Abstract
Enzymes are the proteins responsible for the catalysis of life. Enzymes sharing a common ancestor as defined by sequence and structure similarity are grouped into families and superfamilies. The molecular function of enzymes is defined as their ability to catalyze biochemical reactions; it is manually classified by the Enzyme Commission and robust approaches to quantitatively compare catalytic reactions are just beginning to appear. Here, we present an overview of studies at the interface of the evolution and function of enzymes.
Main Text
The notion of enzymes as biocatalysts was originally presented in 1833 with the discovery of the conversion of starch into sugars catalyzed by diastase (1). However, it was not until the 20th century that scientists realized their full potential in the context of medicine and technology. Major landmarks were the development of methods for enzyme isolation and purification, the realization that enzymes are proteins with biochemical activity and their characterization using x-ray diffraction techniques (2,3). Studies on the dynamic nature of the structure of ribonuclease and efforts to decipher the catalytic mechanism of lysozyme revealed enzymology as an emerging scientific discipline.
Enzymes have many functional attributes. At the molecular level, enzymes catalyze biochemical reactions by accelerating the conversion of substrates into products in a buried pocket within the active site of the enzyme. Without enzyme catalysis, most reactions would be too slow to be useful for life, although not all reactions in nature require catalysis (4). From the pioneering studies by Krebs on the citric acid cycle (5) to the elaboration of comprehensive biochemical wall charts and databases, we have realized that enzymes do not act independently but modulate collectively metabolic pathways and networks. Enzymes perform their molecular function in a particular cell compartment. For instance, hexokinase turns D-glucose into α-D-glucose-6-phosphate in the glycolysis pathway, which takes place in the cytosol. Finally, there is great diversity in the fraction of enzymes in different organisms (6) and variations at the organelle, cell type, and tissue levels have also been observed (7–9).
Classifying the function of enzymes
In a similar way to our present-day data deluge in genomics, the good old days of enzymology and biochemistry witnessed the growing accumulation of vast amounts of enzyme data: biochemical reactions, enzyme kinetics, crystallographic structures, and mechanistic interpretations. However, the means of data storage and dissemination were different at the time, databases did not exist as such and functional data were scattered through the literature making any form of overview analysis challenging. The nomenclature of enzymes was also problematic, enzymes were given trivial names to identify them. Some names were carefully chosen by groups of biochemists, however sometimes names were given to the same enzyme by different scientific schools, likewise different enzymes were named the same way. This led to confusing and ambiguous communication between researchers (10). For example, NADPH dehydrogenase was first known as NADPH diaphorase and old yellow enzyme due to its ability to reduce various dyes, both trivial names still persist today (11,12). Soon after, D-amino acid oxidase was designated as new yellow enzyme and distinction between both enzymes became even more difficult. The remarkable increase in the number of newly discovered enzymes called for the development of a system to name and classify them in a consistent manner.
Just as taxonomic classification proved so useful to identify and dissect the diversity of living organisms during the 18th century, in 1956 biochemists and enzymologists launched an initiative to gather all available information about the overall catalyzed reactions to name and classify enzymes. This was led by experts from the Enzyme Commission (EC) of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) who presented a framework to name old enzymes according to which new enzymes could be classified. Enzymes are now named and identified systematically with an EC number; this code is a four-level description that is used to classify enzymes depending on the overall chemical transformation of substrates into products (13). The first level corresponds to six different classes according to the type of chemistry being carried out. Oxidoreductases catalyze oxidation/reduction reactions (EC 1), transferases transfer a chemical group (EC 2), for example, a methyl or glycosyl moiety; hydrolases perform hydrolysis of chemical bonds (EC 3), lyases also cleave chemical bonds by other means than by oxidation or hydrolysis (EC 4), isomerases catalyze geometric and structural changes between isomers (EC 5) and finally, ligases join two compounds with associated hydrolysis of a nucleoside triphosphate molecule (EC 6). These EC classes are further divided into subclasses and sub-subclasses (second and third level, respectively) in line with a variety of criteria such as the chemical bond cleaved or formed, the reaction center, the transferred chemical group, and the cofactor used for catalysis. The final level of classification defines substrate specificity. For example, alanine racemase is an isomerase (EC 5), in particular a racemase (EC 5.1) acting on the amino acid (EC 5.1.1) alanine (EC 5.1.1.1).
When the EC started to operate, the prevalent view was that enzymes were substrate specific, however as several enzymes were discovered to catalyze more than one reaction (enzyme promiscuity), EC numbers started to list additional reactions catalyzed by the same enzyme. Many studies have characterized promiscuity in detail (14–17) and revealed a possible role of promiscuous enzymes as intermediates in the evolution of enzyme function (18–20). However, strictly speaking, the EC classification is still made on the basis of the main catalyzed reaction therefore rendering a limited categorization to describe the full potential activity of enzymes. Nonetheless, the assignment of EC numbers to enzymes is now a common routine in the functional annotation of proteins and protein-coding genes in databases such as UniprotKB (21) and Ensembl (22) and has been adopted by the widely used Gene Ontology (GO) (23).
The evolution of enzyme function
The ability of organisms to adapt to the changing conditions of their habitat is crucial to guarantee their survival and reproduction. Adaptation to chemical variations in the living environment drives the innovation, exchange, and demise of enzyme function. At the metabolic level, this process of adaptation is related to the ability of enzymes to evolve beneficial functions in an environment of changing chemical conditions (24), e.g., the capability of bacteria to acquire resistance to drugs and pesticides.
The emergence of enzymes
The discovery of nonenzymatic metal-catalyzed metabolic reactions in laboratory experiments reproducing the chemistry of an early ocean suggests a potential abiotic origin of catalytic function (25). As the temperature of the Earth cooled down and stabilized, enzymes might have evolved from nonenzymatic precursors or protoenzymes, which used to operate at higher temperatures (26). This evolution allowed better control of substrate specificity by preventing the synthesis of undesirable products and an enhanced regulation of metabolism (4). Although enzyme biosynthesis is energy demanding compared to its nonenzymatic counterpart, it led to the regulation of core metabolic pathways shared across most living organisms, e.g., glycolysis. An example of the synergy between the abiotic and living worlds is metalloenzymes. Nature has evolved the structure of these enzymes to wrap around metal catalysts endowing the complexes with the ability to harness and control diverse chemistry in biology.
Enzyme functionalization
Multiple research studies have unveiled genetic mechanisms whereby innovation in enzyme function might have emerged. In its simplest form, the genetic diversity driving adaptation relies on the accumulation of point mutations. Although the majority of them are neutral or deleterious, gain-of-function mutations may create a new activity in an existing enzyme. Subsequently, beneficial mutations might either increase the level of the activity or when occurring in regulatory regions, they might enhance gene expression and therefore boost the cellular concentration of the enzyme up to physiological levels (27). The evolution of new activity is modulated by stability-function tradeoffs (20,28), and if it provides a selective advantage to the organism in its ability to perform a fundamental biological process such as the competition for resources, will become fixed within a population with beneficial mutations that enhance this activity gradually strengthening the fitness of the organism. The function then might either remain as a new activity of a multifunctional enzyme or segregate as the primary activity of a new enzyme, through evolutionary processes such as gene duplication and specialization (29). The neo- and subfunctionalization of enzymes is explained by a generally accepted model known as innovation-amplification-divergence (24), which is considered to be iterative so the newly evolved enzyme may develop new functions leading to further adaptive cycles.
Using sequence and structure to generate families of enzymes
The analysis of the sequences and structures of evolutionary related enzymes reveals relationships between catalytic activities. Approaches to capture fine details about the conservation of sequence and structural elements in protein domains use hidden Markov models (30), which facilitate the classification of proteins in families and superfamilies (31,32). Although the three-dimensional location of active sites is frequently conserved within superfamilies (33), variations of the physicochemical properties of the residues lining the pockets and other patterns of structural change have been observed (34,35). Enzymes accommodate alternative chemistries using a combination of chemistry-driven and substrate-driven evolution (36,37). The overall chemical reaction is often changed while conserving at least one mechanistic step (38,39), however binding similar substrates while conserving the reaction chemistry is also observed (40,41). Analogous reactions are sometimes catalyzed by different superfamilies (42) using similar active sites (43) or catalytic mechanisms (44). This illustrates how nature might evolve the same functional outcome independently using different structural solutions—convergent evolution.
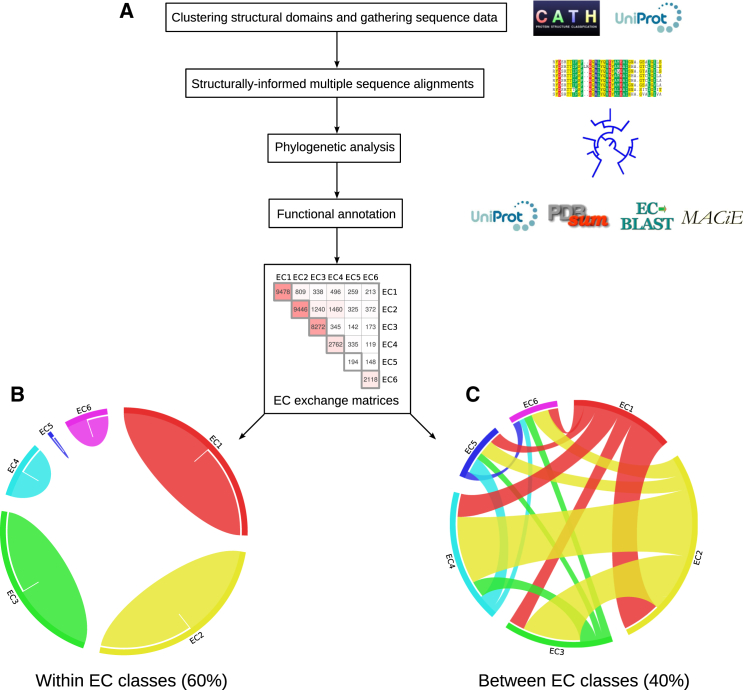
To investigate the evolutionary routes whereby divergence in sequence and structure lead to new enzyme functions in superfamilies, researchers use phylogenetic analysis. For example, Furnham and colleagues developed FunTree (45), a resource containing phylogenetic trees decorated with structural, functional, and mechanistic information that captures the evolution of enzyme function in superfamilies (Fig. 1A).
Figure 1.
Exploring the evolution of enzyme function within 283 multifunctional CATH superfamilies. (A) FunTree approach (45): first, structural clusters of CATH domains involved in enzyme function are created, populated with sequence relatives, and structurally informed multiple sequence alignments are generated. Second, using alignments as the starting point, species-guided phylogenetic trees are created. Finally, functional annotations are retrieved from protein data resources and the frequency of all possible exchanges between different EC numbers within each superfamily is added to an EC exchange matrix. To visualize this matrix, circular diagrams are shown with ribbons representing the frequency of EC changes observed during evolution (bandwidth) (B) within EC classes (diagonal of EC exchange matrix) and (C) between EC classes (off-diagonal). Although the ribbons were colored according to the lowest EC primary class, they are bidirectional and hence the frequency of changes ECX->ECY is the same to ECY->ECX. Data were obtained from CATH version 3.5 (32) and graphics were generated using Circos (56).
Cataloguing enzyme function evolution
During evolution, most enzymes evolve to become enzymes from the same EC class (60% of all EC changes) (Fig. 1B) (e.g., one hydrolase will evolve a new hydrolase function). However, the remaining 40% of changes are between enzymes catalyzing different overall chemistry (Fig. 1C). Remarkably, all possible changes between EC classes are observed. There are some preferences such as transferases (EC 2) becoming oxidoreductases (EC 1), hydrolases (EC 3), and lyases (EC 4). Isomerases (EC 5) are exceptional and evolve new overall chemistry more often than conserving the chemistry of isomerization (46).
Enzyme reaction classification
The functional changes we observed within the same EC class would be expected with the existing classification of enzyme function, e.g., an enzyme could easily change to another catalyzing similar chemistry thus changing the substrate specificity only. However, exchanges between different EC classes suggest that the chemistry of enzymes is more complex than previously classified, with close relationships between enzymes with radically different EC numbers. The chemistry of related enzyme functions can now be explored using robust computational approaches like EC-BLAST (47). This tool searches and compares reactions on the basis of bond changes, reaction centers, and structures of substrates and products. Just like classical studies investigated the evolution of related protein sequences and structures (48), an accurate comparison of enzyme functions might facilitate the interpretation of the evolution of enzymes in the context of their chemistry, which is relevant for enzyme design and genomics (49).
Role of domain composition and allostery in enzyme function
Many enzymes are multidomain and acquire new functionality by changing the domain composition during evolution (50). Enzyme function is assigned on a whole-sequence basis without associating specific functions to the composite domains (51,52). Therefore, cataloguing the functional evolution of each individual domain is a complex process, which can lead to multiple different evolutionary routes. Laboratory experiments do also reveal how amino acid residues located far from the active site often make important contributions to binding and catalysis by inducing conformational changes (53) and acting allosterically (54). More experimental research (55) exploring how enzyme function changes with protein structure and domain architecture is necessary.
Conclusions
Enzymes have both biological and chemical attributes. Their sequences and structures delineate their role in the genome and proteome of all living organisms and their ability to catalyze chemical reactions extends their biological function to metabolic pathways and networks. Many enzymes are promiscuous and perform multiple reactions and as protein sequence evolves, enzymes can change their reaction profile. Combining bond changes and reaction centers with structural information about the substrates, products, and mechanisms is needed to capture the essence of enzyme chemistry in a functional classification. The development of tools to navigate through reaction space (e.g., EC-BLAST) paves a new way for an improved description of enzyme reactions, providing a deeper perspective of biological function.
Acknowledgments
The authors thank the European Molecular Biology Laboratory (EMBL) for funding.
Editor: Ivet Bahar.
References
- 1.Payen A., Persoz J. Mémoire sur la Diastase, les principaux Produits de ses Réactions, et leurs applications aux arts industriels. Ann. Chim. Phys. 1833;53:73–92. [Google Scholar]
- 2.Sumner J. The isolation and crystallization of the enzyme urease. J. Biol. Chem. 1926;69:435–441. [Google Scholar]
- 3.Blake C.C., Koenig D.F., Sarma V.R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206:757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 4.Keller M.A., Piedrafita G., Ralser M. The widespread role of non-enzymatic reactions in cellular metabolism. Curr. Opin. Biotechnol. 2015;34C:153–161. doi: 10.1016/j.copbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krebs H.A. The citric acid cycle and the Szent-Györgyi cycle in pigeon breast muscle. Biochem. J. 1940;34:775–779. doi: 10.1042/bj0340775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freilich S., Spriggs R.V., Thornton J.M. The complement of enzymatic sets in different species. J. Mol. Biol. 2005;349:745–763. doi: 10.1016/j.jmb.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 7.Lewis N.E., Schramm G., Palsson B.Ø. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotechnol. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mintz-Oron S., Meir S., Shlomi T. Reconstruction of Arabidopsis metabolic network models accounting for subcellular compartmentalization and tissue-specificity. Proc. Natl. Acad. Sci. USA. 2012;109:339–344. doi: 10.1073/pnas.1100358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stincone A., Prigione A., Ralser M. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 2014 doi: 10.1111/brv.12140. 000–000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornish-Bowden A. Current IUBMB recommendations on enzyme nomenclature and kinetics. Perspect. Sci. 2014;1:74–87. [Google Scholar]
- 11.Daugherty A.B., Govindarajan S., Lutz S. Improved biocatalysts from a synthetic circular permutation library of the flavin-dependent oxidoreductase old yellow enzyme. J. Am. Chem. Soc. 2013;135:14425–14432. doi: 10.1021/ja4074886. [DOI] [PubMed] [Google Scholar]
- 12.Savignon T., Costa E., Barradas P.C. Prenatal hypoxic-ischemic insult changes the distribution and number of NADPH-diaphorase cells in the cerebellum. PLoS ONE. 2012;7:e35786. doi: 10.1371/journal.pone.0035786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tipton K., Boyce S. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. doi: 10.1093/bioinformatics/16.1.34. [DOI] [PubMed] [Google Scholar]
- 14.Duarte F., Amrein B.A., Kamerlin S.C.L. Modeling catalytic promiscuity in the alkaline phosphatase superfamily. Phys. Chem. Chem. Phys. 2013;15:11160–11177. doi: 10.1039/c3cp51179k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatti-Lafranconi P., Hollfelder F. Flexibility and reactivity in promiscuous enzymes. ChemBioChem. 2013;14:285–292. doi: 10.1002/cbic.201200628. [DOI] [PubMed] [Google Scholar]
- 16.Pandya C., Farelli J.D., Allen K.N. Enzyme promiscuity: engine of evolutionary innovation. J. Biol. Chem. 2014;289:30229–30236. doi: 10.1074/jbc.R114.572990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanal A., Yu McLoughlin S., Copley S.D. Differential effects of a mutation on the normal and promiscuous activities of orthologs: implications for natural and directed evolution. Mol. Biol. Evol. 2015;32:100–108. doi: 10.1093/molbev/msu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aharoni A., Gaidukov L., Tawfik D.S. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 19.Dellus-Gur E., Toth-Petroczy A., Tawfik D.S. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J. Mol. Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 20.Kaltenbach M., Tokuriki N. Dynamics and constraints of enzyme evolution. J. Exp. Zoolog. B Mol. Dev. Evol. 2014;322:468–487. doi: 10.1002/jez.b.22562. [DOI] [PubMed] [Google Scholar]
- 21.UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kersey P.J., Allen J.E., Staines D.M. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic Acids Res. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner M., Ball C.A., Sherlock G., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copley S.D. Toward a systems biology perspective on enzyme evolution. J. Biol. Chem. 2012;287:3–10. doi: 10.1074/jbc.R111.254714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller M.A., Turchyn A.V., Ralser M. Non-enzymatic glycolysis and pentose phosphate pathway-like reactions in a plausible Archean ocean. Mol. Syst. Biol. 2014;10:725. doi: 10.1002/msb.20145228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfenden R. Massive thermal acceleration of the emergence of primordial chemistry, the incidence of spontaneous mutation, and the evolution of enzymes. J. Biol. Chem. 2014;289:30198–30204. doi: 10.1074/jbc.R114.567081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulenburg C., Miller B.G. Enzyme recruitment and its role in metabolic expansion. Biochemistry. 2014;53:836–845. doi: 10.1021/bi401667f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studer R.A., Christin P.-A., Orengo C.A. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proc. Natl. Acad. Sci. USA. 2014;111:2223–2228. doi: 10.1073/pnas.1310811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glasner M.E., Gerlt J.A., Babbitt P.C. Evolution of enzyme superfamilies. Curr. Opin. Chem. Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Eddy S.R. Accelerated profile HMM searches. PLOS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punta M., Coggill P.C., Finn R.D. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sillitoe I., Cuff A.L., Orengo C.A. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41:D490–D498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessailly B.H., Dawson N.L., Orengo C.A. Functional site plasticity in domain superfamilies. Biochim. Biophys. Acta. 2013;1834:874–889. doi: 10.1016/j.bbapap.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson N.L., Studer R., Orengo C. What can comparative genomics reveal about the mechanisms of protein function evolution? In: Wüthrich K., Wilson I.A., Hilvert D., Wolan D.W., editors. New Chemistry and New Opportunities from the Expanding Protein Universe. World Scientific; Singapore: 2015. pp. 16–19. [Google Scholar]
- 35.Brown S.D., Babbitt P.C. New insights about enzyme evolution from large scale studies of sequence and structure relationships. J. Biol. Chem. 2014;289:30221–30228. doi: 10.1074/jbc.R114.569350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babbitt P.C. Definitions of enzyme function for the structural genomics era. Curr. Opin. Chem. Biol. 2003;7:230–237. doi: 10.1016/s1367-5931(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 37.Chiang R.A., Sali A., Babbitt P.C. Evolutionarily conserved substrate substructures for automated annotation of enzyme superfamilies. PLOS Comput. Biol. 2008;4:e1000142. doi: 10.1371/journal.pcbi.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartlett G.J., Borkakoti N., Thornton J.M. Catalysing new reactions during evolution: economy of residues and mechanism. J. Mol. Biol. 2003;331:829–860. doi: 10.1016/s0022-2836(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 39.Gerlt J.A., Babbitt P.C., Almo S.C. Divergent evolution in enolase superfamily: strategies for assigning functions. J. Biol. Chem. 2012;287:29–34. doi: 10.1074/jbc.R111.240945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furnham N., Sillitoe I., Thornton J.M. Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLOS Comput. Biol. 2012;8:e1002403. doi: 10.1371/journal.pcbi.1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bastard K., Smith A.A.T., Salanoubat M. Revealing the hidden functional diversity of an enzyme family. Nat. Chem. Biol. 2014;10:42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 42.Omelchenko M.V., Galperin M.Y., Koonin E.V. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol. Direct. 2010;5:31. doi: 10.1186/1745-6150-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gherardini P.F., Wass M.N., Sternberg M.J.E. Convergent evolution of enzyme active sites is not a rare phenomenon. J. Mol. Biol. 2007;372:817–845. doi: 10.1016/j.jmb.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 44.Almonacid D.E., Yera E.R., Babbitt P.C. Quantitative comparison of catalytic mechanisms and overall reactions in convergently evolved enzymes: implications for classification of enzyme function. PLOS Comput. Biol. 2010;6:e1000700. doi: 10.1371/journal.pcbi.1000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furnham N., Sillitoe I., Thornton J.M. FunTree: a resource for exploring the functional evolution of structurally defined enzyme superfamilies. Nucleic Acids Res. 2012;40:D776–D782. doi: 10.1093/nar/gkr852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez Cuesta S., Furnham N., Thornton J.M. The evolution of enzyme function in the isomerases. Curr. Opin. Struct. Biol. 2014;26:121–130. doi: 10.1016/j.sbi.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rahman S.A., Cuesta S.M., Thornton J.M. EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat. Methods. 2014;11:171–174. doi: 10.1038/nmeth.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabaldón T., Koonin E.V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bashton M., Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Lopez D., Pazos F. Gene ontology functional annotations at the structural domain level. Proteins. 2009;76:598–607. doi: 10.1002/prot.22373. [DOI] [PubMed] [Google Scholar]
- 52.Fang H., Gough J. DcGO: database of domain-centric ontologies on functions, phenotypes, diseases and more. Nucleic Acids Res. 2013;41:D536–D544. doi: 10.1093/nar/gks1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hilvert D., Frère J.-M., Versèes W. Discussions of session 2. In: Wüthrich K., Wilson I.A., Hilvert D., Wolan D.W., editors. New Chemistry and New Opportunities from the Expanding Protein Universe. World Scientific; Singapore: 2014. pp. 94–116. [Google Scholar]
- 54.Makhlynets O.V., Raymond E.A., Korendovych I.V. Design of allosterically regulated protein catalysts. Biochemistry. 2015;54:1444–1456. doi: 10.1021/bi5015248. [DOI] [PubMed] [Google Scholar]
- 55.Baier F., Tokuriki N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014;426:2442–2456. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 56.Krzywinski M., Schein J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]