Abstract
Escherichia coli sequence type 131 (ST131) is a pandemic clone associated with multidrug-resistant, extraintestinal infections, attributable to the presence of the CTX-M-15 extended-spectrum β-lactamase gene and mutations entailing fluoroquinolone resistance. Studies on subclones within E. coli ST131 are critically required for targeting and implementation of successful control efforts. Our study comprehensively analyzed the genomic and functional attributes of the H30-Rx subclonal strains NA097 and NA114, belonging to the ST131 lineage. We carried out whole-genome sequencing, comparative analysis, phenotypic virulence assays, and profiling of the antibacterial responses of THP1 cells infected with these subclones. Phylogenomic analysis suggested that the strains were clonal in nature and confined entirely to a single clade. Comparative genomic analysis revealed that the virulence and resistance repertoires were comparable among the H30-Rx ST131 strains except for the commensal ST131 strain SE15. Similarly, seven phage-specific regions were found to be strongly associated with the H30-Rx strains but were largely absent in the genome of SE15. Phenotypic analysis confirmed the virulence and resistance similarities between the two strains. However, NA097 was found to be more robust than NA114 in terms of virulence gene carriage (dra operon), invasion ability (P < 0.05), and antimicrobial resistance (streptomycin resistance). RT2 gene expression profiling revealed generic upregulation of key proinflammatory responses in THP1 cells, irrespective of ST131 lineage status. In conclusion, our study provides comprehensive, genome-inferred insights into the biology and immunological properties of ST131 strains and suggests clonal diversification of genomic and phenotypic features within the H30-Rx subclone of E. coli ST131.
INTRODUCTION
Extraintestinal infections due to Escherichia coli cause significant morbidity and death and increased health care costs (1, 2). Management of such infections is difficult due to the increasing abundance and wide spectrum of antimicrobial resistance, ranging from fluoroquinolones to extended-spectrum cephalosporins and to the “last resort” antimicrobials (e.g., carbapenems) (3).
Many multidrug-resistant (MDR) bacterial strains are now being reported, especially those belonging to clones capable of rapid global dissemination (4, 5). H30 sequence type 131 (ST131) is one predominant E. coli clone or lineage that is responsible for large proportions of community- and hospital-acquired urinary tract infections (UTIs) and bloodstream infections throughout the world (6–8). The E. coli H30-Rx subclone has emerged with strong associations with fluoroquinolone resistance and CTX-M-15 extended-spectrum β-lactamase (ESBL) production (9); H30 denotes the presence of allele 30 for fimH (type 1 fimbrial adhesin gene), R indicates its association with fluoroquinolone resistance, and x conveys its carriage of blaCTX-M-15. Clonal expansion has now become a dominant mechanism in the proliferation of antimicrobial resistance in ST131 (10). Alarmingly, ST131 strains harboring New Delhi metallo-β-lactamase 1 (NDM-1), which confers carbapenem resistance, have also been reported among E. coli strains in particular and Enterobacteriaceae in general (11). ST131 E. coli strains not only are prevalent in human populations but also demonstrate interspecies transmission (12–14).
The epidemiological success of E. coli ST131 has been attributed to a wide variety of factors, including enhanced virulence and metabolic capabilities, the ability to acquire extensive and newer resistance traits, and better survivability in intestinal and extraintestinal sites (15, 16). However, the exact reasons for the rapid emergence and successful spread of ST131 clones remain largely undefined. Whole-genome sequencing (WGS) has revolutionized the study of bacterial evolution. In order to understand the recent evolutionary history of the most important E. coli strains in circulation, it is necessary to analyze their diversity in terms of the mutations, recombinations, and gene gains and losses in the population, including their profiles of bacteriophage contents (17).
Although ST131 is recognized as a pandemic clone involved in serious extraintestinal pathogenic Escherichia coli (ExPEC) infections, posing grave public health risks (15), it has received meager attention in countries such as India, despite tremendous host diversity, poor sanitation, and widespread abuse of antibiotics with an unclear antibiotic policy. Despite ongoing research on the epidemiological significance of ST131, little is known regarding the host responses and immunological profiles during infection for members of this lineage.
To gain insight into its pathogenicity and to identify distinct traits associated with the high-risk H30-Rx subclone, we sought to investigate the whole-genome sequence of one of the representative ST131 strains (NA097) and to perform comparative genomic analysis using several reference genomes; we characterized the ST131 strain NA097 because of its functional and genomic attributes, including antimicrobial resistance and pathogenicity. Additionally, we performed profiling of innate immune responses to E. coli infections in differentiated THP1 macrophages with an adherent and invasive ST131 E. coli strain (NA097). The results of this study support the idea that expansion of the H30-Rx sublineage has contributed substantially to the dissemination of ST131 E. coli, and they provide a snapshot of the innate immune signatures of the host against ST131 infections.
MATERIALS AND METHODS
ST131 strains and genome sequencing.
The E. coli strain NA097 was isolated from a urine sample from a 29-year-old male patient with septicemia who presented to the D. Y. Patil Hospital (Pimpri, India) in 2009. Strain NA114 was isolated in 2009 and was sequenced and reported previously by our group (18). Strain NA097 was characterized with routine biochemical techniques and was stored at −80°C in Luria-Bertani (LB) broth supplemented with 20% glycerol. High-quality genomic DNA was obtained using the Qiagen DNeasy blood and tissue kit (Qiagen, Germany). Whole-genome sequencing (WGS) was carried out with an Illumina MiSeq sequencer, which generated 1.3 million paired-end reads of 251 bp, with an insert size of 400 to 500 bp. After filtering and trimming of the paired-end reads using the NGS QC Toolkit (http://www.nipgr.res.in/ngsqctoolkit.html), the high-quality reads amounted to genome coverage of approximately 34 times. The reads were then assembled into contigs using the Velvet de novo assembler (19), run with an optimum hash length of 77. The contigs were then screened for the possible presence of plasmids based on their alignment with the online E. coli sequence databases, with parameters such as 70% identity and query coverage, using BLASTn (20). Only chromosomal contigs were ordered and scaffolded using an in-house workbench, C-L-Authenticator, to avoid misscaffolding due to plasmid-related contigs. The generated scaffolds and plasmid contigs were used for further analysis. The draft genome was then submitted to the RAST server (21) for annotation, and the genome statistics from the resulting file were extracted using ARTEMIS (22). Gene prediction was carried out using GeneMarkS (23). The numbers of tRNAs and rRNAs were identified using tRNAscan-SE (24) and RNAmmer (25), respectively.
Genomic data and comparative genomic analysis.
A total of 107 whole-genome sequences, including those of ST131 strains NA097, NA114, JJ1886, and EC958 from the NCBI and other genomes described by Petty et al. (4), were used to construct a core-genome-based phylogenetic tree using Harvest (26), with the complete genome of the SE15 strain taken as a reference. The resulting tree was visualized using the SplitsTree program (http://www.splitstree.org). BRIG (27) was used to visualize the genome variations within clade C, including NA097 along with other reference ST131 genomes from the NCBI. For all of the genomes, predicted protein sequences from GeneMarkS were searched against the database of E. coli virulence genes downloaded from the Virulence Factors Database (VFDB) (28) by using BLASTp. Only hits with 80% identity and 70% query coverage were considered positive hits. A comparative table was created based on the presence or absence of these virulence genes in all of the genomes compared. A similar table was generated for putative resistance-related genes after comparison with the Comprehensive Antibiotic Resistance Database (CARD) (29), using BLASTp. PhiSpy (30) and PHAST (31) were used to identify the putative phage sequences in the genome. Insertion sequence (IS) elements and genomic islands were identified using IS finder (32) and IslandViewer (33), respectively. Predicted protein sequences were also searched against the Conserved Domains Database (CDD) (34) using NCBI Batch CD-Search, for functional categorization. In-house scripts were used to extract the best hits and corresponding cluster of orthologous groups (COG) classes from the results.
Molecular characterization and susceptibility testing.
E. coli phylogenetic groups (A, B1, B2, and D) were determined with a well-established PCR method, as described previously (35). The isolates were preliminarily assessed for ST131 status with an allele-specific PCR assay for pabB gene detection (36). Antimicrobial susceptibility was examined with the standard Kirby-Bauer method, using an Icosa UTI-1 antibiotic ring (HiMedia, Mumbai, India), for 20 different antibiotics. ESBL production was detected using the double-disc synergy test. The results of both tests were interpreted according to Clinical and Laboratory Standards Institute (CLSI) criteria. Metallo-β-lactamase (MBL) production was screened with meropenem and meropenem-EDTA Etest strips (HiMedia, Mumbai, India), according to the manufacturer's directions. Detection of ESBL (blaCTX-M-15, blaTEM, and blaSHV) and MBL (blaNDM-1) genes was carried out by PCR (15). Plasmids were determined using Kado lysis, and replicon types were identified using five multiplex and three single PCR assays for the presence of a total of 18 plasmid types (37).
Phenotypic characterization of strains.
Serum resistance assays were performed in 50% human serum (Pan Biotech) as described previously (38), with some modifications. Briefly, 5 μl of overnight bacterial culture was added to 495 μl of fresh LB medium and grown for 1 h. After centrifugation, the bacteria were resuspended in 1 ml of sterile phosphate-buffered saline (PBS), and 30 μl of culture with 270 μl of 50% human serum was added in triplicate to 96-well microtiter plates. An aliquot of 30 μl of each culture was serially diluted and plated (0-h count), and the rest was incubated for 3 h at 37°C. After incubation, the bacteria were serially diluted and plated (3-h count). Counting was performed after an overnight incubation, and CFU were calculated. The initial CFU value was subtracted from the final count in order to obtain growth in serum, and data were plotted on a graph. Hemagglutination assays were carried out with freshly isolated red blood cells (RBCs) from human blood, and results were interpreted as described elsewhere (39). Biotyping of strains for carbohydrate utilization of 35 sugar sources was performed using the Hi-Carbo kit (HiMedia, Mumbai, India), according to the manufacturer's protocol. Biofilm formation was examined in a total of nine replicates in 96-well microtiter plates with M63 medium, after 48 h, as described previously (8).
Epithelial cell adhesion and invasion assays.
Adhesion and invasion assays were performed as described previously (38), with some modifications. T24 and ACHN cells (human bladder and kidney epithelial cell lines) were grown in RPMI 1640 medium and Ham's F-12 medium (Invitrogen) supplemented with 10% and 15% fetal bovine serum (FBS) (Invitrogen), respectively. Cells were grown in 5% CO2 at 37°C. Cells were seeded into 24-well plates 48 h prior to cell infection assays. Infection was performed at a multiplicity of infection (MOI) of 10. Following 3 h of incubation at 37°C, the monolayer was washed three times using 1× PBS (Gibco) and subsequently lysed with 0.1% Triton X-100 in 1× PBS for 10 min at 37°C. The serially diluted lysates were plated on LB agar plates for determination of CFU of adherent bacteria. For invasion assays, the cells were incubated for 3 h and the monolayer was washed three times with 1× PBS and incubated for 1.5 h with 1 ml of fresh Ham's F-12 medium containing 100 μg/ml gentamicin. After incubation, the cells were lysed with 0.1% Triton X-100 in PBS for 10 min at 37°C. Lysates were serially diluted and CFU were determined as in the adhesion assays described above.
Infection of cultured THP1 cells and RT2 profiling of antibacterial responses.
Monocyte cells were cultured and maintained at 37°C in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS (Invitrogen), in 5% CO2. Prior to infection, about 1 × 106 cells were seeded into 35-mm-diameter petri dishes (Corning) and stimulated for 36 h using phorbol myristate acetate (PMA). After stimulation, cells were incubated for 12 h in antibiotic-free medium. Subsequently, E. coli strains (NA097 and DH5α) were grown overnight, followed by secondary growth to an optical density at 600 nm of 0.95. E. coli was harvested and added to the differentiated THP1 cells at an MOI of 10. Similarly, another set of 1 × 106 cells was treated with 1 μg lipopolysaccharide (LPS). Infected cells were incubated for 3 h. Total RNA was isolated using a GenElute kit (Sigma), and cDNA was synthesized from 1 μg of DNase (Sigma)-treated RNA, using a first-strand cDNA synthesis kit (Qiagen, Germany). Real-time PCR was performed on cDNA using the RT2 Profiler PCR array system (Qiagen), according to the manufacturer's instructions, in an Eppendorf RealPlex 4 system. The experiments were performed in triplicate, and the data were analyzed using the online RT2 Profiler array data analysis tool (version 3.5) from Qiagen.
Statistical analysis.
Statistical analyses for biofilm assays, serum bactericidal assays, adhesion assays, and invasion assays were carried out using the nonparametric Mann-Whitney U test, using GraphPad Prism 5.01. For calculations with quantitative real-time (qRT)-PCR data, custom online software was used as directed in the PAHS-148Z RT2 Profiler manual (Qiagen), and calculations were performed according to the manufacturer's instructions. P values of <0.05 were considered to be significant.
Nucleotide sequence accession numbers.
This whole-genome shotgun project has been deposited in DDBJ/EMBL/GenBank under accession number JSXJ00000000. The version described in this paper is version JSXJ01000000.
RESULTS
Genome statistics and comparative genomic analysis.
The genome of NA097 comprised 187 contigs, constituting an approximate size of 5.1 Mb. The genome was predicted to encode ∼5,164 protein coding sequences, with an average coding sequence length of 885 bp, amounting to a coding capacity of 88%. The genome revealed 51.83% G+C, 73 tRNAs, and 3 rRNA genes. The functional annotation of the NA097 genome using the COG database revealed a majority of genes related to metabolism (37%), followed by cellular processing and signaling (21%) and information storage and processing (17%). A fraction of the genes (14%) belonged to multiple classes, while 10% remained poorly characterized (see Fig. S2 in the supplemental material). In silico multilocus sequence typing (MLST) analysis using the MLST database (http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli) showed that this organism belongs to sequence type 131. We identified 13 families of insertion sequence (IS) elements within the NA097 genome, including at least one copy each of IS21, IS66, IS6, IS1, IS30, Tn3, IS1380, IS110, IS3, ISL3, IS4, IS200, and ISAs1.
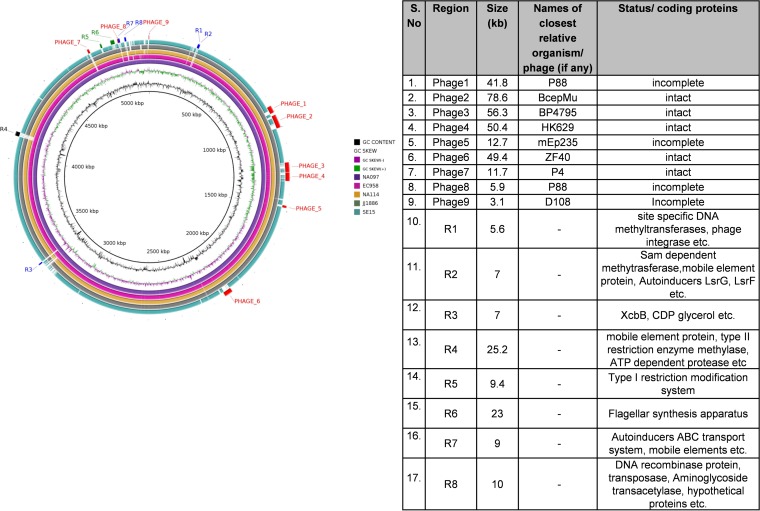
Whole-genome sequence-based phylogenetic analysis of ST131 strains together with ST95 strains revealed three different clades (clade A, clade B, and clade C), as described in Fig. S1 in the supplemental material. The strains NA097 and NA114 clustered within clade C, which predominantly included strains of H30 and H30-Rx subclones of ST131 E. coli. The outcomes of uropathogenic E. coli (UPEC)-specific virulence and resistance gene profiling for five ST131 genomes are summarized in Tables S1 and S2 in the supplemental material. The virulence and resistance repertoires were comparable among the ST131 strains except for SE15, which had few virulence and fewer resistance genes. NA097 primarily revealed the dra operon, in contrast to the pyelonephritis-associated pili (pap) operon in NA114. The strains had similar resistance patterns except that NA097 was also positive for the strA, strB, and aac3′IIC genes. The genomes of EC958 and NA097 contained both blaTEM and blaCTX-M-15 genes; SE15 was negative for both, and the rest of the genomes had one of the two β-lactamase genes. To elucidate the variations among genomes that clustered in clade C, we subjected four well-characterized reference ST131 genomes, i.e., EC958, JJ1886, NA114, and SE15, with NA097 as a reference, to comparative analysis using BRIG. The most common variation observed for SE15 (a commensal ST131 strain) and the four H30-Rx subclonal ST131 strains was the absence of seven phage-associated regions in the SE15 genome (Fig. 1). Three major genomic islands, namely, GI-selC, GI-pheV, and GI-leuX, were also found in the NA097 genome, along with several other genomic islands with sizes ranging from 0.5 kb to 30 kb, as predicted by IslandViewer. The genome of NA097 exclusively contained the lysogenic phage P4 and other genomic regions encoding functions such as autoinduction, capsular polysaccharide synthesis, quorum sensing, aminoglycoside resistance, and others (labeled in blue in Fig. 1). A detailed description of different genomic regions and phages is also provided in Fig. 1. The genome of NA097 shared similarity with the recently published EC958 E. coli ST131 genome sequence, which contains several mobile genetic elements (MGEs) that would account for increased fitness or pathogenicity. Overall, the genomes of four H30-Rx strains were found to be highly clonal in nature, despite their distinct geographical and temporal origins.
FIG 1.
Left, BRIG figure, representing a comparison of NA097 with four reference genomes of ST131 E. coli downloaded from the NCBI. Blue regions (R1, R2, R3, R7, and R8) are present only in NA097, the black region (R4) is present only in NA097 and EC958, green regions (R5 and R6) are absent in SE15, and red regions are phage-associated regions. Right, details of the regions and their relevant sizes.
Molecular characterization and susceptibility testing.
The NA097 strain was positive for typical characteristics of clone ST131, i.e., it belonged to phylogroup B2 and was PCR positive for the pabB allele as well as CTX-M-15 and TEM β-lactamase genes. Phenotypically, NA097 was determined to be an ESBL producer and was MBL negative. The strain was refractory to the following 13 of 20 antimicrobial agents tested: ampicillin, ampicillin-sulbactam, and carbenicillin of the penicillin class; cephalothin (first generation), cefazolin (first generation), cefotaxime (third generation), and cefoperazone (third generation) of the cephalosporin class; norfloxacin, ciprofloxacin, moxifloxacin, and gatifloxacin of the fluoroquinolone class; and streptomycin and co-trimoxazole of the aminoglycoside and trimethoprim-sulfamethoxazole classes, respectively. The strain was completely susceptible only to the macrolide antibiotic class and to five other antibiotics belonging to the aforementioned classes. Kado lysis revealed the presence of four plasmids, but only two plasmids could be identified in PCR-based replicon typing.
Phenotypic characteristics of ST131 strains NA097 and NA114.
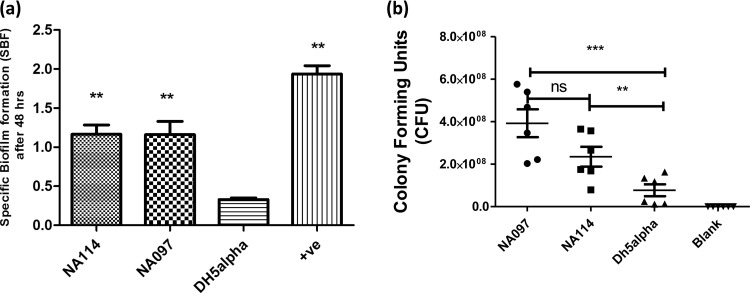
Biofilm formation is considered to be an important virulence property among pathogenic E. coli strains (40), especially those that cause urinary tract infections (UTIs) and sepsis. Therefore, we investigated the ability of two ST131 strains to form biofilms in M63 minimal medium. We observed significant specific biofilm formation (SBF) values for both NA097 and NA114 strains, compared to DH5α, after 48 h of incubation (Fig. 2a). Because serum resistance is reported to be a marker of virulence (41), we examined the survival of the two ST131 strains and DH5α in human serum after 3 h. The mean CFU values for growth of NA097, NA114, and DH5α were 3.9 × 108, 2.6 × 108, and 7.7 × 107 CFU, respectively, while the blank showed no growth (Fig. 2b). NA097 and NA114 were significantly resistant to serum, compared to DH5α. Moreover, the levels of serum resistance for NA097 and NA114 were comparable. Furthermore, NA097 showed mannose-resistant hemagglutination (MRHA) in human RBCs. To assess whether isolates of the ST131 lineage shared specific metabolic characteristics, we investigated the carbohydrate utilization ability of NA097 using 35 API test reagents (HiMedia, Mumbai, India). Of the 35 sugars, NA097 was able to utilize 20 sugars (57%). The metabolic capabilities of NA097 and NA114 for carbohydrate utilization were found to be comparable. However, the strains did not demonstrate enhanced utilization of a panel of metabolic substrates, compared to the data on metabolic utilization by E. coli strains belonging to other sequence types (15).
FIG 2.
Biofilm formation capabilities and serum bactericidal resistance of E. coli strains NA114 and NA097. (a) Specific biofilm formation (SBF) after 48 h of incubation. (b) Resistance to human serum, as determined by CFU counting after 3 h of incubation. Strain DH5α was used as a negative control for both experiments, and NA434 served as a positive control (+ve) for the determination of SBF. The experiment was performed in nine replicates for analysis of biofilm formation and six replicates for the serum assay, and statistical values were obtained with the nonparametric Mann-Whitney U test, using GraphPad Prism 5.01. ns, nonsignificant; **, P < 0.01; ***, P < 0.001.
Adhesion and invasion assays with human epithelial cells.
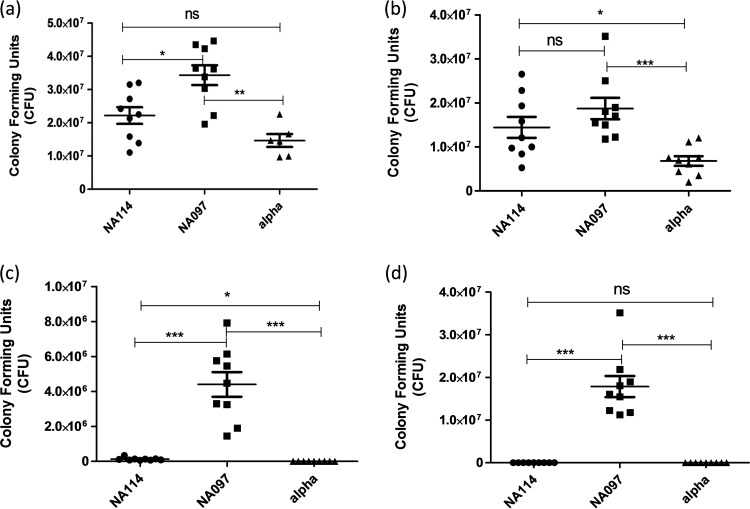
Because of the significance of human kidney and bladder epithelial cells in the development of acute UPEC-related complications and because we wanted to investigate whether the presence or absence of virulence genes affected the pathogenic nature of the bacteria (42), we conducted in vitro cell adhesion and invasion assays using strains NA097 and NA114, along with the negative control (DH5α), with both ACHN and T24 cells. We observed significant levels of adhesion and invasion of NA097 with both cell types, compared to DH5α (Fig. 3). In contrast, NA114 was adherent with both cell types but was less invasive (1.3 × 105 CFU) with T24 cells and was noninvasive with ACHN cells.
FIG 3.
Adhesion and invasion capabilities of E. coli strains NA114 and NA097. (a and b) Adhesion on the human bladder epithelial cell line T24 (a) and the human kidney epithelial cell line ACHN (b) after 3 h of incubation. (c and d) Invasion capabilities in the T24 cell line (c) and the ACHN cell line (d) after 3 h of incubation and additional incubation for 1.5 h in medium containing gentamicin (100 μg/ml). DH5α (alpha) was used as a negative control for the adhesion and invasion assays, experiments were performed three times in triplicate, and statistical values were obtained with the nonparametric Mann-Whitney U test, using GraphPad Prism 5.01. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RT2 gene expression profiling of antibacterial responses in THP1 cells.
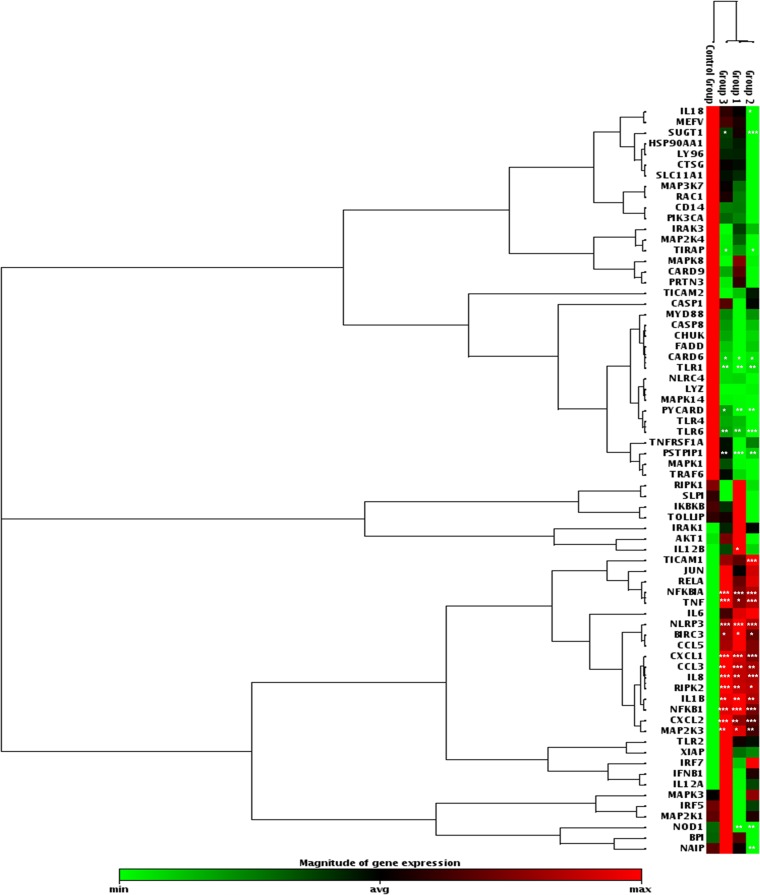
In order to analyze the transcription of antibacterial response genes in differentiated human THP1 macrophages infected with the ST131 E. coli strain NA097, the DH5α strain, and LPS, we performed quantitative PCR to determine the expression of 84 genes, corresponding to inflammatory response factors, pattern recognition receptors (PRRs), chemokines, cytokines, or downstream signaling components. The expression levels for 12 genes were unaltered in all three groups (including the control), while levels for a total of 24 transcripts were identified as being significantly altered. In all three categories, 12 genes were upregulated, while five genes were downregulated. A majority of significantly upregulated genes encoded cytokines such as interleukin 8 (IL-8), IL-1β, and tumor necrosis factor (TNF) and chemokines such as CXCL1, CXCL2, and CCL3; the remaining significantly upregulated transcripts encoded intracellular signaling factors such as BIRC3, MAP2K3, NFKB1, NFKBIA, NLRP3, and RIPK2. In contrast, transcripts corresponding to CARD6, TLR1, TLR6, PSTPIP1, and PYCARD were significantly downregulated (Fig. 4). IL-6 was upregulated more than 100-fold in all cases. Overall, the levels of transcriptionally altered genes were similar (with no significant differences) in all three groups. The rest of the genes did not show any significant fold changes, compared to the untreated control; however, a few, such as IFNB1, NOD1, SUGT1, and NAIP, were upregulated differentially. These data provide a comprehensive picture of transcriptional regulation of macrophage responses to E. coli infections.
FIG 4.
Cluster diagram indicating transcript levels of genes associated with bacterial infections in THP-1 cells. Group 1, LPS-treated cell sample; group 2, cells infected with E. coli strain DH5α; group 3, cells infected with strain NA097. Untreated THP-1 cells were used as the control. Online RT2 Profiler array data analysis custom software (version 3.5) from Qiagen was used to obtain statistical values (Student's t test), according to the RT2 Profiler instructions. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Previous work by our group showed that ST131 E. coli strains in India have emerged as important causes of community-acquired urinary tract infections (8, 15). Molecular epidemiological analysis of these strains demonstrated that they are largely evolving clonally and are associated with several successful phenotypes that are likely involved in ST131 E. coli worldwide dissemination and infections (8, 15, 18, 43). This highlights the urgent need to understand this clonal pathogen, in order to develop better control strategies. In this study, we investigated the comprehensive profile of antibacterial responses of human macrophages upon ST131 E. coli infection. We further investigated the variations in the phenotypes of ST131 strains by analyzing the phenotypic traits versus the whole-genome sequence data for our two ST131 strains (NA097 and NA114). Whole-genome comparisons and phylogenetic comparisons were also performed with other sequenced genomes of ST131 clones.
Our data lend support to observations indicating adaptive diversification and clonal expansion of ST131 as the underlying mechanisms for the evolution of fluoroquinolone-resistant and ESBL-producing ST131 E. coli (44, 45). Our data also reveal that the similarity in phenotypes observed for our two ST131 strains correlates with the observation of the clonal genetic structure due to the evolution of a distinct subclade within ST131, namely, H30-Rx (10). The minor differences observed could be due to variations in virulence gene carriage, as observed by others (46).
ST131 E. coli is now the dominant extraintestinal pathogen throughout the world and is a major etiological agent of bladder and kidney infections (47). This organism represents a classic example of evolution through both horizontal and vertical (clonal expansion) mechanisms. Although ST131 first attracted attention because of its close association with ESBLs, especially the plasmid-mediated CTX-M-15, recent studies involving population genetic analysis of ST131 strains indicated that the global rise in extended-spectrum cephalosporin-resistant ST131 E. coli infections is due to the expansion of a single, highly virulent subclone, H30-Rx (10). Phylogenomic analysis of two of our ST131 strains (NA114 and NA097), together with several ST131 reference genomes (4), revealed that both of the Indian strains NA097 and NA114 belonged to the H30-Rx subclone (see Fig. S1 in the supplemental material). These results might represent only the first of many such findings and therefore warrant attention from clinicians and researchers, as our previous studies have already documented that ST131 E. coli strains represent a major population among clinical ExPEC isolates in India (8, 15).
Earlier studies reported the presence of mobile genetic elements (MGEs) in ST131 E. coli strains, which are not found in non-ST131 UPEC strains (4). Our study confirms this and documents the high level of conservation of seven prophage-associated genomic regions among all of the H30-Rx strains studied (Fig. 1); these seven prophage elements were clearly absent in the commensal non-H30-Rx ST131 SE15 genome (48). The genome of our ST131 strain NA097 revealed greater genetic affinity with the recently published EC958 ST131 genome from England (49), followed by the genome of the Indian ST131 strain NA114. This variation could possibly be due to differences in the arrangements of different genomic regions, arising from variable genomic contents encompassing mobile elements and other auxiliary genomic components. However, such a degree of clonality between two geographically distinct isolates hints at the possibility of their potential to emerge and to expand globally. Other studies have also observed phylogeographic patterns among spatially dispersed ST131 isolates (8).
Given the extensive genomic similarities observed among the strains within the H30-Rx clade, we sought to determine whether there were any phenotypic similarities between the two representative H30-Rx strains, i.e., NA114 and NA097. We compared the abilities of our two H30-Rx strains to form biofilms in vitro over 48 h. There was no significant difference in the levels of specific biofilm formation between the two strains (Fig. 2a). Furthermore, we found that the two strains were equally resistant to serum bactericidal activity (Fig. 2b). We also conducted in vitro cell adhesion and invasion assays with T24 bladder epithelial cells and ACHN kidney epithelial cells. In addition to the virulence-associated phenotypes, identical patterns were observed for the adhesion capabilities of the two strains on both types of epithelial cells (Fig. 3). However, the two strains differed significantly in their abilities to invade the epithelial cells. The NA097 strain was highly invasive, while NA114 showed insignificant invasion (Fig. 3). This observed difference could be partly attributable to the presence of the dra operon (which was found to be absent in the NA114 strain) in the NA097 strain. Goluszko et al. observed similar results, with insertional mutants of dra operon members being unable to invade HeLa cells (50). Finally, we compared the abilities of our two ST131 strains to metabolize 35 carbon sources. We found similar patterns of carbon utilization for the two strains. Overall, our data provide a correlation between the genetic frameworks and the virulence phenotypes of two H30-Rx strains. In contrast, previous studies showed that ST131 strains in general were genetically monomorphic but phenotypically diverse (46). This might be due to the fact that all such studies analyzed ST131 strains without taking into account the recent subclones of ST131, namely, H30-R and H30-Rx. When these subclones are studied separately, they might yield clearer distinct observations.
While several studies have investigated the pathogenesis of ST131 E. coli using in vitro and in vivo assays, the innate immune responses involved and the molecular pathways governing these responses largely remain obscure. Deciphering the innate responses against this pathogen is key to understanding the immunological basis of bacterial clearance from the host. Here we performed transcriptome profiling of innate immune signatures in the context of host responses to ST131 strains, the control strain DH5α, and LPS alone. We observed that only the key inflammatory pathways were activated upon infection with ST131, DH5α, or LPS. Therefore, we could not identify major strain-specific host responses, which implies that, after infection, capsular antigens would be primarily associated with the immune responses and thus pathogenesis could not necessarily be correlated with repertoires of virulence genes or mobile genetic elements. A previous study by Vimont et al. (16) confirmed that the inflammatory responses of kidney cells after infection with ST131 versus non-ST131 were comparable. Also, Horn et al. (51) observed similar histopathological outcomes in lung tissues after infection with pathogenic versus fecal avian E. coli. These observations, together with our results, suggest that the fitness and epidemiological success of ST131 strains are more relevant in the upstream steps involved at the onset of infection.
In conclusion, our study confirmed the presence of H30-Rx ST131 E. coli among Indian ST131 strains and provided more insights into the clonal diversification of genomic and phenotypic features of H30-Rx ST131 strains. The study also shows the immunological basis of the outcomes of infection with ST131 strains. Such expository observations could be of great significance for understanding the emergence of globally disseminated, virulent H30-Rx strains. Further studies are needed to investigate whether this clone undergoes evolutionary selection and whether that has any effect on its virulence and resistance phenotypes.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge funding received from the Department of Biotechnology, Government of India [grant BT/HRD/NBA/34/01/2011(ix)]. We also acknowledge support received from University of Malaya High Impact Research Grant UM.C/625/1HIR/MOHE/02 (A000002-5000 1) and support from Indo-German International Research Training Group GRK1673, an initiative of the German Research Foundation and the University of Hyderabad (India). A.R. acknowledges the Indian Council of Medical Research for the award of a junior research fellowship. N.A. is an adjunct professor at the Academy of Scientific and Innovative Research (AcSIR), New Delhi, India.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01447-15.
REFERENCES
- 1.Tumbarello M, Sanguinetti M, Montuori E, Trecarichi EM, Posteraro B, Fiori B, Citton R, D'Inzeo T, Fadda G, Cauda R, Spanu T. 2007. Predictors of mortality in patients with bloodstream infections caused by extended-spectrum-β-lactamase-producing Enterobacteriaceae: importance of inadequate initial antimicrobial treatment. Antimicrob Agents Chemother 51:1987–1994. doi: 10.1128/AAC.01509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO. 2001. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis 32:1162–1171. doi: 10.1086/319757. [DOI] [PubMed] [Google Scholar]
- 3.Hong T, Moland ES, Abdalhamid B, Hanson ND, Wang J, Sloan C, Fabian D, Farajallah A, Levine J, Thomson KS. 2005. Escherichia coli: development of carbapenem resistance during therapy. Clin Infect Dis 40:e84–e86. doi: 10.1086/429822. [DOI] [PubMed] [Google Scholar]
- 4.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Canica MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. [DOI] [PubMed] [Google Scholar]
- 7.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Canton R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain A, Ewers C, Nandanwar N, Guenther S, Jadhav S, Wieler LH, Ahmed N. 2012. Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-β-lactamase-producing lineage. Antimicrob Agents Chemother 56:6358–6365. doi: 10.1128/AAC.01099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4(6):e00377-13. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Miller S, Johnston B, Clabots C, Debroy C. 2009. Sharing of Escherichia coli sequence type ST131 and other multidrug-resistant and urovirulent E. coli strains among dogs and cats within a household. J Clin Microbiol 47:3721–3725. doi: 10.1128/JCM.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob Agents Chemother 55:3782–3787. doi: 10.1128/AAC.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 15.Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. 2014. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother 58:7240–7249. doi: 10.1128/AAC.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon JM, Garry L, Clermont O, Denamur E, Arlet G, Vandewalle A. 2012. The CTX-M-15-producing Escherichia coli clone O25b:H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One 7:e46547. doi: 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed N, Dobrindt U, Hacker J, Hasnain SE. 2008. Genomic fluidity and pathogenic bacteria: applications in diagnostics, epidemiology and intervention. Nat Rev Microbiol 6:387–394. doi: 10.1038/nrmicro1889. [DOI] [PubMed] [Google Scholar]
- 18.Avasthi TS, Kumar N, Baddam R, Hussain A, Nandanwar N, Jadhav S, Ahmed N. 2011. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J Bacteriol 193:4272–4273. doi: 10.1128/JB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F. 2012. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 23.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes: implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langille MG, Brinkman FS. 2009. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppe E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 64:274–277. doi: 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 37.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Nandanwar N, Janssen T, Kuhl M, Ahmed N, Ewers C, Wieler LH. 2014. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol 304:835–842. doi: 10.1016/j.ijmm.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Siegfried L, Kmetova M, Puzova H, Molokacova M, Filka J. 1994. Virulence-associated factors in Escherichia coli strains isolated from children with urinary tract infections. J Med Microbiol 41:127–132. doi: 10.1099/00222615-41-2-127. [DOI] [PubMed] [Google Scholar]
- 40.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr Top Microbiol Immunol 322:249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolan LK, Horne SM, Giddings CW, Foley SL, Johnson TJ, Lynne AM, Skyberg J. 2003. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet Res Commun 27:101–110. doi: 10.1023/A:1022854902700. [DOI] [PubMed] [Google Scholar]
- 42.Bien J, Sokolova O, Bozko P. 2012. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. Int J Nephrol 2012:681473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, Wieler LH, Ewers C, Ahmed N. 2011. Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One 6:e18063. doi: 10.1371/journal.pone.0018063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paul S, Linardopoulou EV, Billig M, Tchesnokova V, Price LB, Johnson JR, Chattopadhyay S, Sokurenko EV. 2013. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J Bacteriol 195:231–242. doi: 10.1128/JB.01524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee R, Johnson JR. 2013. Escherichia coli ST131: variations on a theme of clonal expansion. Enferm Infecc Microbiol Clin 31:355–356. doi: 10.1016/j.eimc.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark G, Paszkiewicz K, Hale J, Weston V, Constantinidou C, Penn C, Achtman M, McNally A. 2012. Genomic analysis uncovers a phenotypically diverse but genetically homogeneous Escherichia coli ST131 clone circulating in unrelated urinary tract infections. J Antimicrob Chemother 67:868–877. doi: 10.1093/jac/dkr585. [DOI] [PubMed] [Google Scholar]
- 47.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan MD, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. doi: 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goluszko P, Popov V, Selvarangan R, Nowicki S, Pham T, Nowicki BJ. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J Infect Dis 176:158–167. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 51.Horn F, Corrêa AMR, Barbieri NL, Glodde S, Weyrauch KD, Kaspers B, Driemeier D, Ewers C, Wieler LH. 2012. Infections with avian pathogenic and fecal Escherichia coli strains display similar lung histopathology and macrophage apoptosis. PLoS One 7:e41031. doi: 10.1371/journal.pone.0041031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.