Abstract
A comparative molecular dynamics analysis of the pyruvate kinase from Leishmania mexicana is presented in the absence and presence of the allosteric effector fructose 2,6-bisphosphate. Comparisons of the simulations of the large 240 kDa apo and holo tetramers show that binding of fructose 2,6-bisphosphate cools the enzyme and reduces dynamic movement, particularly of the B-domain. The reduced dynamic movement of the holo form traps the pyruvate kinase tetramer in its enzymatically active state with the B-domain acting as a lid to cover the active site. The simulations are also consistent with a transition of the mobile active-site α6′ helix, which would adopt a helical conformation in the active R-state and a less structured coil conformation in the inactive T-state. Analysis of the rigid body motions over the trajectory highlights the concerted anticorrelated rigid body rocking motion of the four protomers, which drives the T to R transition. The transitions predicted by these simulations are largely consistent with the Monod-Wyman-Changeux model for allosteric activation but also suggest that rigidification or cooling of the overall structure upon effector binding plays an additional role in enzyme activation.
Introduction
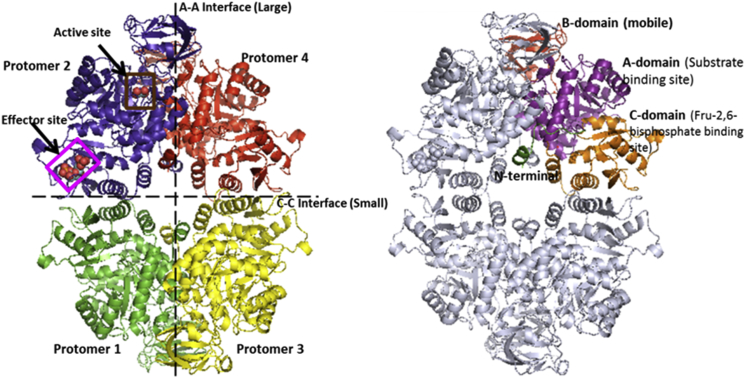
Pyruvate kinases (PYKs) are homotetrameric enzymes that catalyze the final reaction of glycolysis in which phosphoenolpyruvate and ADP are converted into pyruvate and ATP. The single chain PYK protomers are 50–60 kDa depending on species and are composed of four distinct domains. The active site is located in a cleft between the mobile B-domain and the end of the -barreled A-domain of each of the four protomers (Fig. 1). The enzymatically active PYK tetramer has approximate dihedral symmetry (point group D2) with three orthogonal twofold axes relating the four protomers to each other. Crystallographic analyses of mammalian, bacterial, fungal, and trypanosomatid PYKs all show a similar architecture (1) with the tetramer comprising a dimer of dimers resulting in one large A-A′ interface (between the A-domains) and one small C-C′ interface between the C-domains (see Fig. 1).
Figure 1.
Molecular architecture of PYK. (Left) Tetrameric form of LmPYK showing the four single chain protomers. Oxalate and F26BP are shown as spheres occupying the active site and effector site, respectively. The vertical and horizontal lines show the large (A-A) and small (C-C) interfaces. (Right) The N-terminal domain (green), B-domain (gray), A-domain (purple), and C-domain (orange) are shown for one of the protomers. To see this figure in color, go online.
The allosteric behavior of PYK is regulated by the effector molecule fructose bisphosphate (FBP), which binds to the C-domain of each protomer close to the small C-C′ interface of the tetramer. The question we address in this work is how binding of FBP at a site over 40 Å from the active site results in a 7- to 10-fold increase in activity (depending on species). Despite the structural similarities between the tetrameric PYK enzymes from the different families, there are very distinct differences in allosteric mechanisms. For example, the effector molecule for the trypanosomatid PYKs is fructose-2,6-bisphosphate (F26BP), whereas the effector for bacterial and mammalian PYKs is fructose-1,6-bisphosphate. Such structural differences result in lower amino acid sequence identity around the effector binding site. Furthermore, there is biophysical evidence to show that regulatory mechanisms can vary and human M2PYK; for example, dissociates from active tetramer to inactive monomers as a control mechanism (2). There is no evidence that any of the other PYKs use this as a regulatory control mechanism.
In this work we present a comparative molecular dynamics (MD) analysis of the PYK from Leishmania mexicana, LmPYK to study the effect of ligand binding on local and global dynamics of the tetramer. LmPYK was chosen as a model to study the allosteric activation of PYK as it is the only example of an allosteric enzyme where x-ray structures for all four relevant states are available; namely a T-state apo structure with no ligands bound; a structure in which only the effector molecule (F26BP) is bound; a substrate-only structure and a fully liganded structure with both substrate analog and effector bound (1). Overlays of the tetramers for each of these structures showed that only the unliganded T-state tetramer adopted a significantly different conformation from the other three R-state structures that have very similar (RMSFIT < 1 Å) conformations. The conformational transitions observed on going from the T to R state preserved the approximate D2 symmetry of the tetramer but involved changes in the relative orientations of the protomers and also changes in the interprotomer interactions across the A-A′ and C-C′ interfaces. The major differences between the T and R state structures can be described as rigid-body rotations of the protomer domains and led to a description of the a rock-and-lock mechanism for allosteric activation of LmPYK (1).
The rock-and-lock mechanism fits with the original Monod-Wyman-Changeux ideas of an essentially two-state equilibrium between inactive T-states and active R-states as observed experimentally in a number of protein x-ray structures (3). Despite the insight obtained from the R and T state LmPYK x-ray structures, there are still some unanswered questions about the mechanism by which the enzyme shifts between the off and on states and in particular how the activity of the enzyme is altered by the binding of regulatory molecules. In this regard it is intriguing to note from the x-ray structures that the bound F26BP effector molecules do not make interprotomer contacts. Instead, they bind to a flexible loop region. Experimentally, it was found from thermal denaturation studies (1) that binding of the effector to LmPYK significantly increased the thermal melting temperature of the protein and the suggestion has been made that the ∼sevenfold increase of the catalytic rate of LmPYK upon F26BP binding is caused by a rigidification of the enzyme. MD provides an ideal tool to explore this possible mechanism.
Here, we describe two MD simulations for the complete tetramer; the first in the absence of F26BP (the apo structure); the second with F26BP bound, (the holo structure). MD simulations of the isolated monomer are also described. The term protomer refers here to a PYK chain when it is bound in the tetramer, whereas isolated monomer refers to a single PYK chain surrounded by solvent. The comparison of the apo and holo MD simulations identifies allosterically regulated movements of the tetramer as induced by FBP binding. Comparison of correlation of residue fluctuations from the tetramer and monomer trajectories show the role of thermal fluctuations on the individual domains of the protomers as well as providing insight into conformational transitions of the tetramer. The cooling effect of FBP binding, which causes a general reduction in protein flexibility, is also analyzed. These thermodynamic effects along with specific structural rearrangements provide a detailed picture of the allosteric mechanism of LmPYK.
Materials and Methods
The MD simulations were carried out using starting models from the Protein Data Bank x-ray structures 3HQQ. The apo and holo tetrameric structures were created from identical crystal coordinates apart from removing the FBP molecule to provide the apo structure. Comparisons of the tetramer conformations of the starting structures are presented in the Supporting Material. To understand the extent of neighboring protomer effects, additional simulations were performed for isolated monomers from the apo and holo structures. Any monomer simulations used protomers extracted from the relevant tetramer x-ray structure.
All the MD simulations were carried out using GROMACS (GROningen MAchine for Chemical Simulations) package version 4.5 (4) with the AMBER99sb-ildn (5) force-field parameter set. The starting structures were solvated in a dodecahedron box placed at a distance of 0.9 nm from the box boundary. Simple point charge water molecules were used to fill the box, followed by the addition of sodium (Na+) and chloride (Cl−) ions to neutralize the system. The final systems contained 84,000 (monomer) and 300,000 atoms (tetramer) with 76,000 (monomer) and 272,000 to 282,000 (tetramer) water molecules, respectively (Table 1). To maintain a constant temperature of 318 K, the protein and nonprotein atoms were coupled to their own temperature baths using the V-rescale thermostat. This was followed by an NPT equilibration, in which the pressure was maintained isotropically at 1 bar using the Berendsen thermostat (6) with a coupling constant of 0.1 ps. For the apo simulation, the NPT equilibration was carried out for 250 ps with 50,000 steps of 5 fs (4). The holo equilibration was also carried out for 250 ps but using 62,500 steps of 4 fs. The NVT equilibrations were performed in the same way for 250 ps but with 50,000 steps at 5 fs for the apo structure and 62,500 steps at 4 fs for the holo structure. It is known that the Berendsen thermostat does not generate a proper canonical ensemble; however, such discrepancies are insignificant for large systems (7).
Table 1.
Description of the simulation parameters
| Structure | Water Atoms | Calpha Atoms | System Atoms | Time Step (fs) | Number of Frames | Simulation Time (ns) |
|---|---|---|---|---|---|---|
| Holo tetramer | 272,010 | 1,992 | 305,354 | 3 | 15,642 | 48 |
| Apo tetramer | 272,043 | 1992 | 305,251 | 5 | 16,114 | 78 |
| Holo monomer | 76,353 | 498 | 84,689 | 3 | 21,764 | 65 |
| Apo monomer | 76,374 | 498 | 84,676 | 5 | 10,766 | 53 |
For all simulations the water molecules and bond lengths were restrained using the SETTLE (8) and LINCS (9) algorithms, respectively. A single cut-off of 1 nm was used for the treatment of Van der Waals interactions. Long-range electrostatic interactions were simulated using the particle mesh Ewald method with 0.16 FF grid spacing and 4th order B-spline interpolation for the reciprocal sum space. The systems were relaxed by 1000 steps of steepest descent energy minimization procedure before the simulations. The snapshots were saved every 2 ps, thereby yielding 10,000–20,000 frames (see Table 1). The use of the LINCS algorithm has been shown to allow simulations with an increased time step of up to 5 fs (4), and this was used in the 78 ns simulation of apo Lm PYK. See Table 1 for a summary of details of the time steps and simulation times.
Periodic boundary conditions were applied in all the directions. In the analysis of trajectories, the initial conformation for the tetramer, protomer, and the isolated monomer were fixed and taken as the standard and all other conformations were expressed relative to this initial reference structure. Root mean-square deviation plots for the apo and the holo structures are given in Figs. S2–S5. Rigid body displacements and rotations were removed from all trajectories. Acpype (10) from AMBER was used to generate the topology of the ligand and PyMol (11) was used for generating the tetrameric structure. The computing resource used for this simulation was HecToR (12) Phase 3 service of Cray XE6 compute nodes consisting of two 2.3 GHz 16-core AMD Interlagos chips with a total of 2816 nodes.
Results and Discussion
Selection of starting structure for MD simulations
PKYs from mammals through to bacteria are all active as tetramers and share similar overall architectures though the protomers can vary a little in length between species. In the Leishmania mexicana parasite, the four more or less conserved domains are the N-terminal domain that is composed of residues 1–17, the A-domain that comprises residues 18–88, and 187–356; the B-domain, which forms the mobile lid comprises residues 89–186 and the C-domain that incorporates the effector site and comprises residues 357–498.
A detailed comparison of the low resolution 3HQQ structure with 3HQN (unliganded LmPYK in the T-state) and 3HQP (LmPYK in complex with F26BP, ATP, and oxalate) is given in the Supporting Material. It can be seen from Fig. S1 and Table S1 that despite the low resolution of 3HQQ, the structure provides clear experimental evidence for the R-like conformational state of the F26BP effector bound tetramer. The conformation of the 3HQQ tetramer was taken as a starting point for both an effector bound (LmPYK-F26BP) tetramer simulation and (by removing F26BP) a ligand-free LmPYK simulation. Comparison of these two simulations provides a useful insight into the nature of the effector-regulated T to R transitions.
Analysis of calculated and experimental B-factors
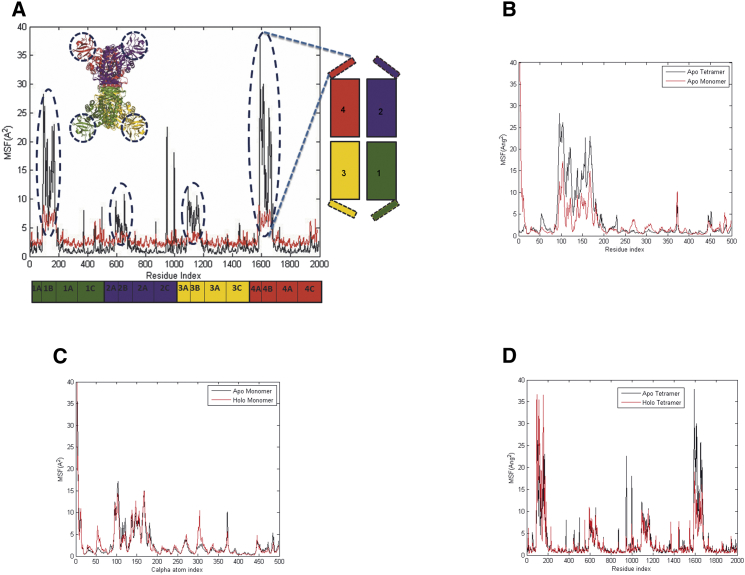
The motion of each atom appears as erratic fluctuations in the MD trajectory. However, there are strong correlations between the motions of subsets of atoms and these collective protein motions may be extracted from the elements of the correlation matrix Each element of the matrix Cij is obtained from the dot product where and ΔRj are the instantaneous fluctuation vectors of atoms i and j from their respective average positions over the time of the simulation. Only the Cα atoms are considered in this analysis. When the index i is equal to j, we obtain the mean square fluctuations (MSF), , of the residues, which may be compared with fluctuations obtained from the experimental B-factors determined by x-ray crystallographic refinement. See Fig. 2A for the comparison between the measured B-factors from the apo x-ray structure (3HQN) and the calculated B-factors from the MD simulation of the apo tetramer. The experimental B-factors of 3HQN were transformed to mean square displacement values using the expression: . The resulting values were multiplied by the factor 318/100 to account for the temperature difference between experiment (100 K) and simulation (318 K).
Figure 2.
Atomic fluctuations in PYK. (A) Comparison of experimental B-factors (red) from apoPYK (3HQN) with the MSFs of the Cα atoms of the LmPYK tetramer calculated from MD simulation (black). Both the representation of the tetramer (right) and the molecular cartoon (inset) highlight the mobile B-domains with dotted lines. (B) Comparison of MSFs of the Cα atoms of the apo LmPYK tetramer (black) with the apo isolated monomer (red) calculated from MD simulation showing the enhanced mobility of the B-domain (residues 89–186) is even more exaggerated in the tetramer than the isolated monomer. (C) MSF comparison of apo LmPYK tetramer (average value 3.18 Å2) and holo (F26BP)-bound tetramer (average value 2.81 Å2). (D) Differences in mean fluctuations for the isolated apo monomer (no FBP bound; black) and the isolated holo monomer (with FBP bound; red). To see this figure in color, go online.
Comparison of the x-ray data and simulation results shows that the latter agrees quantitatively with the experiment except the B-domain cap region, residues 89–186 (Figs. 1 and 2A), where simulation results show a much larger amplitude of motion compared to the x-ray structure. The larger fluctuation of the B-domain may plausibly be attributed to the aqueous environment in simulations in contrast to a more restrictive crystal environment. Indeed, in a number of different crystal forms of PYK the B-domain is found to be totally disordered showing no interpretable electron density, whereas the rest of the structure is well defined (1), suggesting that the B-domain can adopt multiple conformations even in crystal form.
What effect does constraining the PYK as a symmetrical tetramer have on the molecular flexibility? To answer this question we carried out MD simulations on an isolated monomer immersed in a water bath. A comparison of MSF values with the corresponding protomer chain locked in as part of a tetramer highlights the highly mobile B-domain movement in the tetramer simulation (Fig. 2B). The enhanced mobility of the B-domain (residues 89–186) is also present in the isolated monomer simulation but is much reduced with average MSF values for the B-domains alone of 13.8 Å2 and 6.4 Å2 for tetramer and monomer, respectively. This rather unexpected result suggests that the B-domains behave differently (and are more mobile) as part of a tetramer.
We also compared molecular motion of the tetramer with and without the FBP bound (Fig. 2C). This showed that the apo structure fluctuates slightly more than the FBP-bound holo tetramer with MSF values of 3.2 Å2 and 2.8 Å2, respectively. The simulation also shows an interesting asymmetry of movement of the B-domains that breaks the D2 symmetry of the tetramer; the B-domains (residues 89–186) of protomers 1, 2, 3, and 4 have MSF values of 13.8 Å2, 4.7 Å2, 6.6 Å2, and 16.5 Å2, respectively. On binding the F26BP effector the MSF values change to 16.7 Å2, 5.3 Å2, 5.6 Å2, and 9.4 Å2, respectively. However, as observed experimentally in the x-ray structure (Fig. 2A), the asymmetry of movement of the B-domains, with protomers 1 and 4 fluctuating more than protomers 2 and 3, is preserved in the apo and holo MD simulations.
Comparable simulations showing the effect of FBP on binding to the isolated monomer were also carried out (Fig. 2D). MD simulations (∼70 ns) of the isolated monomer (residues 1–498) showed that the holo (FBP bound) isolated monomer had small reduction of 0.08Å2 over the 498 Cα atoms in MSFs compared with the isolated apo monomer.
Analysis of correlation matrices
The B-factors yield information on the fluctuations of the individual residues but do not contain information about the correlations between fluctuations of two different residues. The measure of correlation between the fluctuations and of ith and jth alpha-carbons can be assessed by calculating the projection of one on the other, i.e., at every instant and averaging over the full trajectory. The average, , if positive, indicates that the two residues move, on average, in the same direction. A negative correlation, or anticorrelation, indicates that the two atoms move in opposite directions. If two residues are displaced equally along the same direction, their motions will then be positively correlated and the distance between them will not change. Positively correlated motions represent rigid body motions. If, on the other hand, two atoms move in opposite directions, their motion will be negatively correlated and the distance between them will either increase or decrease.
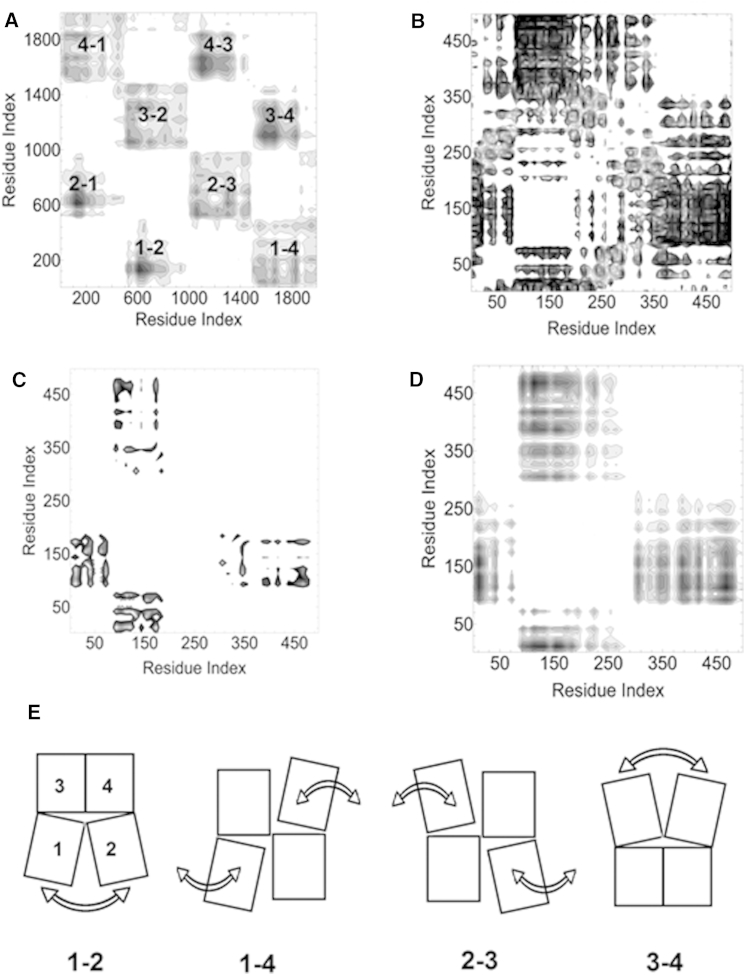
Anticorrelated residue motions and their changes upon effector binding provide significant information on the allosteric behavior of the LmPYK tetramer. The negative correlations of the apo-tetramer are visualized in Fig. 3A by shading each anticorrelated residue pair. Shading is proportional to the magnitude of the anticorrelation and darker shaded regions correspond to groups or domains of anticorrelated residues. The white regions in Fig. 3A correspond to protomer pairs with positive correlations. See Fig. 3E for the numbering and the pairwise motions of the protomers for each shaded region. The tetramer consists of protomer 1 (residues 1–498), protomer 2 (499–997), protomer 3 (998–1496), and protomer 4 (1497–1995). The clear block-like pattern in Fig. 3A shows that the protomers are moving in an anticorrelated fashion, which is consistent with a concerted rocking motion (see Fig. 3E).
Figure 3.
Correlated atomic motions in PYK. (A) Negative correlations for the apo tetramer. Anticorrelations of all pairs of residues in the four protomers are plotted: protomer 1 (residues 1–498), protomer 2 (499–997), protomer 3 (998–1496), and protomer 4 (1497–1995). The block-like pattern shows that the protomers are moving in an anticorrelated fashion. The dark regions result from fluctuations of the system with fixed centroid of the tetramer. Fluctuations of a protomer obtained by fixing its centroid shows correlations internal to the protomer, as depicted below in (B)–(D). (B) Negatively correlated regions of the apo protomer. Strongest anticorrelations are between the B-domain (residues (89–186)) and the N-terminal (1–89) and the C-domains (340–498). (C) Differences in negative correlations of apo and holo protomer expressed as Dark regions show the decrease of negative correlations upon binding of the effector. This is consistent with effector binding dampening the motion of the B-domain. (D) Differences in negative correlations of the isolated holo and apo monomer expressed as . Dark regions show that negative fluctuations of the apo structure are much stronger than those of the holo structure. These responses are much less, however, than those observed in the protomer. (E) Cartoon of the anticorrelated rocking motions of the four PYK protomers (with residue numbering defined as in A).
A more detailed analysis of the matrix showing anticorrelations in a single protomer within the tetramer is presented in Fig. 3B, where the dark regions show the negatively correlated regions. The fluctuations of the centroid of the protomer have been removed from the trajectory of the monomer, thus the calculated fluctuations are internal to the monomer. Strongest anticorrelations are between the B-domain (residues 89–186) and the N-terminal (1–89) and the C-domains (340–498). The change of correlations upon binding of the effector is of interest. In Fig. 3C, the shaded regions identify those residues of the protomer where anticorrelations are either weakened or became positive upon binding of the effector. The weakened anticorrelated movements between the B-domain and the N-terminal residues and between the B- and C-domain residues are consistent with the interpretation from the temperature factor analysis showing that effector binding dampens the motion of the B-domain. As an example, the negative correlation between residues Asn-17 in the N-terminal and Lys-118 in the B-domain is −15.0 Å2 in the apo structure and reduces to –4.4 Å2 upon effector binding. As another example, the negative correlation of −15.4 Å2 between Glu-156 in the B-domain and Asn-340 in the C-domain of the apo structure reduces to −8.8 Å2 upon effector binding. Because negative correlations result from the opening and closing of the B-domain cap, their decrease suggests that binding of the effector reduces the amplitude of the B-domain oscillations.
To study the effect of neighboring protomers on the dynamics of the tetramer, simulations of the isolated holo and apo monomers in solution were also analyzed. The differences of negative correlations between the isolated monomer holo and isolated monomer apo LmPYK chains are shown in Fig. 3D. The shaded regions show that the anticorrelations of the apo structure are stronger than those of holo showing that binding of the effector decreases the negative correlations of the cap region and the effector binding region. This is a similar effect as shown from the tetramer simulation analysis (Fig. 3B) but now with a significantly reduced magnitude. We conclude that allosteric activity resulting from the anticorrelated behavior of the B-domain cap and the effector binding regions is already built into the isolated monomer, but is amplified by tetramer formation.
Pairwise distance distributions identify differences in behavior between the Apo and Holo structures
MD trajectories of the apo and the holo structures give information on how much time the system spends in a given conformational state (13). Structural and dynamic differences in the apo and holo structures are analyzed here using a plot of pairwise distance distributions. To track the interaction between a pair of residues over the course of an MD simulation the interatom distance is calculated at each time point and the frequency distribution of the distance is plotted over the time course of the simulation. In many cases, as expected, the interresidue distances show a normal distribution though frequently bimodal distributions or skewed distributions are observed. This graphical summary provides a convenient way of comparing the behavior of holo and apo forms of LmPYK.
Activator binding changes residue fluctuations around the effector binding region
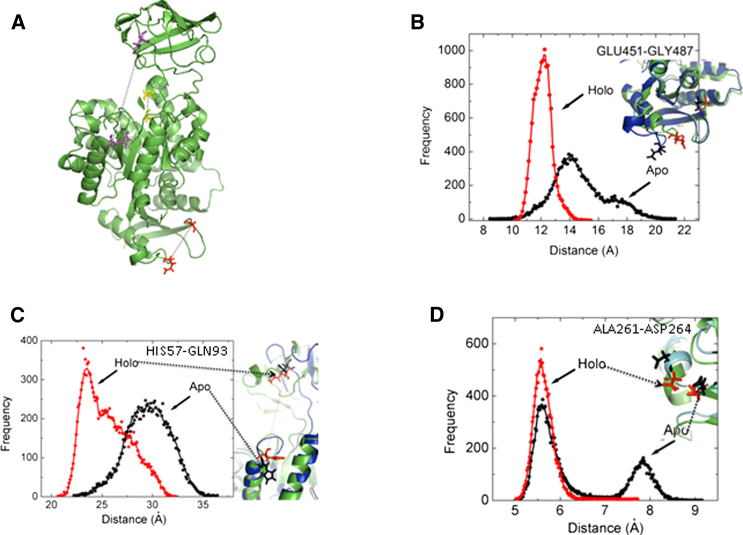
The ligand F26BP sits between two turn regions and interacts with Glu-451 and Gly-487. The distance between these two residues with F26BP bound is 12.0 Å as measured in the crystal structure. The MD simulation with F26BP removed (apo) shows a relaxation of the loop as shown by the increased inter-Cα distance between these two residues compared with the holo structure (Fig. 4, A and B), Bound F26BP in the holo structure keeps the two residues close to the experimental x-ray distance with a mean distance of 12.5 Å, whereas in the more flexible apo structure there is a broader almost bimodal distribution with preferred distances of 14.2Å and 17.5 Å (Fig. 4B).
Figure 4.
Selected pairwise distance distributions within a PYK protomer showing conformational changes induced by FBP binding. (A) Cartoon of a PYK protomer highlighting the residue pairs. 451–487; red, 261–264; yellow, 57–93; purple. (B) Distribution of the distance between GLU-451 and GLY-487 for the apo (black circles) and the holo (red circles) obtained from the MD trajectory. Inset shows conformations of the apo and holo tetramer. Apo is shown as a blue cartoon and the two residues are in stick representation in black. Holo is shown as a green cartoon with residues shown as red sticks. (C) Distribution of the distance between His-57 and Gln-93 for apo (black dots) and holo (red dots) obtained from the MD trajectory. Inset shows conformations of the apo and holo tetramer. Apo is shown as a blue cartoon and the two residues are in stick representation in black. Holo is shown as a green cartoon with residues shown as red sticks. (D) Distribution of the distance between Asp-261 and Ala-264 for apo (black dots) and holo (red dots) obtained from the MD trajectory. Inset shows conformations of the apo and holo tetramer. Apo is shown as a blue cartoon and the two residues are in stick representation in black. Holo is shown as a green cartoon with residues shown as red sticks. To see this figure in color, go online.
B-domain motion closes the active site
The largest movements in the enzyme activity of PYK is the opening and closing of the B-domain. This has been monitored by the interresidue distance changes between residues of A (His-57; red) and B (Gln-93; green) domains for each protomer in the tetramer (Fig. 4C). The mean distance in the apo tetramer is 30Å consistent with an open conformation of the B-domain (Fig. 4C). However, for the holo tetramer we see a bimodal distribution with peaks at 23Å and 28 Å, which suggests that the bound F26BP is affecting the motion and open or closed state of the B-domain.
Monitoring distortions in the active site α6 helix
Asp-261 and Ala-264 are one helical turn apart in the short α6 helix, which provides a crucial geometrical template for binding the substrate. The switch from the T to R states in LmPYK involves the rigid body rotation of the AC core (1), which enables Arg-310 on one protomer to form specific hydrogen bonds to the backbone carbonyls of arginine and glycine residues located in the α6′ helix of the adjacent protomer. The short α6′ helix (260-VARGDLGVEIP-270) is unusual in that it contains two glycines. The MD simulation supports the idea that the allosteric mechanism may involve a transition between an ordered (R-state) helix able to bind substrate and a disordered (T-state) conformer. We observe a single, well-defined peak for the holo tetramer, which corresponds with the ordered (R-state) helix. The apo tetramer however shows a bimodal distribution for the apo tetramer with peaks at 5.5 Å and 7.8Å (Fig. 4D). These two peaks in apo tetramer suggest that the helix unwinds in the absence of F26BP.
Conclusions
The MD simulations of tetrameric LmPYK in the apo and holo (F26BP-bound) states provide insight into the allosteric effector mechanism triggered by F26BP. By studying the dynamic behavior of the tetramer compared with isolated monomer chains the simulations also show how the major allosteric effect is linked to a concerted motion of the complete tetramer. The analysis of correlated motion is consistent with a synchronized rocking of the protomers. The differences in the magnitudes of the B-factors on effector binding suggests that the F26BP binding dampens molecular motion throughout the tetramer and effectively cools down the tetramer and traps the tetramer in the active R-state conformation. This hypothesis is also consistent with the different correlation plots, which show that the magnitude of the anticorrelated movements, particularly involving the B-domain, are reduced on F26BP binding. Interestingly, the behavior of an isolated monomer PYK chain shows almost no effect on the overall B-factor upon F26BP binding, suggesting that the coupled movement of the protomers is required for this damping effect.
These observations largely fit with the classical Monod-Wyman-Changeux model of allostery, which suggests that oligomeric enzymes undergo symmetrical transitions (classically between the T- and R-states) that can be stabilized by ligand binding (14). Our observations showing a lowering of B-factors on effector binding might further suggest the effector is not only locking the R-state using local van der Waals and hydrogen bond interactions (as suggested previously (1)), but is also acting as a general heat-sink to cool down the whole tetramer. More recent descriptions of allostery highlight the role of entropy changes on effector binding (15,16) as well as the importance of the intrinsic dynamic nature of the protein (16,17). Our observations on the effect of F26BP binding on ligand flexibility suggest that protein rigidity (related to melting temperature) and intrinsic heat capacity are important factors in stabilizing the R-state of PYK, and other allosteric proteins.
Another insight about the flexibility of PYK comes from a comparison of tetramer with the isolated monomer, which shows that the B-domain in the tetramer is more mobile than in the isolated monomer. Interestingly, the B-domain movements in the tetramer have been frequently found to be asymmetric in a large number of PYK x-ray structures (e.g., Protein Data Bank codes 1PKY, 1PKN, 1AQF, and 1F3W) (18). The asymmetry in these structures is such that B-domains of protomer pair 1 and 4 have similar B-factors and relative orientations, which are frequently different from those of protomers 2 and 3 (protomer numbering as defined in Fig. 3E). The asymmetry is sometimes imposed by the tetramer lying on a crystallographic twofold axis, but there are also many examples showing the same behavior when there is no crystallographic constraint. The enhanced B-domain movement in the tetramer compared with the isolated monomer again suggests that interprotomer interactions regulate the pairwise B-domain movements. The strong anticorrelation signals within each protomer (Fig. 3) highlights the flapping of the B-domains though it is not clear mechanistically how the binding of the effector dampens the motion or how the synchronous movement of the B-domain pairs is regulated.
The analysis of pairwise distance distributions is consistent with the results from the analysis of correlated motions in the tetramer and shows that F26BP binding not only restricts the mobility of the B-domain (as shown in Fig. 4C) but also keeps the active site helix tight and the enzyme in an active R-state conformation (Fig. 4D). This proposed helical order-disorder transition between the T- and R-states of LmPYK is key to explaining the on-off state of the enzyme as the phosphoenolpyruvate substrate can only bind in the active site when the short α6′-helix (260-VARGDLGVEIP-270) adopts an ordered conformation. These MD simulations appear to show this transition for the unliganded apo form within the 80 ns simulation indicates the relative instability of this glycine-rich helix. It is also significant in the simulations that the holo state is in the ordered (active R) state over the complete simulation while the apo (unliganded) can sample both disordered and ordered conformations (Fig. 1D). These results suggest that the apo-enzyme can sample a range of conformational states, some of which would be similar to the active R-state. This dynamic description of PYK fits with the ensemble representation and population shift ideas of allosteric activation (16,19).
The MD analysis presented in this work therefore supports a model in which concerted domain movements, as described in the MYC model, dominate the allosteric mechanism of LmPYK. However, the analysis of specific order-disorder transitions and the heat-sink effect of the bound effector highlight the importance of entropic and vibrational movements in regulating the allosteric effect of effector binding on the enzyme activity of Leishmania PYK.
Author Contributions
A.N., P.T., and B.E. performed computational studies and analyzed data. B.E. and M.D.W. designed the research, analyzed data, and wrote the article.
Acknowledgments
We thank the British Council for grants to B.E. and M.W. The computational work was supported by the Edinburgh Parallel Processing Facility and the Centre for Translational and Chemical Biology. A.N. was supported by the Darwin Trust.
Editor: Amedeo Caflisch.
Footnotes
Supporting Results and Discussion, five figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00606-2.
Contributor Information
Burak Erman, Email: berman@ku.edu.tr.
Malcolm D. Walkinshaw, Email: m.walkinshaw@ed.ac.uk.
Supporting Material
References
- 1.Morgan H.P., McNae I.W., Walkinshaw M.D. Allosteric mechanism of pyruvate kinase from Leishmania mexicana uses a rock and lock model. J. Biol. Chem. 2010;285:12892–12898. doi: 10.1074/jbc.M109.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan H.P., O’Reilly F.J., Walkinshaw M.D. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proc. Natl. Acad. Sci. 2013;110:5881–5886. doi: 10.1073/pnas.1217157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daily M.D., Gray J.J. Local motions in a benchmark of allosteric proteins. Proteins. 2007;67:385–399. doi: 10.1002/prot.21300. [DOI] [PubMed] [Google Scholar]
- 4.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 5.Lindorff-Larsen K., Piana S., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 7.van der Spoel D., E. Lindahl, B. Hess, and the GROMACS development team. 2014. GROMACS User Manual version 4.6.6.
- 8.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 9.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 10.Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res. Notes. 2012;5:367–374. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano W. DeLano Scientific; San Carlos, CA: 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- 12.Edinburgh Parallel Computing Centre. 2007–2014. HECToR: UK National Supercomputing Service.
- 13.Wieligmann K., Pineda De Castro L.F., Zacharias M. Molecular dynamics simulations on the free and complexed N-terminal SH2 domain of SHP-2. In Silico Biol. (Gedrukt) 2002;2:305–311. [PubMed] [Google Scholar]
- 14.Monod J., Wyman J., Changeux J.-P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 15.Cooper A., Dryden D.T. Allostery without conformational change. A plausible model. Eur. Biophys. J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 16.Cui Q., Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motlagh H.N., Wrabl J.O., Hilser V.J. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wooll J.O., Friesen R.H.E., Czerwinski E.W. Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase. J. Mol. Biol. 2001;312:525–540. doi: 10.1006/jmbi.2001.4978. [DOI] [PubMed] [Google Scholar]
- 19.Chung-Jung T., Nussinov R. A unified view of “how allostery works”. PLOS Comput. Biol. 2014;10:e1003394. doi: 10.1371/journal.pcbi.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.