Abstract
Streptomyces bacteria are renowned for their ability to produce bioactive secondary metabolites. Recently, synthetic biology has enabled the production of intermediates and shunt products, which may have altered biological activities compared to the end products of the pathways. Here, we have evaluated the potential of recently isolated alnumycins and other closely related pyranonaphthoquinone (PNQ) polyketides against Staphylococcus aureus biofilms. The antimicrobial potency of the compounds against planktonic cells and biofilms was determined by redox dye-based viability staining, and the antibiofilm efficacy of the compounds was confirmed by viable counting. A novel antistaphylococcal polyketide, alnumycin D, was identified. Unexpectedly, the C-ribosylated pathway shunt product alnumycin D was more active against planktonic and biofilm cells than the pathway end product alnumycin A, where a ribose unit has been converted into a dioxane moiety. The evaluation of the antibiofilm potential of other alnumycins revealed that the presence of the ribose moiety in pyranose form is essential for high activity against preformed biofilms. Furthermore, the antibiofilm potential of other closely related PNQ polyketides was examined. Based on their previously reported activity against planktonic S. aureus cells, granaticin B, kalafungin, and medermycin were also selected for testing, and among them, granaticin B was found to be the most potent against preformed biofilms. The most active antibiofilm PNQs, alnumycin D and granaticin B, share several structural features that may be important for their antibiofilm activity. They are uncharged, glycosylated, and also contain a similar oxygenation pattern of the lateral naphthoquinone ring. These findings highlight the potential of antibiotic biosynthetic pathways as a source of effective antibiofilm compounds.
INTRODUCTION
Most bacteria can switch between two lifestyles and exist either as free-living, planktonic cells or as bacterial biofilms that are surface-associated communities of cells embedded in a self-produced matrix. The matrix consists of extracellular polymeric substances (EPS), and it is one of the factors contributing to increased tolerance to antibiotics associated with bacterial biofilms (1).
The Gram-positive bacterial species Staphylococcus aureus is a common pathogen and the most commonly isolated bacterial species from chronic wounds and ocular infections. It is also frequently responsible for infections of indwelling medical devices and hospital-acquired infections in general (2). A great deal of literature has been dedicated to methicillin-resistant S. aureus (MRSA) strains and the threat they pose, but another alarming fact is that the methicillin-susceptible S. aureus strains can switch to a biofilm state that is remarkably tolerant to antibiotics (3, 4). The antibiotic concentration necessary to eliminate biofilms often exceeds the highest deliverable concentration, as 10- to 1,000-fold concentrations of the antibiotics are typically needed against biofilms compared with planktonic bacteria (4, 5). Thus, there is an urgent need for effective antibiofilm drugs against pathogens like S. aureus.
The discovery earlier this year of a new potent broad-spectrum antibiotic, teixobactin, which was isolated from previously unculturable soil bacteria (6), serves as an inspirational example in support of the importance of resurrecting antimicrobial discovery from natural sources. The suggestion in the literature by top authors that the lack of discovery of antibiotics may be overcome by a systematic application of Waksman discovery strategies and the renaissance of exploration of natural sources, such as soil bacteria (7, 8), is in our opinion remarkably valid for the discovery of antibiofilm compounds as well. Since biofilms are the most common bacterial lifeform in nature, it seems only logical that, along with antibiotics that are effective against planktonic bacteria, natural antibiofilm compounds with optimized structures also would have evolved in bacteria. The recent emergence of synthetic biology as a means to expand the chemical space of natural products that is available for exploration of biological activities holds great promise for increasing our chances of identifying more and better bioactive metabolites in the future (9), in particular antibiofilm compounds.
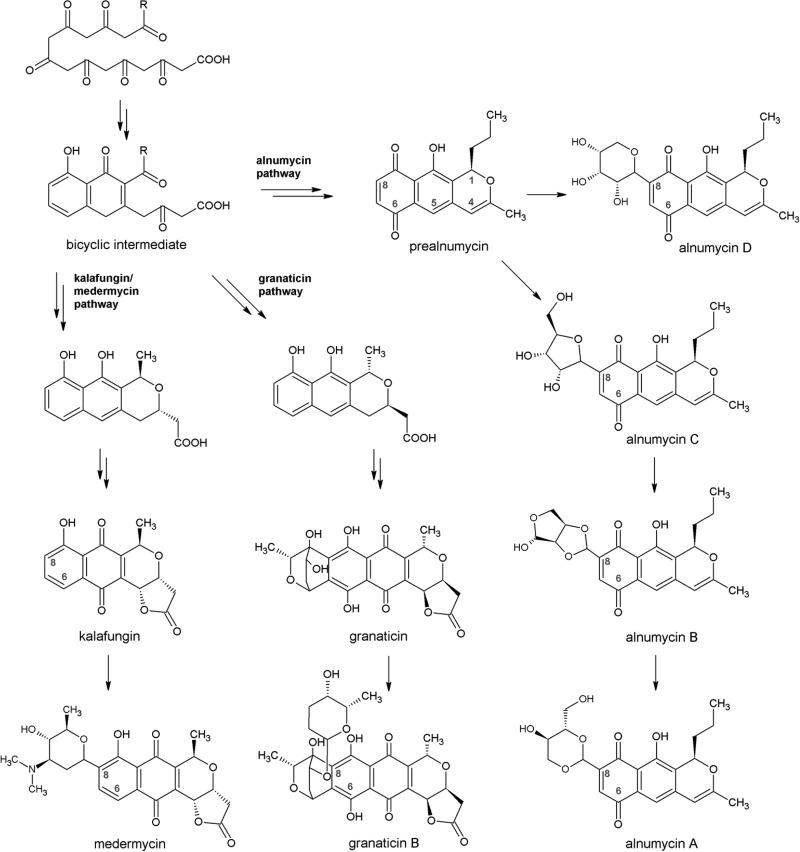
The soil-dwelling bacterial species of the genus Streptomyces are well known as important producers of antibiotics and other medicinally useful compounds. It has been estimated that streptomycin is found in about 1% of random soil actinomycetes, whereas tetracycline and actinomycin are present at an occurrence of about 0.1% (10). The pyranonaphthoquinone (PNQ) polyketides are a subclass of bacterial type II polyketides that have also been successful as antimicrobials, as exemplified by the aforementioned tetracycline (11). The PNQ polyketides share a biosynthetic origin; the carbon backbone chain of the compounds is synthesized from common acetate precursor units by homologous polyketide synthase enzymes, and the formed polyketide chain is then enzymatically folded and further modified into a fused three-ring aglycone unit that is composed of a pyran, a quinone, and a benzene ring. The p-quinone is typically found as a central ring, like in actinorhodin, granaticin, and medermycin, but in the case of alnumycin A, the quinone structure has an atypical position as a lateral ring (Fig. 1). On several PNQ pathways, the formed aglycone unit is enzymatically C-glycosylated (12).
FIG 1.
Structures of the PNQ polyketides analyzed in this study and a simplified representation of their biosynthetic origin, starting from the polyketide chain synthesized by a minimal polyketide synthase. The numbering of the atoms of the aglycone unit is shown for prealnumycin, and the positions C-6 and C-8 are marked for all tested compounds. R = CH3 on the kalafungin/medermycin and granaticin pathways, and R = CH2CH2CH3 on the alnumycin pathway.
The goal of this contribution was to resurrect the exploration of the antimicrobial potential of the PNQ polyketides via the evaluation of their antibiofilm effects. Previously, several PNQs have been reported to possess antibacterial activity against Gram-positive species, including S. aureus. Of these, granaticin B and medermycin (lactoquinomycin; Fig. 1) were found to be active against multiresistant S. aureus strains (13, 14). Research in the 1960s also showed that the nonglycosylated wide-spectrum antibiotic kalafungin (Fig. 1) displayed activity against S. aureus strains in the lower micromolar range (15). In addition, recent investigations into the biosynthesis of alnumycin A (K1115 B1) (16, 17) have provided a small library of structurally related metabolites (Fig. 1) (18, 19), which have not been tested for their antistaphylococcal activity to date. Furthermore, none of the PNQs have been specifically investigated for their antibiofilm activity. In this contribution, we show that PNQs can, apart from their effects on single-cell bacteria, act on bacterial biofilms at low concentrations, reach the metabolically active core of the staphylococcal biofilms, and induce their killing.
MATERIALS AND METHODS
Production and purification of the PNQs.
The alnumycins were isolated from the cultures of recombinant Streptomyces albus strains, purified to a minimum of 95% purity exactly as previously described (18, 19), and stored in CHCl3 at −20°C. Alnumycin A was isolated from the cultures of an S. albus strain that contained the intact alnumycin gene cluster on a cosmid pAlnuori. Other alnumycins were isolated from the deletion mutant strains S. albus/pAlnuoriΔind (prealnumycin) (18), S. albus/pAlnuoriΔaln3 (alnumycin B), and S. albus/pAlnuoriΔaln6 (alnumycins C1 and D) (19).
Granaticin B, kalafungin, and medermycin were isolated from the cultures of known Streptomyces producer strains and purified as described below. The granaticin B-producing strain Streptomyces violaceoruber Tü22 (DSM-40701) was ordered from the Deutche Sammlung von Microorganismen und Zellkulturen (DSMZ, Germany). Granaticin B was produced in a total of 1 liter of NL 19 medium (20) divided into 50-ml aliquots in 250-ml Erlenmeyer flasks, and 1 g of the adsorbent Amberlite XAD7HP (Rohm and Haas) was added prior to sterilization. Each flask was inoculated with 1 ml of a 3-day-old preculture in tryptic soy broth (TSB; Sigma-Aldrich). After 4 days at 300 rpm and 28°C, the XAD resin was collected by repeated decanting and washing with cold tap water. The metabolites were extracted into methanol, which was followed by neutral methanol-chloroform extraction in phosphate-buffered saline (PBS; Lonza). The deep pink extract was then subjected to preparative high-performance liquid chromatography (HPLC) with a Waters Sunfire prep C18, 5-μm, 10- by 250-mm column, and the metabolites were eluted with a 2- to 25-min gradient from 50% to 100% methanol. The main fraction was further purified by a second preparative HPLC step with a Phenomenex Luna Phenyl-Hexyl 100, 10-μm, 250- by 10-mm column and a 2- to 25-min gradient from 40% to 100% methanol. The final main fraction (purity 94%) (see Fig. S1 in the supplemental material) was stored in methanol at −20°C.
The kalafungin-producing strain Streptomyces tanashiensis Kala (DSM-731) was also obtained from the DSMZ, Germany. A 3-day-old preculture in GYM Streptomyces medium (consisting of 4 g glucose, 4 g yeast extract, 10 g malt extract, and 12 g agar per liter adjusted at pH 7.2 before agar addition) was used to inoculate 1.5 liters of the fermentation medium, which consisted of 25 g glucose monohydrate, 5 g peptone, and 5 g calcium carbonate per liter at pH 7.2 (15). After 4 days of incubation as 50-ml batches in 250-ml Erlenmeyer flasks at 29°C and 300 rpm, the supernatant was collected by centrifugation, and the pH was adjusted to 7 to 7.5. The supernatant was extracted with chloroform, and the extract washed once with ultrapure water. The Waters Sunfire column and a 2- to 30-min gradient from 30% to 100% methanol were used for the preparative HPLC. The main fraction (purity 96%) (see Fig. S1 in the supplemental material) was stored in chloroform at −20°C.
Medermycin was isolated from a total of 2 liters of Streptomyces coelicolor CH999/pIK340 cultures in R5MS medium (21). The 500-ml Erlenmeyer flasks containing 50 ml of the medium were inoculated with 1 ml of a 3-day-old preculture. After 5 days at 250 rpm and 29°C, the supernatant was collected by centrifugation and the pH was adjusted to 7.4 with 1 M HCl. After two repeated extractions with ethyl acetate and washing of the extract with ultrapure water, the extract was dried and applied onto a silica (Kieselgel 100, 0.063 to 0.200; Merck) column in 4:1 chloroform/methanol. The main yellow fraction from the isocratic elution was further purified by preparative HPLC using the Waters SunFire column and a 2- to 30-min gradient from 30% methanol in 10 mM ammonium acetate to 100% methanol. The main fraction (purity 92%) (see Fig. S1 in the supplemental material) was stored in chloroform at −20°C.
A Merck Hitachi L-6200A system was used for all preparative HPLC steps, while the analytical HPLC was performed using a Phenomenex Kinetex 2.6-μm, C18, 100-Å, 100- by 4.60-mm column attached to a Shimadzu SCL-10Avp system equipped with an SPD-M10Avp diode array detector.
Mass spectrometry analysis.
The identities of the main products isolated from the Streptomyces cultures were established by observed antistaphylococcal activity, extraction behavior, retention times in HPLC, matching UV-visible (UV-Vis) spectra (see Fig. S1 in the supplemental material), and mass spectrometry (MS) analysis of the purified compounds. The liquid chromatography-mass spectrometry (LC-MS) analysis was performed with an Agilent 1260 Infinity 6120 Quadropole LC-MS system with the Phenomenex Kinetex column and a 2- to 20-min gradient from 30% methanol in 0.1% (vol/vol) HCOOH to 100% methanol. Expected m/z ratios were observed for the purified compounds: granaticin B 557.2 ([M–H]−, calculated 557.17), kalafungin 301.1 ([M+H]+, calculated 301.07), and medermycin 458.2 ([M+H]+, calculated 458.18).
Antibiofilm and antibacterial assays.
The biofilm-forming S. aureus control strains ATCC 25923 and Newman were grown in TSB at 37°C and 200 rpm in flat-bottom Nunclon Delta surface 96-well polystyrene plates (Thermo Fisher Scientific), as previously described (22), with the exception of using 10% (vol/vol) dimethyl sulfoxide (DMSO, minimum 99.9%; Sigma-Aldrich) in TSB in place of a default concentration (2% to 5%) due to the apparent aggregation of granaticin B on the biofilm material during the postexposure experiments. TSB with 10% DMSO was used in the solvent control wells, and TSB alone was used in additional maximum viability control wells. To confirm that the DMSO concentration did not affect the activity of the compounds, biofilm postexposure inhibitory concentration values were recorded for alnumycin D in 2% and 10% DMSO.
In the preexposure experiments, 20 μl of serial compound dilutions in DMSO or pure DMSO as a control and 180 μl of a 1:100 diluted bacterial preculture corresponding to ca. 106 CFU/ml were added into each well. After an 18-h-long growth period, the planktonic and biofilm phases were analyzed by redox-dye-based viable staining. The established resazurin-based method was used (22). Briefly, a 400 μM resazurin (sodium salt; Sigma-Aldrich) stock solution was prepared in sterile PBS and stored at 4°C in the dark. The planktonic phase was stained by adding 10 μl of the stock solution, and the biofilms were stained by adding 200 μl of 20 μM resazurin dilution in PBS per well. The plates were incubated in the dark at 200 rpm for 20 min, and the fluorescence was then measured at λex = 560 nm and λem = 590 nm with a Varioskan Flash multimode plate reader (Thermo Fisher Scientific, Finland).
In the postexposure experiments, 20 μl of serial compound dilutions or DMSO were only added after the planktonic phase was removed from the 18-h-old TSB cultures, where the biofilms had already formed. After the addition of 180 μl of fresh TSB, the biofilms were left to grow for another 24 h. Finally, the treated and control biofilms were stained with resazurin as in the preexposure experiments described above (22). Rifampin (minimum 97%; Sigma-Aldrich) was used as a control antibiotic in all viability assays.
The efficacy of the compounds was confirmed by a logarithmic reduction (log R) assay. The log R was calculated by subtracting the average logarithmic viable counts (CFU/ml) in treated wells from the average logarithmic viable counts in solvent control (10% DMSO) wells (23). After the removal of the planktonic phase, a final concentration of 40 μM of each test compound (20 μl) and TSB (180 μl) was added onto the 18-h-old biofilms in two separate wells, and the 96-well plates were incubated further for 24 h. The planktonic phase was then removed, and the biofilms at the bottom of the wells were individually suspended by scraping, using the stem of an inverted, plastic inoculation loop, in 100 μl of TSB and rinsing with another 100 μl of TSB. The two TSB suspensions were combined and further homogenized by 5 min of ultrasound in a Sonorex Digitec water bath sonicator (Bandelin, Switzerland) at 25°C and 35 kHz. The number of viable bacteria in each suspension was then determined by plating a range of dilutions on tryptic soy agar (Sigma-Aldrich).
Cytotoxicity assays.
Two human cell lines were utilized: human lung (HL) and human adenocarcinoma (HeLa 229) cells. The HL epithelial cells (24) were grown in RPMI 1640 (Biowhittaker; Lonza) supplemented with 7% inactivated fetal bovine serum (FBS), 2 mM l-glutamine, and 20 μg/ml gentamicin. The HeLa 229 cells (ATCC CCL-2.1) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% inactivated FBS, 1% (2 mM) l-glutamine, and 20 μg/ml gentamicin. The two cell lines were routinely cultured in 75-cm2 cell culture flasks at 37°C in 5% CO2 in an air-ventilated humidified incubator to around 90% confluence. Harvesting was done by adding 0.05% (vol/vol) trypsin and 0.02% (wt/vol) EDTA in PBS (for HeLa cells) or 0.25% trypsin in PBS (for HL cells). Cell suspensions (200 μl) of 4 × 105 cells/ml were added into 96-well microtiter plates (Nunc Delta surface) and incubated at 37°C for 24 h. After that, 20 μl of culture medium was removed and replaced with a similar volume of compound dilutions (0.05 to 36 μM), and plates were incubated for an additional 24 h. Then, culture medium was replaced with a solution of resazurin in PBS at a concentration of 20 μM and maintained for 2 h at 37°C in incubator conditions. Reduced resazurin signal as indication of cell viability was measured with the Varioskan flash plate reader (λex = 570 nm and λem = 590 nm). Cells without test compounds were included as positive controls, wells containing only media as negative controls, and 0.5% DMSO as solvent controls. Percentages of cell viability were calculated in relation to the untreated cells.
Data analysis.
The performance of the antibiofilm activity assays was monitored by calculating the statistical parameters Z' (screening window coefficient), S/B (signal-to-background), and S/N (signal-to-noise), as previously described (4). For the determination of the 50% inhibitory concentrations (IC50s) against planktonic bacteria and biofilms, 12 different serial compound dilutions covering at least a 3-log concentration range between 10 nM and 100 μM were analyzed per compound. Furthermore, each IC50 was confirmed by an independent experiment that resulted in a value within the same 95% confidence interval. In the cytotoxicity assays, nine different concentrations were analyzed per compound (50 nM to 36 μM). Half inhibitory concentrations (IC50) or half lethal concentrations (LC50) and 95% confidence intervals were calculated via nonlinear regression analysis (sigmoidal dose-response with variable slope) using GraphPad Prism v. 5.00 for Windows (GraphPad Software, USA). The therapeutic index was calculated as the ratio of the LC50 values (as a measure of cytotoxicity on mammalian cells) and the IC50 values obtained in the postexposure assay (as a measure of the antimicrobial effect of the compounds on preformed biofilms).
RESULTS
Inhibition of biofilm formation by the alnumycins.
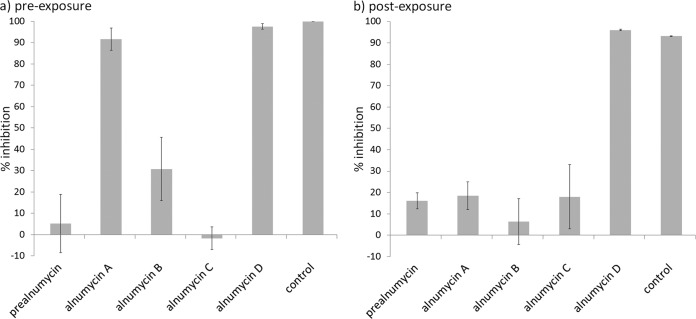
For direct identification of the antibiofilm alnumycins that are active in the low micromolar range, the initial antibiofilm assays were performed at a 40 μM assay concentration. The results revealed that the pathway end product alnumycin A and the pathway side product alnumycin D inhibited the formation of the biofilms in a preexposure assay (Fig. 2a). The pathway intermediates prealnumycin (18), alnumycin B, and alnumycin C did not appear to harbor antibiofilm activity at the assay concentration (Fig. 2a). The structural difference of these compounds is that the alnumycins A and B carry dioxane and dioxolane moieties at C-8, respectively, and the alnumycins C and D carry a ribose unit, which is in furanose form in the former and in pyranose form in the latter (Fig. 1).
FIG 2.
The antibiofilm activity screening results with alnumycins against S. aureus ATCC 25923 presented as inhibition of biofilm viability in preexposure (a) and postexposure (b). The concentration of test compounds and the control antibiotic rifampin was 40 μM. The average value and standard deviation are shown for three biological replicates. The S/N values were 8.87 and 9.21, and the Z' values were 0.73 and 0.67 for the preexposure and postexposure assays, respectively. Compounds resulting in >50% inhibition were considered active.
For a more precise evaluation of the potential of the alnumycins A and D in the inhibition of biofilm formation, inhibitory concentration (IC) values were determined. The obtained IC50s indicated that alnumycin A was only moderately active in the preexposure, with an IC50 of 39.3 μM, and its activity against planktonic S. aureus ATCC 25923 cells was in the same range. Alnumycin D, on the other hand, was ca. 12- and 22-fold more active than the pathway end product alnumycin A against planktonic cells and in the inhibition of biofilm formation, with IC50s in the low micromolar range in the preexposure assay (Table 1). Prior to this study, the recently characterized pathway shunt product was not known to possess any bioactivity.
TABLE 1.
Preexposure IC50s were determined against the planktonic versus biofilm cells of S. aureus ATCC 25923 using the resazurin-based viability assay
| Compound | Planktonic phase IC50 (μM)a | Biofilm preexposure IC50 (μM)a |
|---|---|---|
| Alnumycin A | 32.4 (30.0–35.0) | 39.3 (37.4–41.3) |
| Alnumycin D | 2.66 (2.13–3.32) | 1.75 (1.48–2.08) |
| Granaticin B | 2.61 (2.06–3.31) | 2.76 (2.17–3.49) |
| Kalafungin | 1.11 (0.81–1.53) | 3.87 (3.50–4.28) |
| Medermycin | 2.81 (2.55–3.09) | 2.50 (2.22–2.81) |
The 95% confidence intervals are shown in parenthesis.
Effect of the alnumycins on preformed biofilms.
In the postexposure assay with the alnumycins, only the shunt product alnumycin D showed activity and resulted in 100% killing of the biofilm cells at a 40 μM assay concentration. Neither the nonglycosylated prealnumycin nor the structural analogs of alnumycin D—alnumycins A, B, and C—were active against preformed biofilms at the assay concentration (Fig. 2b). Similar IC50 values were recorded for alnumycin D against the preformed biofilms of two different S. aureus strains. Of note, the compound was almost equally active in inhibiting preformed biofilms and planktonic growth as indicated by an IC50 ratio of 1.5 (Table 2).
TABLE 2.
Activity and efficacy of the compounds against preformed, 18-h-old biofilms of S. aureus ATCC 25923a
| Compoundb | Biofilm postexposure IC50 (μM)c | IC50 (biofilm postexposure)/IC50 (planktonic phase)c | Strain Newman biofilm postexposure IC50 (μM)c | Log R, log (CFU/ml) |
|---|---|---|---|---|
| Alnumycin D | 4.02 (3.11–5.19) | 1.5 | 3.18 (2.81–3.60) | 3.48 |
| Granaticin B | 3.72 (2.21–6.25) | 1.4 | 5.21 (3.95–6.88) | 2.66 |
| Kalafungin | 27.8 (25.1–30.8) | 25 | n.a.d | 1.17 |
| Medermycin | 24.6 (22.2–27.2) | 8.8 | n.a. | 0.65 |
The efficacy against the preformed ATCC 25923 biofilms was confirmed by a log R assay, which involved a 24-h-long incubation in 40 μM compounds.
In the case of alnumycin D and granaticin B, the IC50s were also determined against S. aureus strain Newman.
All IC50 values were determined using the resazurin-based viability assay, and the 95% confidence intervals are shown in parenthesis.
n.a., not analyzed.
Antibacterial and antibiofilm activity of other PNQs.
Granaticins, represented by granaticin B in this study, kalafungin, and medermycin have previously been identified as antibiotics against single-cell Staphylococcus spp. Thus, these PNQ metabolites were selected for comparative analysis of their antibacterial and antibiofilm potential against S. aureus, and the compounds were isolated and purified from cultures of the known Streptomyces producer strains.
The previously reported MIC values for PNQs against S. aureus strains range from 0.9 to 3.6 μM for granaticin B (13), from 7 to 53 μM for kalafungin (15), and from 0.4 to 1.7 μM for medermycin (14). In accordance with this data, antistaphylococcal activity of the PNQs was also observed here as activity against planktonic cells and as effective inhibition of biofilm formation. The biofilm preexposure IC50 values were below 4 μM for all three compounds (Table 1).
Granaticin B was also highly active against preformed biofilms, as indicated by the determined postexposure IC50 values of 3.72 μM and 5.21 μM against strains ATCC 25923 and Newman, respectively. Analogously to alnumycin D, granaticin B was almost equally active against biofilm and planktonic cells. Kalafungin and medermycin were less active against preformed biofilms, as indicated by postexposure IC50 values of 27.8 μM and 24.6 μM, respectively (Table 2).
The efficacy of all PNQs against preformed biofilms was examined by a log reduction (log R) assay (Table 2). The decrease in CFU by 2.7 and 3.5 logarithmic units caused by 40 μM granaticin B and alnumycin D, respectively, verified that the two are effective against preformed biofilms. Conversely, the lower antibiofilm activity of kalafungin and medermycin was verified by the assay (Table 2).
Cytotoxicity of alnumycin D and granaticin B against human cell lines.
The in vitro cytotoxicity of the most active antibiofilms was evaluated against two human cell lines: the HL cell line that originates from the human respiratory tract and the HeLa adenocarcinoma cell line. The LC50s of alnumycin D against the HL cells and HeLa cells were 8.9 μM and 9.1 μM, respectively (95% confidence intervals, 8.0 to 9.9 μM and 7.8 to 10.5 μM, respectively). Based on these cytotoxicity results and the measured potencies (IC50s) on the two S. aureus strains (Table 2), the therapeutic window (ratios of LC50 and IC50) of alnumycin D against preformed S. aureus biofilms was estimated to be ca. 2.2 to 2.9.
Granaticin B was found to be ca. 10-fold more cytotoxic than alnumycin D, as its LC50 against the HL cells was 0.84 μM, and its LC50 against the HeLa cells was 0.83 μM (95% confidence intervals, 0.76 to 0.94 μM and 0.70 to 0.98 μM, respectively). Relatively similar LC50s against cancer cell lines have been reported for granaticin B recently (25).
DISCUSSION
Natural products continue to be an important source of drug leads. During the period between 1981 and 2010, more than half of the small molecules approved for use as drugs were natural products or directly derived from them, and only 36% of the new chemical entities originated from truly synthetic sources without any natural inspiration. The impact of natural products is even more marked in the field of anti-infectives, covering antibacterial, antifungal, antiparasitic, and antiviral agents, where 69% of all drugs are naturally derived or inspired (26).
Our findings indicate that even the known classes of antimicrobial natural products may contain compounds that have specifically evolved against bacterial biofilms. Granaticin B and alnumycin D were found to be almost equally active against biofilm cells as they are against planktonic cells. Unexpectedly, the pathway shunt product alnumycin D, which is produced by a gene knockout strain, was found to be more active against planktonic and biofilm cells than the pathway end product alnumycin A. To our knowledge, the observed high bioactivity of a shunt product of an antibiotic biosynthetic pathway is a rare finding. The result suggests that pathway manipulation and other synthetic biology tools that may result in an increased production of shunt products can be utilized more extensively for drug discovery purposes.
The presence of the C-ribosyl unit in an unusual pyranose form was found to be essential for the antibiofilm activity of the alnumycins (Fig. 2b). From a structural point of view, all alnumycins are relatively similar, as they share the same aglycone unit, and differences can only be seen in the moiety attached at C-8 (Fig. 1). It is also interesting to note that of the other tested PNQs, only granaticin B was as active as alnumycin D against preformed biofilms (Table 2), and that the two compounds are glycosylated. While alnumycin D is C-glycosylated at position C-8 with a rare ribopyranosyl unit, the sugar units of granaticin B are glucose-derived and the first one is connected to the aglycone unit via two C–C bonds at positions C-7 and C-8 (Fig. 1). Alnumycin D and granaticin B also share a similar oxygenation pattern of the lateral naphthoquinone ring; the former carries a carbonyl oxygen atom and the latter carries a hydroxyl group at position C-6. The less potent antibiofilms kalafungin and medermycin lack the oxygen atom at position C-6 (Fig. 1), which suggests that the oxygenation pattern also may have an impact on the antibiofilm activity of the PNQs.
In contrast to the alnumycins, the presence of a sugar moiety did not result in the higher antibiofilm activity of medermycin compared with that of the aglycone form kalafungin (Fig. 1). This unexpected result might, at least in part, be due to the fact that the amino sugar unit of medermycin is charged at neutral pH, although other structural features may also contribute to the lower antibiofilm activity. The more potent antibiofilm granaticin B contains a γ-lactone moiety with an opposite stereochemistry (Fig. 1), but it seems unlikely that the lower activity of kalafungin and medermycin would be due to the orientation of the lactone moiety. It has been shown previously that an opposite stereochemistry of the lactone moiety does not affect other antimicrobial properties of these PNQs (27).
The preliminary in vitro toxicity evaluation revealed that of the most active antibiofilm PNQs, granaticin B was more toxic to human cell lines, while alnumycin D displayed selective toxicity against planktonic and biofilm bacteria. Very recently, less cytotoxic dihydrogranaticin A and B analogs with an alnumycin-like oxygenation pattern of the aglycone units have been isolated from Streptomyces sp. CPCC 200532 (25), and it would be very interesting to evaluate the antibiofilm activity of these compounds. In the future, the natural antibiofilm potential of the PNQs may be more efficiently exploited via the engineered biosynthesis of novel C-glycosylated compound analogs with improved bioactivity profiles.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Drug Discovery and Chemical Biology (DDCB) network of Biocenter Finland, the Tor, Joe, and Pentti Borgs Memorial Fund, and the Academy of Finland (decisions 136060, 282981, and 272266).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00991-15.
REFERENCES
- 1.Kiedrowski MR, Horswill AR. 2011. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci 1241:104–121. doi: 10.1111/j.1749-6632.2011.06281.x. [DOI] [PubMed] [Google Scholar]
- 2.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amorena B, Gracia E, Monzón M, Leiva J, Oteiza C, Pérez M, Alabart JL, Hernández-Yago J. 1999. Antibiotic susceptibility assay for Staphylococcus aureus in biofilms developed in vitro. J Antimicrob Chemother 44:43–55. doi: 10.1093/jac/44.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Sandberg M, Määttänen A, Peltonen J, Vuorela PM, Fallarero A. 2008. Automating a 96-well microtitre plate model for Staphylococcus aureus biofilms: an approach to screening of natural antimicrobial compounds. Int J Antimicrob Agents 32:233–240. doi: 10.1016/j.ijantimicag.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen TB, Givskov M. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 8.Wright GD. 2014. Something old, something new: revisiting natural products in antibiotic drug discovery. Can J Microbiol 60:147–154. doi: 10.1139/cjm-2014-0063. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell W. 2011. Natural products from synthetic biology. Curr Opin Chem Biol 15:505–515. doi: 10.1016/j.cbpa.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 10.Baltz RH. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. 2007. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat Prod Rep 24:162–190. doi: 10.1039/B507395M. [DOI] [PubMed] [Google Scholar]
- 12.Metsä-Ketelä M, Oja T, Taguchi T, Okamoto S, Ichinose K. 2013. Biosynthesis of pyranonaphthoquinone polyketides reveals diverse strategies for enzymatic carbon-carbon bond formation. Curr Opin Chem Biol 17:562–570. doi: 10.1016/j.cbpa.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 13.Elson AL, Box SJ, Gilpin ML. 1988. New quinone antibiotics of the granaticin type, isolated from Streptomyces lateritius. I. Production, isolation and properties. J Antibiot (Tokyo) 41:570–572. [DOI] [PubMed] [Google Scholar]
- 14.Takano S, Hasuda K, Ito A, Koide Y, Ishii F. 1976. A new antibiotic, medermycin. J Antibiot (Tokyo) 29:765–768. doi: 10.7164/antibiotics.29.765. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LE, Dietz A. 1968. Kalafungin, a new antibiotic produced by Streptomyces tanashiensis strain Kala. Appl Microbiol 16:1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieber B, Nüske J, Ritzau M, Gräfe U. 1998. Alnumycin a new naphthoquinone antibiotic produced by an endophytic Streptomyces sp. J Antibiot (Tokyo) 51:381–382. doi: 10.7164/antibiotics.51.381. [DOI] [PubMed] [Google Scholar]
- 17.Naruse N, Goto M, Watanabe Y, Terasawa T, Dobashi K. 1998. K1115 A, a new anthraquinone that inhibits the binding of activator protein-1 (AP-1) to its recognition sites. II. Taxonomy, fermentation, isolation, physico-chemical properties and structure determination. J Antibiot (Tokyo) 51:545–552. [DOI] [PubMed] [Google Scholar]
- 18.Oja T, Palmu K, Lehmussola H, Leppäranta O, Hännikäinen K, Niemi J, Mäntsälä P, Metsä-Ketelä M. 2008. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem Biol 15:1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 19.Oja T, Klika KD, Appassamy L, Sinkkonen J, Mäntsälä P, Niemi J, Metsä-Ketelä M. 2012. Biosynthetic pathway toward carbohydrate-like moieties of alnumycins contains unusual steps for C–C bond formation and cleavage. Proc Natl Acad Sci U S A 109:6024–6029. doi: 10.1073/pnas.1201530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham LH, Vater J, Rotard W, Mügge C. 2005. Identification of secondary metabolites from Streptomyces violaceoruber Tü22 by means of on-flow LC-NMR and LC-DAD-MS. Magn Reson Chem 43:710–723. doi: 10.1002/mrc.1633. [DOI] [PubMed] [Google Scholar]
- 21.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. 2003. Cloning, sequencing and heterologous expression of the medermycin biosynthetic cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 149:1633–1645. [DOI] [PubMed] [Google Scholar]
- 22.Sandberg ME, Schellmann D, Brunhofer G, Erker T, Busygin I, Leino R, Vuorela PM, Fallarero A. 2009. Pros and cons of using resazurin staining for quantification of viable Staphylococcus aureus biofilms in a screening assay. J Microbiol Methods 78:104–106. doi: 10.1016/j.mimet.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Pitts B, Hamilton MA, Zelver N, Stewart P. 2003. A microtiter-plate screening method for biofilm disinfection and removal. J Microbiol Methods 54:269–276. doi: 10.1016/S0167-7012(03)00034-4. [DOI] [PubMed] [Google Scholar]
- 24.Kuo CC, Grayston JT. 1990. A sensitive cell line, HL cells, for isolation and propagation of Chlamydia pneumoniae strain TWAR. J Infect Dis 162:755–758. doi: 10.1093/infdis/162.3.755. [DOI] [PubMed] [Google Scholar]
- 25.Jiang B, Li S, Zhao W, Li T, Zuo L, Nan Y, Wu L, Liu H, Yu L, Shan G, Zuo L. 2014. 6-Deoxy-13-hydroxy-8,11-dione-dihydrogranaticin B, an intermediate in granaticin biosynthesis, from Streptomyces sp. CPCC 200532. J Nat Prod 77:2130–2133. doi: 10.1021/np500138k. [DOI] [PubMed] [Google Scholar]
- 26.Cragg GM, Newman DJ. 2013. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta 1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatsuta K, Ozeki H, Yamaguchi M, Tanaka M, Okui T, Nakata M. 1991. Total synthesis and biological evaluation of unnatural (-)-medermycin [(-)-lactoquinomycin]. J Antibiot (Tokyo) 44:901–902. doi: 10.7164/antibiotics.44.901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.