β-ol | Ergosta-7,22,24(28)-trien-3β-ol | +0.03 |
| Ergosta-8,24(28)-dien-3β-ol | Ergosta-8,22,24(28)-trien-3β-ol | +0.03 |
RRT = retention time for sterol TMS ether/retention time for internal standard (cholestane). ΔRRT = RRT for sterol 1 − RRT for sterol 2. RRTs were taken from the literature (16).
Sterol 5 had a RRT of 1.47 (calculated RRT, 1.49); significantly shorter RRTs were calculated for the other sterols, i.e., 1.36 for sterol 7 and 1.40 for sterol 11. Consequently, sterol 5 was determined to be 14-methylergosta-7,24(28)-dien-3β,6α-diol.
The RRTs of Δ7,22- and Δ7,24-sterols are significantly different (16). Sterol 7 showed exactly the calculated RRT of 1.36. For the only alternative based on MS data (sterol 11), a RRT of 1.40 was calculated. Therefore, sterol 7 was identified as 14-methylergosta-8,22-dien-3β,6α-diol.
RRTs allow the correct allocation of double bonds in the sterol backbone (Tables 2 and 3). Therefore, sterol 6 was determined to be definitely 14-methylergosta-8,22,24(28)-trien-3β,6α-diol (calculated RRT, 1.41; detected RTT, 1.39), and the alternative structure 14-methylergosta-7,22,24(28)-trien-3β,6α-diol (compound 9) (calculated RRT, 1.44) was excluded.
For the observed 4,4,14-trimethyl structure, three sterol structures were considered, namely, 4,4,14-trimethylergosta-7,22-dien-3β-ol (compound 16), 4,4,14-trimethylergosta-7,24(28)-dien-3β-ol (compound 18, not shown in Fig. 2), and 4,4,14-trimethylergosta-8,22-dien-3β-ol (compound 15). The detected RRT of 1.48 fit very well for structure 16 (calculated RRT, 1.49); structures 15 (calculated RRT, 1.45) and 18 (calculated RRT, 1.55) were excluded on the basis of their calculated RRTs. Consequently, sterol 16 was identified as 4,4,14-trimethylergosta-7,22-dien-3β-ol.
The novel sterols (compounds 5, 6, and 7) and sterol 16 were integrated into an extended ergosterol biosynthesis pathway under Erg11p inhibition (Fig. 2). All detected sterols are shown in blue in Fig. 2, whereas postulated sterols that are obvious but not detected intermediates are shown in black.
Effects of EMC120B12, fluconazole, and nocodazole on sterol patterns.
The benzimidazole derivative nocodazole (7.5 μg/ml) showed no effect in the postlanosterol part of ergosterol biosynthesis, as expected. No accumulation of ergosterol precursors or aberrantly generated sterols was detected (Fig. 4A2, B2, and C2), compared to untreated control samples (Fig. 4A1, B1, and C1).
The chromatograms in Fig. 4A3 to A6 show the effects of fluconazole and EMC120B12 treatment on wild-type (WT) C. albicans (SC5314, azole sensitive). In the absence of an antifungal compound (Fig. 4A1), mainly ergosterol (compound 13), with small amounts of lanosterol (compound 1) and 4,4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol, compound 2), could be detected. As expected, in the presence of fluconazole at 0.5 μg/ml (Fig. 4A4) or 2 μg/ml (Fig. 4A6), depletion of ergosterol (compound 13) was observed and the levels of the ergosterol precursors compound 1, compound 2, and 14-methyl-ergosta-8,24(28)-dien-3β,6α-diol (compound 4) increased significantly. Sterol 4 is not normally present in cells and is considered a marker sterol for C4-demethylase inhibition (16, 34, 35).
For EMC120B12 (Fig. 4A3 and A5), we found changes in the sterol pattern of C. albicans SC5314 very similar to those observed in the presence of fluconazole (Fig. 4A4 and A6). With identical inhibitor concentrations of 0.5 μg/ml (Fig. 4A3 and A4), EMC120B12 treatment led to significantly greater accumulation of compound 4 than did fluconazole treatment. Only at a higher concentration of 2 μg/ml did fluconazole show the same quantitative results as observed for EMC120B12 at both 0.5 μg/ml and 2 μg/ml. In general, lower levels of ergosterol (compound 13) were detected in the samples treated with EMC120B12. Furthermore, both inhibitors led to slight accumulation of other C14-methylated sterols, including 14-methylergosta-8,24(28)-dien-3β-ol (compound 3), 14-methylergosta-8,22-dien-3β,6α-diol (compound 7), and 4,4,14-trimethylergosta-7,22-dien-3β-ol (compound 16). Sterol 7 is one of three novel sterols (compounds 5, 6, and 7) with a 3β,6α-diol structure identified in this investigation, and sterol 16 is similar to 4,4,14-trimethylergosta-8,24(28)-dien-3β-ol (eburicol, compound 2) (see above) (Fig. 2 and 3). Fan et al. (51) detected two ergosterol precursors with 4,4,14-trimethyl structures during the course of characterization of the sterol C14-demethylase of Fusarium graminearum; one of those described sterols probably is compound 16, but no MS data were provided by the authors.
For the azole-resistant C. albicans strain Can741 (Fig. 4B3 to B6), much higher concentrations of fluconazole (32 μg/ml) (Fig. 4B6) and EMC120B12 (16 μg/ml) (Fig. 4B5) were necessary to produce visible changes in the sterol pattern. Treatment with 2 μg/ml EMC120B12 (Fig. 4B3) led to an ∼50% reduction in the level of ergosterol (compound 13) but only small increases in the levels of C14-methylated sterols (compounds 2 and 4). The amounts of lanosterol (compound 1) were almost unchanged in all samples (Fig. 4B3 to B6).
The chromatogram in Fig. 4B5 shows strong accumulation of compound 4 and weak accumulation of compounds 16 and 2, compared to that in Fig. 4B3. The sterol ratio pattern in Fig. 4B5 is similar to that in Fig. 4A3. With fluconazole treatment at 32 μg/ml (Fig. 4B6), sterol biosynthesis was suppressed almost completely but no shift to 3β,6α-diol sterols occurred, compared to Fig. 4B4. Only weak formation of sterol 16 was observed.
For WT C. krusei (CK201) (Fig. 4C1 to C6), much higher concentrations of fluconazole were required to reduce the amounts of ergosterol (compound 13) in the cells, as expected. Interestingly, already at a concentration of 2 μg/ml EMC120B12, accumulation of nonphysiological sterols was observed in C. krusei (Fig. 4C3), corresponding to the MIC of 1 μg/ml. The corresponding fluconazole incubation had almost no impact on the sterol composition in the C. krusei isolate used (Fig. 4C4). However, fluconazole yielded a sterol pattern comparable to that in Fig. 4C3 at a very high concentration (256 μg/ml) (Fig. 4C6), beyond pharmacological relevance. Also, with a concentration of 16 μg/ml EMC120B12, the amount of ergosterol (compound 13) was clearly reduced in Fig. 4C5, compared to Fig. 4C3, C4, and C6. Coelution of 14-methylergosta-8,22,24(28)-trien-3β,6α-diol (compound 6) and sterol 3 could not be excluded for Fig. 4C5. The main component of peak 3 in Fig. 4C3 and C5 is the 3β,6α-diol sterol 6, a sterol not found in C. albicans.
Interestingly, Candida krusei (CK242) showed one additional 3β,6α-diol sterol with EMC120B12 at >4 μg/ml, namely, 14-methylergosta-7,24(28)-dien-3β,6α-diol (compound 5) (Fig. 4 and 5). The identification of the novel sterols 5, 6, and 7 and the rarely observed sterol 16 is described in detail above. For all characteristic peaks in the chromatograms in Fig. 4 and 5, the respective mass spectra are given in Fig. 3.
FIG 5.
Total ion current chromatogram (m/z 100 to 650) of the sterol fraction of WT Candida krusei (CK242) after treatment with EMC120B12 (4 μg/ml). Sterol 1, lanosterol; sterol 2, eburicol; sterol 4, 14-methylergosta-8,24(28)-dien-3β,6α-diol; sterol 5, 14-methylergosta-7,24(28)-dien-3β,6α-diol; sterol 6, 14-methylergosta-8,22,24(28)-trien-3β,6α-diol; sterol 7, 14-methylergosta-8,22-dien-3β,6α-diol; sterol 13, ergosterol; sterol 16, 4,4,14-trimethylergosta-7,22-dien-3β-ol.
The results given in Fig. 4 and their analysis show that the sterol patterns resulting from incubation with EMC120B12 and fluconazole are very similar; they differ only in the inhibitor concentrations required for the individual strains. This strongly indicates that the two compounds have the same target, Erg11p. Consequently, the benzimidazole EMC120B12 is an inhibitor of the enzyme sterol C14-demethylase, like the triazole fluconazole and the new triazoles posaconazole and voriconazole.
DISCUSSION
In this work, we analyzed the mode of action of a novel potential antifungal drug, (S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl)benzimidazole (EMC120B12). This compound was identified from a compound library, using a cell-based assay (activity-selectivity assay) that simultaneously analyzed both the antifungal activity and the compatibility with human cells (11). EMC120B12 showed high levels of antifungal activity against pathogenic yeasts (including C. glabrata and C. krusei), which are known to be highly resistant to antifungals, especially the commonly used azoles. Initial studies based on transcriptional profiling of C. albicans in the presence and absence of EMC120B12 indicated that the compound might affect the ergosterol biosynthesis pathway. A precise target was not identified, however, and comparisons with other antimycotics were not performed previously. The fact that EMC120B12 belongs to the benzimidazole class would point to targeting of microtubule formation. To clarify this, we performed both biochemical and genome-wide studies to pinpoint the target of EMC120B12.
Comparison of the transcriptional profiles of C. albicans in the presence of EMC120B12 (benzimidazole), fluconazole (triazole), and nocodazole (benzimidazole) clearly showed that the profiles for EMC120B12 and fluconazole were very similar, including activation of 13 genes with direct functions in the ergosterol biosynthesis pathway and the regulator of the ergosterol pathway, UPC2. Very similar responses to fluconazole and EMC120B12 also were observed for differentially regulated genes not connected to the ergosterol pathway. Only minor overlap was noted for nocodazole, however, mostly reflecting common stress response genes (Table 1), which indicates different modes of action. This was supported by the observation that incubation with EMC120B12 did not show an effect on microtubule formation, similar to fluconazole, whereas nocodazole completely blocked the generation of tubulin structures (Fig. 1). Therefore, the benzimidazole EMC120B12 clearly does not target microtubular structures, and the results of the transcriptional profiling strongly suggest a target within the ergosterol biosynthesis pathway.
To analyze further the mode of action of EMC120B12 and to identify the target enzyme of this lead compound, we performed a biochemical analysis of the sterol patterns in Candida cells treated with EMC120B12, fluconazole, and nocodazole. The data obtained provide evidence that the enzyme sterol C14-demethylase, Erg11p, is the target of EMC120B12, since the compound generates a sterol pattern in Candida almost identical to that observed for Candida treated with the known C14-demethylase inhibitor fluconazole.
Previous studies on the mode of action of azoles showed that lanosterol (compound 1) and especially 14-methylergosta-8,24(28)-dien-3β,6α-diol (compound 4) are marker sterols for inhibition of the enzyme sterol C14-demethylase, encoded by ERG11 (34). 14-Methylergosta-8,24(28)-dien-3β,6α-diol has been reported as fungistatic (35). Decreases in ergosterol as an essential cellular membrane component and increases in aberrantly formed sterols (e.g., compounds 1 and 4) have been shown to interfere significantly with the fitness of fungi (15, 45). The formation of compound 4 is catalyzed by the enzyme sterol C5-desaturase (encoded by ERG3) (38). In fungi, defective sterol C5-desaturase leads to resistance to azoles (15, 34, 45). Most likely, production of fungistatic 3β,6α-diols does not take place in resistant strains.
In previous projects, we analyzed the sterol patterns of other azole-treated fungi (e.g., Aspergillus fumigatus, Aspergillus flavus, Candida glabrata, Saccharomyces cerevisiae, and Yarrowia lipolytica) (16). In this study, we used the same procedures for sample preparation, with no chemical modifications during sample preparation. Our results showed that EMC120B12 and fluconazole changed the sterol patterns in the same manner. Both lanosterol (compound 1) and 14-methylergosta-8,24(28)-dien-3β,6α-diol (compound 4) strongly accumulated in SC5314 cells, with low MICs for azoles (Fig. 4A3 to A6), during treatment with EMC120B12 or fluconazole, whereas nocodazole had no effect on the sterol pattern (Fig. 4A2, B2, and C2). In addition, eburicol (compound 2), a precursor of compound 4 generated from accumulating lanosterol (compound 1) by the action of the enzyme sterol C24-methyltransferase (encoded by ERG6), was detected. Most interestingly, in addition to the known set of sterols, we identified three novel sterols that were detected for the first time in C. albicans and C. krusei. Sterols 5 and 6 were exclusively detected in C. krusei. These sterols might contribute to the improved efficacy of EMC120B12, at moderate concentrations, against this resistant yeast.
The effects of EMC120B12 on sterol biosynthesis, on both a transcriptional level and a biochemical level, mimic those of fluconazole, strongly indicating that EMC120B12 targets Erg11p, as do the azoles. This is manifested also by the presence of the marker sterol 4, which in other species has been shown to result from a block of Erg11p. In addition, three novel atypical 3β,6α-diol sterols (compounds 5, 6, and 7) could be identified; to our knowledge, these sterols have not been characterized previously. Although EMC120B12 targets the same enzyme as do the azole antifungals, it is worth exploiting due to its high level of efficacy against resistant yeasts such as C. krusei and C. glabrata. Therefore, these results support EMC120B12 as a promising candidate for further development as a potential antifungal agent.
ACKNOWLEDGMENT
This work was supported within the BMBF program Basisinnovationen in der Genombasierten Infektionsforschung (grant 0315221).
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–136. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tortorano AM, Kibbler C, Peman J, Bernhardt H, Klingspor L, Grillot R. 2006. Candidaemia in Europe: epidemiology and resistance. Int J Antimicrob Agents 27:359–366. doi: 10.1016/j.ijantimicag.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Castanheira M, Messer SA, Jones RN, Farrell DJ, Pfaller MA. 2014. Activity of echinocandins and triazoles against a contemporary (2012) worldwide collection of yeast and moulds collected from invasive infections. Int J Antimicrob Agents 44:320–326. doi: 10.1016/j.ijantimicag.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Odds FC. 2010. Resistance to antifungal agents. Fungal Genet Biol 47:190. doi: 10.1016/j.fgb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ, Rinaldi MG, Barnes R, Hu B, Veselov AV, Tiraboschi N, Nagy E, Gibbs DL, Global Antifungal Surveillance Group. 2005. Results from the ARTEMIS DISK global antifungal surveillance study: a 6.5-year analysis of susceptibilities of Candida and other yeast species to fluconazole and voriconazole by standardized disk diffusion testing. J Clin Microbiol 43:5848–5859. doi: 10.1128/JCM.43.12.5848-5859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karthaus M, Cornely OA. 2007. Treatment options in candidaemia. Mycoses 50:44–49. doi: 10.1111/j.1439-0507.2007.01379.x. [DOI] [PubMed] [Google Scholar]
- 7.Pasqualotto AC, Denning DW. 2008. New and emerging treatments for fungal infections. J Antimicrob Chemother 61:i19–i30. doi: 10.1093/jac/dkm428. [DOI] [PubMed] [Google Scholar]
- 8.Cornely OA, Maertens J, Winston DJ, Perfect J, Ullmann AJ, Walsh TJ, Helfgott D, Holowiecki J, Stockelberg D, Goh Y, Petrini M, Hardalo C, Suresh R, Angulo-Gonzalez D. 2007. Posaconazole vs. fluconazole or itraconazole prophylaxis in patients with neutropenia. N Engl J Med 356:348–359. doi: 10.1056/NEJMoa061094. [DOI] [PubMed] [Google Scholar]
- 9.Ullmann AJ, Cornely OA, Donnelly JP, Akova M, Arendrup MC, Arikan-Akdagli S, Bassetti M, Bille J, Calandra T, Castagnola E, Garbino J, Groll AH, Herbrecht R, Hope WW, Jensen HE, Kullberg BJ, Lass-Florl C, Lortholary O, Meersseman W, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Cuenca-Estrella M. 2012. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: developing European guidelines in clinical microbiology and infectious diseases. Clin Microbiol Infect 18(Suppl 7):1–8. doi: 10.1111/1469-0691.12037. [DOI] [PubMed] [Google Scholar]
- 10.Bauer J, Kinast S, Burger-Kentischer A, Finkelmeier D, Kleymann G, Abu Rayyan W, Schropple K, Singh A, Jung G, Wiesmuller KH, Rupp S, Eickhoff H. 2011. High-throughput-screening-based identification and structure-activity relationship characterization defined (S)-2-(1-aminoisobutyl)-1-(3-chlorobenzyl)benzimidazole as a highly antimycotic agent nontoxic to cell lines. J Med Chem 54:6993–6997. doi: 10.1021/jm200571e. [DOI] [PubMed] [Google Scholar]
- 11.Burger-Kentischer A, Finkelmeier D, Keller P, Bauer J, Eickhoff H, Kleymann G, Abu Rayyan W, Singh A, Schröppel K, Lemuth K, Wiesmüller KH, Rupp S. 2011. A screening assay based on host-pathogen interaction models identifies a set of novel antifungal benzimidazole derivatives. Antimicrob Agents Chemother 55:4789–4801. doi: 10.1128/AAC.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Nagy E, Dobiasova S, Rinaldi M, Barton R, Veselov A, Global Antifungal Surveillance Group. 2008. Candida krusei, a multidrug-resistant opportunistic fungal pathogen: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J Clin Microbiol 46:515–521. doi: 10.1128/JCM.01915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Katiyar SK, Edlind TD. 1996. Site-directed mutagenesis of Saccharomyces cerevisiae β-tubulin: interaction between residue 167 and benzimidazole compounds. FEBS Lett 385:7–10. doi: 10.1016/0014-5793(96)00334-1. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel W. 1980. Effects of the antimicrotubular cancerostatic drug nocodazole on the yeast Saccharomyces cerevisiae. Z Allg Mikrobiol 20:315–324. doi: 10.1002/jobm.3630200503. [DOI] [PubMed] [Google Scholar]
- 15.Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S. 2002. Molecular basis of resistance to azole antifungals. Trends Mol Med 8:76–81. doi: 10.1016/S1471-4914(02)02280-3. [DOI] [PubMed] [Google Scholar]
- 16.Müller C, Staudacher V, Krauss J, Giera M, Bracher F. 2013. A convenient cellular assay for the identification of the molecular target of ergosterol biosynthesis inhibitors and quantification of their effects on total ergosterol biosynthesis. Steroids 78:483–493. doi: 10.1016/j.steroids.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Vanden Bossche H, Warnock DW, Dupont B, Kerridge D, Sengupta S, Improvisi L, Marichal P, Odds FC, Provost F, Ronin O. 1994. Mechanisms and clinical impact of antifungal drug resistance. J Med Vet Mycol 32:189–202. doi: 10.1080/02681219480000821. [DOI] [PubMed] [Google Scholar]
- 18.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect 14:398–405. doi: 10.1111/j.1469-0691.2007.01935.x. [DOI] [PubMed] [Google Scholar]
- 20.Kilmartin JV, Adams AEM. 1984. Structural rearrangements of tubulin and actin during the cell-cycle of the yeast Saccharomyces. J Cell Biol 98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team. 2009. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 22.Smyth GK. 2005. Limma: linear models for microarray data, p 397–420. In Gentleman R, Carey VJ, Huber W, Irizzary RA, Dudoit S (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY. [Google Scholar]
- 23.Lönnstedt I, Speed TP. 2002. Replicated microarray data. Stat Sin 12:31–46. [Google Scholar]
- 24.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article 3. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK, Michaud J, Scott HS. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 27.Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 28.Dudoit S, Popper Schaffer J, Boldrick JC. 2003. Multiple hypothesis testing in microarray experiments. Stat Sci 18:71–103. doi: 10.1214/ss/1056397487. [DOI] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. [Google Scholar]
- 30.Burbiel J, Bracher F. 2003. Azasteroids as antifungals. Steroids 68:587–594. doi: 10.1016/S0039-128X(03)00080-1. [DOI] [PubMed] [Google Scholar]
- 31.Krauss J, Müller C, Kiessling J, Richter S, Staudacher V, Bracher F. 2014. Synthesis and biological evaluation of novel N-alkyl tetra- and decahydroisoquinolines: novel antifungals that target ergosterol biosynthesis. Arch Pharm (Weinheim) 347:283–290. doi: 10.1002/ardp.201300338. [DOI] [PubMed] [Google Scholar]
- 32.Renard D, Perruchon J, Giera M, Muller J, Bracher F. 2009. Side chain azasteroids and thiasteroids as sterol methyltransferase inhibitors in ergosterol biosynthesis. Bioorg Med Chem 17:8123–8137. doi: 10.1016/j.bmc.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 33.Alcazar-Fuoli L, Mellado E, Garcia-Effron G, Lopez JF, Grimalt JO, Cuenca-Estrella JM, Rodriguez-Tudela JL. 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73:339–347. doi: 10.1016/j.steroids.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun 207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 35.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Kelly DE, Kelly SL. 2010. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother 54:3578–3583. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakanishi S, Nishino T, Nagai J, Katsuki H. 1987. Characterization of nystatin-resistant mutants of Saccharomyces cerevisiae and preparation of sterol intermediates using the mutants. J Biochem 101:535–544. [DOI] [PubMed] [Google Scholar]
- 37.Osumi T, Taketani S, Katsuki H, Kuhara T, Matsumoto I. 1978. Ergosterol biosynthesis in yeast; pathways in the late stages and their variation under various conditions. J Biochem 83:681–691. [DOI] [PubMed] [Google Scholar]
- 38.Shimokawa O, Kato Y, Kawano K, Nakayama H. 1989. Accumulation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol in 14α-demethylation mutants of Candida albicans: genetic evidence for the involvement of 5-desaturase. Biochim Biophys Acta 1003:15–19. doi: 10.1016/0005-2760(89)90092-1. [DOI] [PubMed] [Google Scholar]
- 39.Ebert E, Gaudin J, Muecke W, Ramsteiner K, Vogel C, Fuhrer H. 1983. Inhibition of ergosterol biosynthesis by etaconazole in Ustilago maydis. Z Naturforsch 38C:28–34. [Google Scholar]
- 40.Quail MA, Arnold A, Moore DJ, Goosey MW, Kelly SL. 1993. Ketoconazole-mediated growth inhibition in Botrytis cinerea and Saccharomyces cerevisiae. Phytochemistry 32:273–280. doi: 10.1016/S0031-9422(00)94980-X. [DOI] [Google Scholar]
- 41.Griffiths KM, Bacic A, Howlett BJ. 2003. Sterol composition of mycelia of the plant pathogenic ascomycete Leptosphaeria maculans. Phytochemistry 62:147–153. doi: 10.1016/S0031-9422(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 42.Loeffler RST, Hayes AL. 1990. Sterols of the plant pathogenic fungi Botrytis cinerea and Pyrenophora teres. Phytochemistry 29:3423–3425. doi: 10.1016/0031-9422(90)85250-J. [DOI] [Google Scholar]
- 43.Zheng WF, Liu T, Xiang XY, Gu Q. 2007. Sterol composition in field-grown and cultured mycelia of Inonotus obliquus. Yao Xue Xue Bao 42:750–756. [PubMed] [Google Scholar]
- 44.Giera M, Plossl F, Bracher F. 2007. Fast and easy in vitro screening assay for cholesterol biosynthesis inhibitors in the post-squalene pathway. Steroids 72:633–642. doi: 10.1016/j.steroids.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Kelly SL, Lamb DC, Kelly DE, Manning NJ, Loeffler J, Hebart H, Schumacher U, Einsele H. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett 400:80–82. doi: 10.1016/S0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 46.Parks LW, Casey WM. 1995. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol 49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 47.Fryberg M, Unrau AM, Oehlschl AC. 1973. Biosynthesis of ergosterol in yeast: evidence for multiple pathways. J Am Chem Soc 95:5747–5757. doi: 10.1021/ja00798a051. [DOI] [PubMed] [Google Scholar]
- 48.Stein SE, Babushok VI, Brown RL, Linstrom PJ. 2007. Estimation of Kovats retention indices using group contributions. J Chem Inf Model 47:975–980. doi: 10.1021/ci600548y. [DOI] [PubMed] [Google Scholar]
- 49.Gassiot M, Fernandez E, Firpo G, Carbo R, Martin M. 1975. Empirical quantum chemical approach to structure-gas chromatographic retention index relationship. I. Sterol acetates. J Chromatogr 108:337–344. doi: 10.1016/S0021-9673(00)84677-4. [DOI] [PubMed] [Google Scholar]
- 50.Gerst N, Ruan B, Pang J, Wilson WK, Schroepfer GJ Jr. 1997. An updated look at the analysis of unsaturated C27 sterols by gas chromatography and mass spectrometry. J Lipid Res 38:1685–1701. [PubMed] [Google Scholar]
- 51.Fan J, Urban M, Parker JE, Brewer HC, Kelly SL, Hammond-Kosack KE, Fraaije BA, Liu X, Cools HJ. 2013. Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol 198:821–835. doi: 10.1111/nph.12193. [DOI] [PubMed] [Google Scholar]