Abstract
The glucocorticoid receptor (GR) is a steroid-hormone-activated transcription factor that modulates gene expression. Transcriptional regulation by the GR requires dynamic receptor binding to specific target sites located across the genome. This binding remodels the chromatin structure to allow interaction with other transcription factors. Thus, chromatin remodeling is an essential component of GR-mediated transcriptional regulation, and understanding the interactions between these molecules at the structural level provides insights into the mechanisms of how GR and chromatin remodeling cooperate to regulate gene expression. This study suggests models for the assembly of the SWI/SNF-A (SWItch/Sucrose-NonFermentable) complex and its interaction with the GR. We used the PRISM algorithm (PRotein Interactions by Structural Matching) to predict the three-dimensional complex structures of the target proteins. The structural models indicate that BAF57 and/or BAF250 mediate the interaction between the GR and the SWI/SNF-A complex, corroborating experimental data. They further suggest that a BAF60a/BAF155 and/or BAF60a/BAF170 interaction is critical for association between the core and variant subunits. Further, we model the interaction between GR and CCAAT-enhancer-binding proteins (C/EBPs), since the GR can regulate gene expression indirectly by interacting with other transcription factors like C/EBPs. We observe that GR can bind to bZip domains of the C/EBPα homodimer as both a monomer and dimer of the DNA-binding domain. In silico mutagenesis of the predicted interface residues confirm the importance of these residues in binding. In vivo analysis of the computationally suggested mutations reveals that double mutations of the leucine residues (L317D+L335D) may disrupt the interaction between GR and C/EBPα. Determination of the complex structures of the GR is of fundamental relevance to understanding its interactions and functions, since the function of a protein or a complex is dictated by its structure. In addition, it may help us estimate the effects of mutations on GR interactions and signaling.
Introduction
The glucocorticoid receptor (GR) is a member of the nuclear receptor superfamily of transcription factors, which play critical roles in a variety of biological processes, including homeostasis and metabolism. It consists of a hypervariable N-terminal domain, a DNA-binding domain (DBD), a hinge region, and a ligand-binding domain (LBD) (1). GR carries two transcriptional activation functions (AFs), one in the N-terminal domain and the other within the LBD, for the transactivation of gene expression (2). Upon ligand binding, GR translocates from the cytoplasm to the nucleus, where it homodimerizes and produces transcriptional activation or repression, often by direct high-affinity binding to glucocorticoid-responsive elements (GREs) in DNA. However, not all glucocorticoid-modulated genes contain a DNA-binding site for GR. Thus, GR interacts with other transcription factors such as CCAAT-enhancer-binding proteins (C/EBPs), NF-κB, and AP-1 to alter their function and mediate the responses to glucocorticoids (3).
Eukaryotic genes are highly organized into a multilayered chromatin architecture that has a repressive effect on gene transcription by inhibiting access of regulatory factors to recognition sequences within target promoters (4–6). Thus, modulation of chromatin structure by ATP-dependent chromatin-remodeling complexes plays a fundamental role in gene expression. GR makes direct interactions with several subunits of chromatin-remodeling complexes upon conformational change induced by hormone binding (7). Chromatin-remodeling complexes manipulate the nucleosome architecture to allow binding of transcription factors to chromatin (8–10). Cells contain multiple distinct chromatin-remodeling complexes, including SWI/SNF (SWItch/Sucrose-NonFermentable) (11). All SWI/SNF complexes include an SWI2/SNF2-type ATPase, which uses the energy from ATP hydrolysis to alter the structures of nucleosomes (5,12). The SWI/SNF family of chromatin remodelers has been very well studied, especially in the prototype yeast model (13–15). Unlike the yeast SWI/SNF complex, mammalian SWI/SNF is composed of a heterogeneous mixture of subunits that contain BRG1 or BRM ATPase in addition to BRG1-associated factors (BAFs). Human SWI/SNF contains 10–12 BAFs. Although some BAF proteins (BAF155, BAF57, BAF47, and BAF53) are present in all SWI/SNF complexes, others are interchangeable. Human SWI/SNF can be further grouped into two subfamilies, BAF (BRG1- or HRBM-associated factor, also called SWI/SNF-A) and PBAF (polybromo-associated BAF, also called SWI/SNF-B). Though members of these subfamilies include common subunits, their subunit composition is not entirely the same: BAF250 is only associated with the BAF complex, whereas BAF200 and BAF180 are exclusively present in the PBAF complex (16–18).
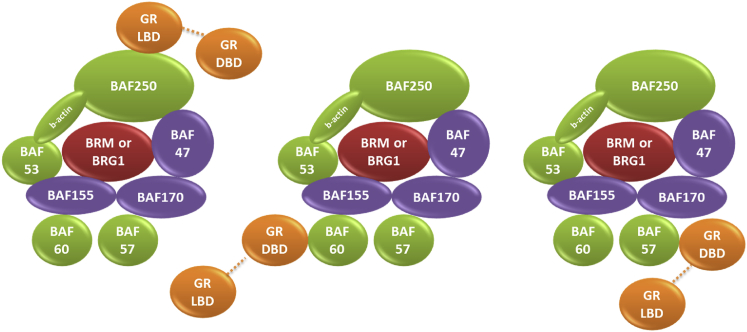
The SWI/SNF chromatin remodeling complex has been implicated in glucocorticoid-stimulated transcription by interacting with GR (19). Binding of the receptor to GREs leads to recruitment of the BRG1-associated remodeling complex and results in changes in gene expression. Multiple interactions between GR and BAF subunits are involved in recruitment and stabilization of the SWI/SNF remodeling complex at target promoters. In vitro assays have previously demonstrated that GR lacking the LBD can interact with BAF60a and BAF57, but not with the core components BRG1, BAF170, or BAF155 (20). Nie et al. (17) showed that BAF250 enhances GR-mediated transcriptional activation by about sixfold. Pull-down experiments suggest a direct interaction between GR and the BAF250 C-terminal region. These results suggest that BAF250 and/or BAF60a of SWI/SNF mediate the interaction between GR and BRG1 (20). By utilizing protein-protein interactions to recruit remodelers to binding sites, agonist-activated GR produces chromatin remodeling, allowing neighboring transcription factor binding sites to be accessed by their relevant proteins in addition to GR (21).
CCAAT-enhancer-binding proteins α and β (C/EBPα and C/EBPβ), belonging to the C/EBP family of transcription factors, have also been shown to recruit the SWI/SNF complex to target promoters. C/EBP proteins regulate the differentiation of several cell types, such as adipocytes, neutrophil granulocytes, eosinophils, and hepatocytes (22–24). They are composed of six members (C/EBPα–C/EBPζ). These proteins share ∼90% sequence identity in the C-terminal 55–65 amino acid residues, which contains the basic-leucine zipper (bZIP) domain. This domain is responsible for dimerization and DNA binding (25). In contrast to the highly conserved C-termini, the N-terminal domains of C/EBP proteins are widely divergent, except for three short subregions. These subregions represent the activation domains involved in transcriptional activation (TADs) (26), and involvement in chromatin remodeling has been demonstrated. For example, biochemical and immunoaffinity experiments showed that the C/EBPβ TAD interacts directly with the hBrm/BRG1 central ATPase (27), and the C/EBPα transactivation element III (TE-III) has been shown to interact with Brm ATPase and the BAF155 core SWI/SNF subunit (28).
Functional genomics experiments in intact liver tissue and differentiating 3T3-L1 preadipocytes have demonstrated cooperative interplay between GR and C/EBPβ at the level of chromatin accessibility (29–31). Depending on the genetic composition and chromatin configuration of the binding site, GR can assist loading of C/EBPβ or C/EBPβ can assist loading of GR (29). Yet direct protein-protein interaction (known as tethering) can also be utilized to deliver both factors to the genome even when a DNA binding site is only present for one (32,33). Thus, GR and C/EBP access to chromatin is regulated by a) the ability to recruit chromatin remodelers, leading to modulation of the local chromatin environment, and/or b) direct interaction between the transcription factors.
Here, we suggest models of how SWI/SNF-A subunits interact structurally with one another and with GR to induce chromatin remodeling and facilitate GR-mediated transcription, respectively. We used the PRISM (34,35) algorithm (PRotein Interactions by Structural Matching) to predict the binary interactions between these molecules. There are different computational methods for the prediction of protein and domain interactions (36). One approach involves using the sequence data of the query proteins and assessing the likelihood of the interaction based on the identification of structural features by sequence similarity (37). More than half of the methods, however, incorporate evolutionary information (38,39) and/or protein structure to improve the accuracy of the predictions (40). PRISM is a powerful template-based protein-protein complex-structure-prediction algorithm that utilizes the structural similarity between template interfaces and target surfaces. In a recent study, Vreven et al. (41) compared the performance of template-based (including PRISM) and template-free (ZDOCK) protein-protein interaction-prediction methods. According to their results, the template-based methods show similar performance to ZDOCK and in fact are more successful in cases where the proteins undergo conformational changes upon binding.
Exploiting PRISM, we built models for the core and variant BAF units based on the predicted binary interactions and determined the complex structures of the subunits capable of interacting directly with the GR. In addition, we model the interaction between GR and C/EBP to describe how these transcription factors can facilitate each other’s recruitment to DNA through tethering. We tested the GR-C/EBP models by in silico mutation of the predicted interface residues, observing an elimination of the predicted GR-C/EBP interaction and supporting the significance of these residues for the complex formation. Identical interface-residue mutations were made in vivo with a view to further validating the structural predictions. The proposed models for GR-SWI/SNF and GR-C/EBP interactions conceptually illustrate two of the many arrangements by which GR may modify transcriptional responses in association with other proteins.
Materials and Methods
Computational prediction of the structures of GR with SWI/SNF components and C/EBP
In this study, we used PRISM (34,35) to model the interactions between GR and the SWI/SNF chromatin-remodeling complex and between GR and C/EBPs. PRISM predicts the structures of protein complexes by utilizing the structural similarity between template experimental interfaces and target surfaces, and it generates a couple of putative models for the query interaction using these template interfaces. In the last step, rigid docking solutions are flexibly refined to resolve steric clashes and ranked according to their global energy. The one with the lowest binding energy score (BES) is considered to be the best solution. In this way, geometric complementarity is incorporated into the docking procedure. When PRISM yields more than one putative model, we first search the literature to see whether there are any experimental findings that corroborate our models; if not, we choose the one with the lowest BES.
Structural data for GR and C/EBP proteins were obtained from the Protein Data Bank (PDB). We used both the DBD and the LBD of GR as our targets. We obtained model structures of SWI/SNF subunits using I-TASSER (42), since there were no available structures in the PDB. We chose the models that have a percentage sequence identity of the templates in the aligned region with the query sequence >30%. The sequence alignments and the threading results of the modeled structures are shown in Fig. S1 and Table S1, respectively, in the Supporting Material. Table 1 lists the proteins used in the predictions. Predictions were made using the BRG1 complex and core BAF units (BAF155, BAF47, and BAF170) and BAF250, BAF57, and BAF60a in addition to the BRG1 ATPase as our target proteins. We predicted the complex structure of GR with C/EBP using the DBD and bZip domain, respectively, then identified the contact regions using HotRegion (43). HotRegion is a database of predicted hot-spot clusters. Using predicted hot-spot residues that are major contributors to the binding energy, it identifies the regions that are important for the stability of protein complexes. We built putative models for the multiprotein complexes based on predicted binary interactions and available literature data. We performed a detailed analysis of interfaces to investigate the stability of the predicted complexes using the tool Protein Interfaces, Surfaces, and Assemblies (PDBePISA) (44). We used the default parameters for calculations. PDBePISA provides information about the total solvent-accessible surface area, and about the interface area, which is the difference between the total accessible surface areas of monomeric and complex structures divided by two. It also calculates the number of potential hydrogen bonds and salt bridges across the interface. Finally, we used EPPIC Server (45) and NOXclass (46) to distinguish biologically relevant interactions from nonbiological interactions resulting from crystal-packing contacts. EPPIC uses the number of core residues and two evolutionary indicators based on the sequence entropy of homolog sequences to determine the biological character of interfaces. It uses the following cutoff values: six core residues for geometry, 0.75 for the entropy core/rim ratio, and −1.0 for core-versus-surface scores. NOXclass utilizes interface properties such as interface area, interface area ratio, and area-based amino acid composition of the protein-protein interface to determine whether an interaction is biological or crystal packing. It uses the following cut-off values: 650 A2 for the interface area and 0.07 for the interface-area ratio.
Table 1.
List of proteins used in PRISM predictions
| PDB Structures |
Model Structures |
|||
|---|---|---|---|---|
| Protein | Domain/Region | PDB ID | Protein | Domain/Region |
| GR | DBD | 1GLU, 3G9M | BAF155 | SWIRM and coiled-coil |
| GR | LBD | 1P93, 3E7C | BAF170 | SWIRM and coiled-coil |
| C/EBPα | bZip | 1NWQ | BAF60a | C-terminal (residues between 140 and 435) |
| BRG1 | bromodomain | 2GRC | BAF250 | C-terminal (residues between 1640 and 2250) |
| BAF47 | full length | |||
| BAF57 | full length | |||
In silico construction of C/EBP mutants
To confirm the residues predicted as structurally important for complex formation between GR and C/EBPs, Leu residues at positions 315, 317, 335, and 341 were mutated in silico. We first mutated these residues to Asp and performed energy minimization using the FoldX computational algorithm (47,48). Then, we used PRISM to predict the effects of mutations on GR-C/EBP interactions.
Cell culture and reagents
Dexamethasone (Dex) was purchased from Sigma-Aldrich (St. Louis, MO). Cells from the 3617 mouse mammary adenocarcinoma cell line were routinely cultured in Dulbecco’s modified Eagle’s medium high glucose supplemented with 10% fetal bovine serum (Hyclone, South Logan, UT) and 1% L-glutamine (Life Technologies, Grand Island, NY). The 3617 cell line contains a large tandem array (∼200 copies) of a mouse mammary tumor virus, Harvey viral ras (MMTV-v-Ha-ras) reporter integrated into chromosome 4 (49). At all times, 3617 cells were grown in the presence of 5 μg/mL tetracycline (Sigma-Aldrich) to prevent expression of a stably integrated GFP-GR (49,50). Before glucocorticoid treatments, cells were seeded to two-well Lab-Tek chamber slides (Thermo Fisher, Waltham, MA) and incubated for at least 18 h in Dulbecco’s modified Eagle’s medium containing 10% charcoal-stripped fetal bovine serum (Hyclone) and 1% L-glutamine.
Plasmids and transient transfection
mCherryFP-GR and mCherry-NF-1 were described previously (50,51). mCherryFP-GR contained rat GR with a C656G change separated from mCherry fluorescent protein by the linker 5′-YKSGLRSRGAGAGAGAGA-3′. pEGFP-C/EBPα (a kind gift from Fred Schaufele, University of California San Francisco, San Francisco, CA) contained rat C/EBPα tagged on the N-terminal with a L65F and L232H variant of the enhanced green-fluorescent protein (EGFP). A 16-amino-acid linker, 5′-QASTMDYKDDDDKDYA-3′, bridges EGFP and C/EBP. C/EBP mutations were generated using a QuikChange II XL site-directed mutagenesis kit according to the manufacturer’s instructions (Stratagene, La Jolla, CA). Mutagenesis primers were designed through http://www.genomics.agilent.com/primerDesignProgram.jsp, and new plasmids were sequenced across the entire protein coding region to verify the desired mutations. Transient transfection of 3617 cells was performed with jetPRIME reagent (PolyPlus, New York, NY) according to the manufacturer’s instructions.
Subcellular localization and N&B analysis
The 3617 cells were transiently transfected with 1 μg of pEGFP-CEBPα or its mutant variants, combined with 1 μg mCherryGR or mCherry-NF-1 as indicated. Cells were incubated for at least 20 min with 100 nM Dex and imaged at the CCR Confocal Microscopy Core Facility (NIH, Bethesda, MD) on a LSM 780 laser scanning microscope (Carl Zeiss, Inc., Thornwood, NY) fitted with a 63X oil immersion objective (NA=1.4). The excitation source was a multi-line Ar laser tuned at 488 nm and/or a 594 nm laser. Fluorescence was detected with a GaAsP detector in photon-counting mode. Number and brightness (N&B) measurements were performed as previously described (52).
Results
Prediction of GR complex structures with SWI/SNF components
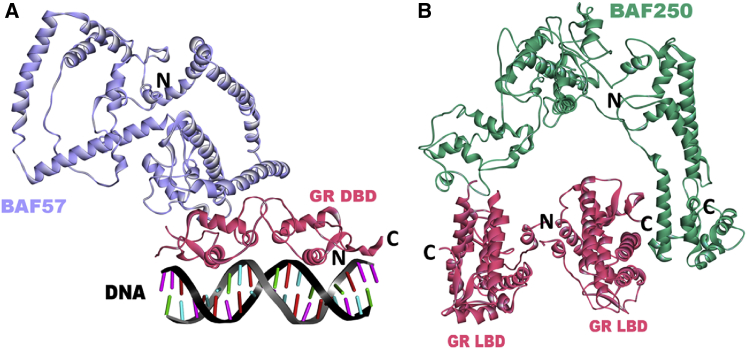
GR has been shown experimentally to interact with BAF60a and BAF57 through its DBD domain and with BAF250 through its LBD domain, suggesting that in vivo, GR has at least three BAF proteins with which to associate (Fig. 1) (17,20). Here, we focused on these interactions and modeled the complex structures using PRISM. Our results confirmed that GR DBD can bind with high affinity to BAF57 (Fig. 2A). The BES for the prediction is −57.67. On the basis of our previous studies (34,53–55), we set the energy-score cutoff value as −10. Thus, predictions with BES values <−10 are considered to be favorable interactions. Pull-down assays previously demonstrated that GR interacts strongly with the N-terminus of BAF60a, but not with the C-terminus. We generated structural models of the BAF60a N-terminal (residues between 4 and 140) and C-terminal regions (residues between 140 and 435). Unlike the C-terminal region, modeling of the BAF60a N-terminal did not yield good-quality models. The sequence identity between our protein and the template for the model was <30% (Table S2 and Fig. S3), so we did not use it in our predictions. Consequently, we were not able to see whether BAF60a can interact with GR DBD through its N-terminus. Instead, using PRISM, we tested whether BAF60a can interact with GR DBD through its C-terminal region, but we did not observe an interaction between these two proteins, supporting the experimental results. Deletion experiments demonstrated that the C-terminal region of BAF250 can directly bind to GR (17). We generated a model structure of the BAF250 C-terminal region (residues between 1641 and 2250) and predicted putative models for the GR-BAF250 interaction. Our results demonstrated that GR can form stable complexes with BAF250 through its LBD, but not through its DBD (Fig. 2B). The BES for the prediction is −24.01. The predicted interface involves residues between 2149 and 2164, close to the C-terminus of BAF250, corroborating the experimental results. Table 2 illustrates the results of a detailed interface analysis of both predictions (GR DBD/BAF57 and GR LBD/BAF250). We also tested whether these suggested interfaces (Fig. 2) are considered to be biological interactions or crystal-packing contacts. Both interactions are classified as biological according to EPPIC and NOXclass. Interface features calculated by EPPIC and NOXclass are tabulated in Tables S3 and S4 and Fig. S4.
Figure 1.
The interaction of GR with the components of the SWI/SNF-A (BAF) complex. SWI/SNF complexes consist of a single ATPase (BRM or BRG1, red), evolutionarily conserved core proteins (BAF155, BAF170, and BAF47, purple), and variant subunits (BAF60a, b, or c; BAF57; BAF53a or b; BAF250a or b; and β-actin, green). Pull-down experiments indicate that there are at least three proteins in the BAF complex that can associate with GR (orange). The dashed lines represent the hinge region (40 residues long) between the DBD and LBD of GR. BAF250 binds to the GR LBD, and BAF57 and BAF60 bind to the GR DBD. To see this figure in color, go online.
Figure 2.
Predicted complex structures of GR with SWI/SNF components. (A) Predicted complex structure of DNA-bound GR DBD (PDB ID 1glu, pink) with the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (SMARCE1/BAF57) (model, lilac). The BES for the prediction is −57.67 (template interface 3usuGH). The interface residues of GR are M434, K435, P439, L441, V442, C443, S444, R479, N480, I483, I484, D485, R488, R498, K499, Q502, A503, G504, and R510. The interface residues of BAF57 are D184, G185, F186, S187, H190, Q232, V236, H237, L241, E242, L246, E249, E250, H252, Q253, K256, Q295, K298, R299, and E302. N and C represent the N- and C-termini, respectively, of the modeled regions. (B) Predicted complex structure of the GR LBD (PDB ID 1p93, pink) with AT-rich interactive-domain-containing protein 1A (ARID1A/BAF250) (model, green). The BES for the prediction is −24.01 (template interface, 3zrjAB). The interface residues of the GR are D641, H726, E727, E730, N731, L732, N734, Y735, F737, Q738, K771, and L773. The interface residues of BAF250 are S509, R513, D517, N520, P521, V522, C523, and R524. N and C represent the N- and C-termini, respectively, of the modeled regions. To see this figure in color, go online.
Table 2.
Interface analysis of GR DBD/BAF57 and GR LBD/BAF250 complexes
| No. of Residues (Nres) |
Surface Area (Å2) | Interface Area (Å2) | No. of Hydrogen Bonds (NHB) | No. of Salt Bridges (NSB) | ||
|---|---|---|---|---|---|---|
| Interface (%) | Total (%) | |||||
| GR DBD/BAF57 Complex (BES −57.67) | ||||||
| GR DBD | 20 (24.7) | 81 (100) | 5929 | |||
| BAF57 | 21 (5.1) | 411 (100) | 30,384 | |||
| Complex | 1176.3 | 11 | 10 | |||
| GR LBD/BAF250 Complex (BES −24.01) | ||||||
| GR LBD | 12 (2.4) | 492 (100) | 37,299 | |||
| BAF250 | 8 (1.3) | 610 (100) | 24887 | |||
| Complex | 691.4 | 1 | 3 | |||
The number of interface residues for both monomers and complexes are shown together with their percentage.
Prediction of complex structures of core BAF units
SWI/SNF core subunits include BAF155, BAF170, and BAF47 in addition to the BRG1 catalytic ATPase. The structure of this multiprotein complex was constructed by superimposing the predicted binary interactions between BAF155 and BRG1, BRG1 and BAF47, and BAF47 and BAF170 (Fig. 3A).
Figure 3.
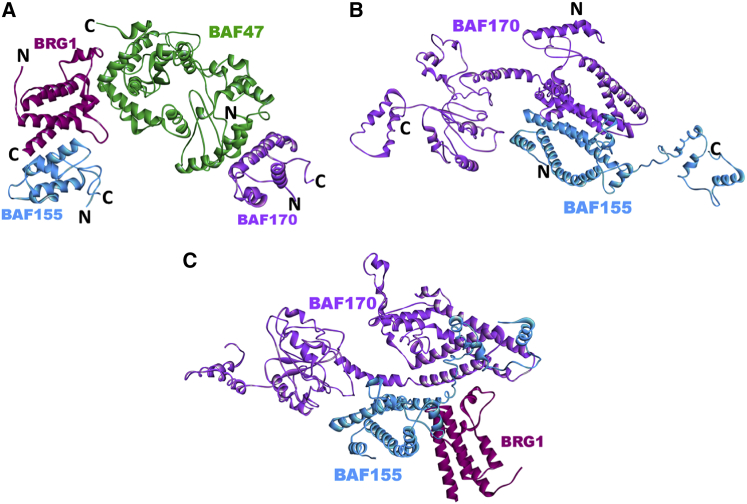
Predicted complex structures of core SWI/SNF components. (A) Core BAF subunits include the SWIRM domain of SWI/SNF complex subunit SMARCC1 (BAF155) (model, light blue); the bromodomain of transcription activator BRG1 (SMARCA4) (PDB ID 2grc, magenta); the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 (SMARCB1/BAF47) (model, green), and the SWIRM domain of SWI/SNF complex subunit SMARCC2 (BAF170) (model, violet). The structure of the core complex is built via superimposition of the predicted binary interactions between BAF155 and BRG1 (BES −13.15; template interface, 3sn6AR), BRG1 and BAF47 (BES −10.81; template interface, 3krtAC), and BAF47 and BAF170 (BES −10.02; template interface, 1q85AB). N and C represent the N- and C-termini, respectively, of the modeled regions. (B) Coiled-coil heterodimer of BAF155 (model, light blue) with BAF170 (model, violet). The BES for the prediction is −28.99 (template interface, 2ieqAC). N and C represent the N- and C-termini, respectively, of the modeled regions. (C) Predicted complex structure of the BAF155/BAF170 heterodimer with BRG1. The BES for the prediction is −12.61 (template interface, 1ylmAB). To see this figure in color, go online.
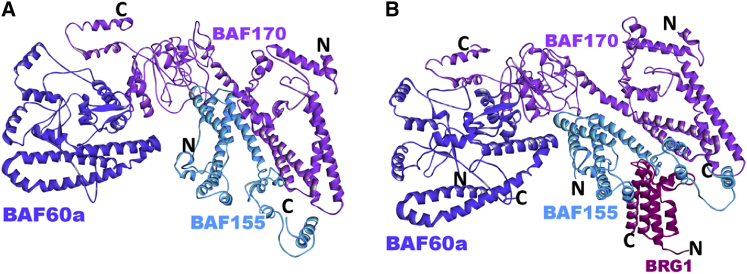
The BAF155 and BAF170 proteins are highly homologous and likely exist either as heterodimers (BAF155/BAF170) or as homodimers (BAF155/155 or BAF170/170) through a leucine zipper motif in the cell (56). Fig. 3 B shows the predicted coiled-coil heterodimer structure (BAF155/BAF170). Our modeling results indicate that the BAF155/BAF170 heterodimer can interact with BRG1 through the BAF155 coiled-coil region (Fig. 3C). However, neither BAF155 nor BAF170 can bind BAF47 through their coiled-coil regions. Thus, the BAF170 SWIRM domain might be involved to mediate the interaction between BAF47 and the other core subunits.
The structural interaction of core BAF units with variant subunits
Pull-down assays with BRG1, BAF170, BAF155, BAF57, and BAF60a performed previously showed direct interactions between the C-terminus of BAF60a and BAF170 and BAF155 and no interactions between BAF60a and BAF57 (20). Our modeling results indicated that variant subunit BAF60a can form stable complexes through its C-terminal domain with the BAF155 or BAF170 SWIRM domain in the core; supporting existing experimental findings (20) (Fig. 4, A and B). The C-terminal domains of BAF60a and BAF57 were not predicted by PRISM to interact. To test whether BAF60a can interact with BAF155 and/or BAF170 in the presence of other core subunits, we superimposed the predicted binary interactions onto our core model (Fig. 4, C and D). No clashes were observed from our protein models. Predictions suggest that BAF60a can also interact with the BAF170 coiled-coil region (Fig. 5A). Using these binary interactions, we constructed the structure of the complex composed of the BAF155/BAF170 heterodimer, BRG1, and BAF60a (Fig. 5B). Taken together, we present two possible ways for the core-variant subunit to interact: BAF155/BAF170 proteins can use their SWIRM domains to interact with both core and variant subunits or mediate the interaction between core and variant subunits using both SWIRM domains and coiled-coil regions.
Figure 4.
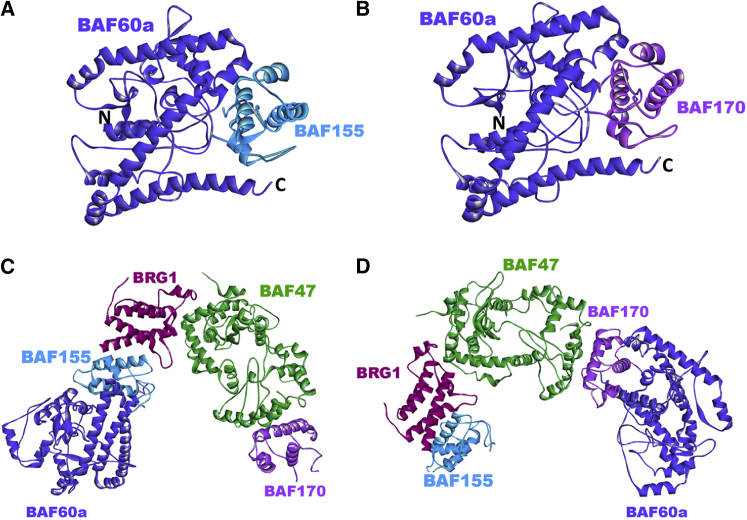
Predicted complex structures of core BAF units with the BAF60a variant subunit. (A) Predicted complex structure of the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SMARCD1/BAF60a) (model, blue) with core subunit BAF155 (light blue) (BES −49.56; template interface, 3ephAB). The modeled structure of BAF60a represents only the region close to the C-terminal domain, so N and C represent the N- and C-termini, respectively, of the BAF60a C-terminus. (B) Predicted complex structure of BAF60a (model, blue) with core subunit BAF170 (BES −45.45; template interface, 3ephAB). (C and D) BAF60a is predicted to join the SWI/SNF core complex through interactions with BAF155 (C) and/or BAF170 (D). To see this figure in color, go online.
Figure 5.
Predicted complex structures of core BAF units with BAF60a variant subunit. (A) Predicted complex structure of BAF60a (model, blue) with the BAF170 coiled-coil region (model, violet) of the BAF155/BAF170 heterodimer (BES −33.31; template interface, 3griAB). N and C represent the N- and C-termini, respectively, of the modeled regions. (B) The structure of the complex is built via superimposition of the predicted binary interactions between the BAF155/BAF170 heterodimer and BRG1, and BAF170 and BAF60a. N and C represent the N- and C-termini, respectively, of the modeled regions. To see this figure in color, go online.
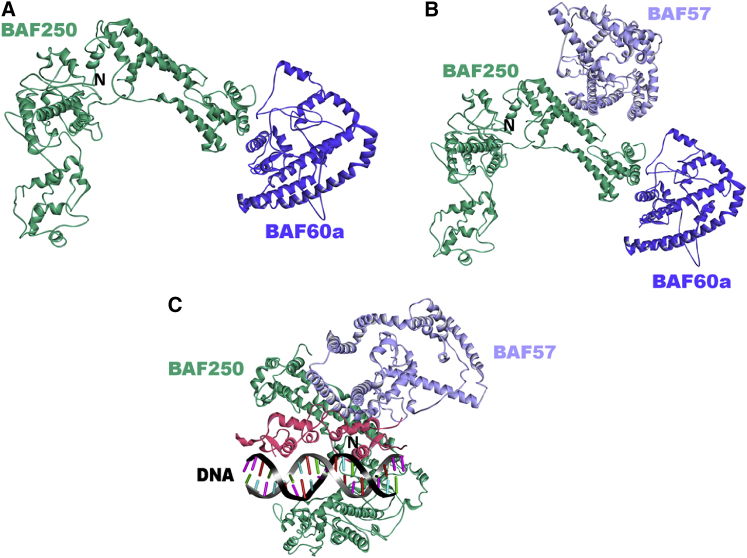
We further predicted the interactions between BAF60a and other variant subunits. Our results indicate that BAF60a can interact with BAF250 (Fig. 6A) through an interface different from the BAF155/BAF170 binding site. Fig. 5B shows the multiprotein complex between BAF250, BAF60a, and BAF57. BAF250 has been shown to interact with the helicase-SANT-associated domain of BRG1 (57). Predictions confirm this interaction and further suggest that BAF250 can bind to both BRG1 and the GR LBD (not shown). Taken together, our results suggest that variant subunits BAF60a and BAF250 may coordinate with the core SWI/SNF complex via multiple core subunits, including BAF155/BAF170, BAF57, and BRG1. To test whether GR, BAF57, and BAF250 can form a ternary complex, we superimposed the BAF57/GR and BAF57/BAF250 interactions (Fig. 6C). The model suggests that the two interactions can co-occur and that BAF57 can interact with GR and BAF250 simultaneously.
Figure 6.
Predicted complex structures between variant SWI/SNF subunits. (A) Predicted binary interaction of BAF250 (model, green) with BAF60a (model, blue). The BES for the prediction is −25.91 (template interface, 3s28BC). (B) Predicted complex structure between BAF250 (green), BAF60a (blue), and BAF57 (lilac). The structure of the complex is built via superimposition of the predicted binary interactions between BAF250 and BAF60a, and BAF57 and BAF250. (C) The structure of the complex is built via superimposition of the predicted binary interactions between GR and BAF57, and BAF57 and BAF250 (BES −27.56; template interface, 1jm0BE). To see this figure in color, go online.
Prediction of GR complex structures with C/EBP
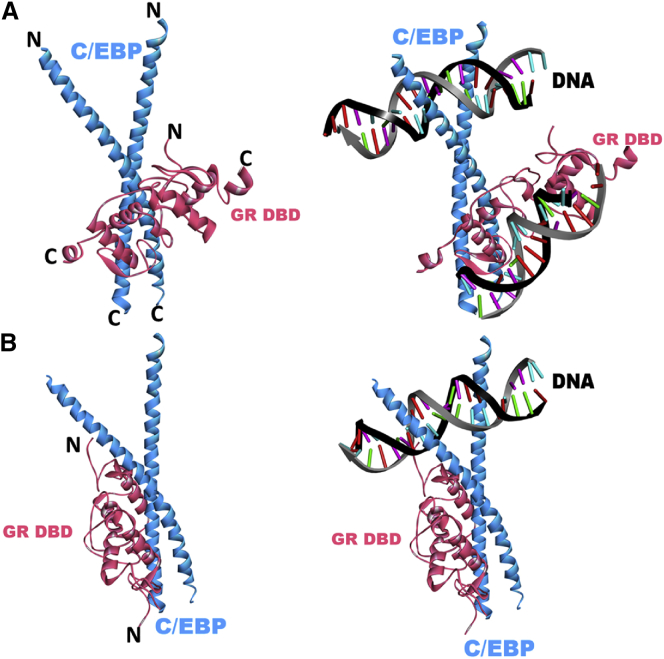
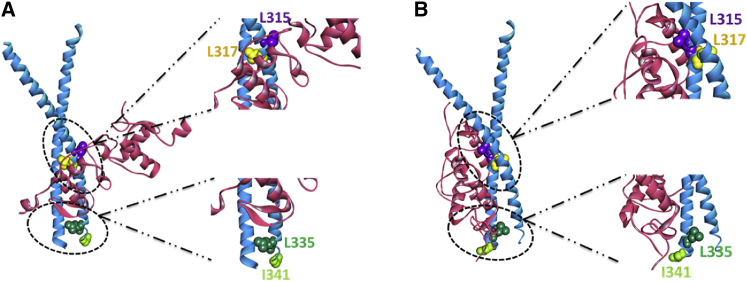
Experimental studies suggest a complex formation between the DBD of GR and the bZip domain of C/EBPα that can bind DNA (32). Using PRISM, we predicted putative models for this interaction. Our models suggest that both monomeric and dimeric GR can bind to a C/EBPα homodimer through the GR DBD domain (Fig. 7). The binding sites are positioned in the region close to the dimerization interface of C/EBPα. Table 3 illustrates the results of a detailed interface analysis of both predictions (GR DBD/CEBP models). To further validate our models, we performed in silico mutations for the predicted interface residues and compared the binding affinities of the wild-type and mutant C/EBPs in terms of BES values. Table 4 summarizes the effects of different mutations on C/EBP dimerization and GR-C/EBP interaction. Although the mutations weakened C/EBP dimerization, they did not completely disrupt it. On the other hand, some C/EBP mutants were predicted in silico to not bind GR. The results indicate that L315D and L317D mutations have more profound effects than L335D, since these mutations disrupted the interaction rather than weakening it as L335D did. As expected, I341D had no effect on either interaction; it did not change the BES significantly, since I341 is not located at the interface (Fig. 8). Overall, the results suggested that these mutations affect GR-C/EBP association, verifying the predicted interface residues for binding.
Figure 7.
Predicted complex structures of GR with C/EBP. (A) First model for the complex structure between the GR DBD (PDB ID 3g9m, pink) and C/EBPα (PDB ID 1nwq, blue). The BES for the prediction is −16.64 (template interface, 3zylAB). The interface residues of GR are H438, M439, L441, V442, C443, S444, A447, L455, R479, N480, D481, D485, R498, K499, Q502, and A503. The interface residues of C/EBP are Q312, L315, E316, L317, S319, D320, N321, R323, L324, K326, R327, V328, Q330, L331, S332, E334, and L335. N and C represent the N- and C-termini, respectively, of the modeled regions. (B) Second model for the complex structure between the GR DBD (PDB ID 1glu, pink) and C/EBPα (PDB ID 1nwq, blue). The BES for the prediction is −27.16 (template interface, 1s7oBC). Although GR is a dimer of DBDs, only one monomer interacts with dimeric C/EBPα. The interface residues of GR are M434, K435, E446, G458, S459, V462, F463, R466, N473, Y474, L475, K490, and P493. The interface residues of C/EBP are A295, V296, K298, S299, R306, Q311, Q312, K313, L315, E316, L317, S319, D322, and K326. N and C represent the N- and C-termini, respectively, of the modeled regions. To see this figure in color, go online.
Table 3.
Interface analysis of GR DBD-C/EBPα complexes
| No. of Residues (Nres) |
Surface Area (Å2) | Interface Area (Å2) | No. of Hydrogen Bonds (NHB) | No. of Salt Bridges (NSB) | ||
|---|---|---|---|---|---|---|
| Interface (%) | Total (%) | |||||
| Model 1 (BES −16.64) | ||||||
| Target 1 (GR DBD) | 16 (10.3) | 156 (100) | 11,006 | |||
| Target 2 (C/EBP) | 17 (14.2) | 120 (100) | 11,774 | |||
| Complex | 1059.6 | 4 | 6 | |||
| Model 2 (BES −27.16) | ||||||
| Target 1 (GR DBD) | 13 (8.1) | 160 (100) | 11,761 | |||
| Target 2 (C/EBP) | 13 (10.8) | 120 (100) | 11,690 | |||
| Complex | 739.9 | 2 | 5 | |||
The number of interface residues for both monomers and complexes are shown together with their percentage.
Table 4.
Effects of mutations on C/EBP dimerization and GR binding
| Target 1 | PDB ID | Target 2 | PDB ID | BES (Wild-Type) | Mutation | BES (Mutant) |
|---|---|---|---|---|---|---|
| C/EBP | 1NWQ | C/EBP | 1NWQ | −88.91 | L315D | −56.86 |
| L317D | −43.78 | |||||
| L335D | −83.29 | |||||
| I341D | −88.39 | |||||
| L317D+L335D | −42.72 | |||||
| GR | 3G9M | C/EBP | 1NWQ | −16.64 (model 1) | L315D | 10.55 |
| L317D | 21.45 | |||||
| L335D | −11.4 | |||||
| I341D | −16.24 | |||||
| L317D+L335D | 41.24 | |||||
| GR | 1GLU | C/EBP | 1NWQ | −27.16 (model 2) | L315D | 25.4 |
| L317D | 59.73 | |||||
| L335D | −23.58 | |||||
| I341D | −27.1 | |||||
| L317D+L335D | 72.40 |
Figure 8.
Residues that are mutated in silico. Residues that are located at (L315 (purple) and L317 (yellow)) and away from the interface (L335 (green) and I341 (light green)) are mutated in models 1 (A) and 2 (B). To see this figure in color, go online.
In vivo testing of GR-C/EBP interactions
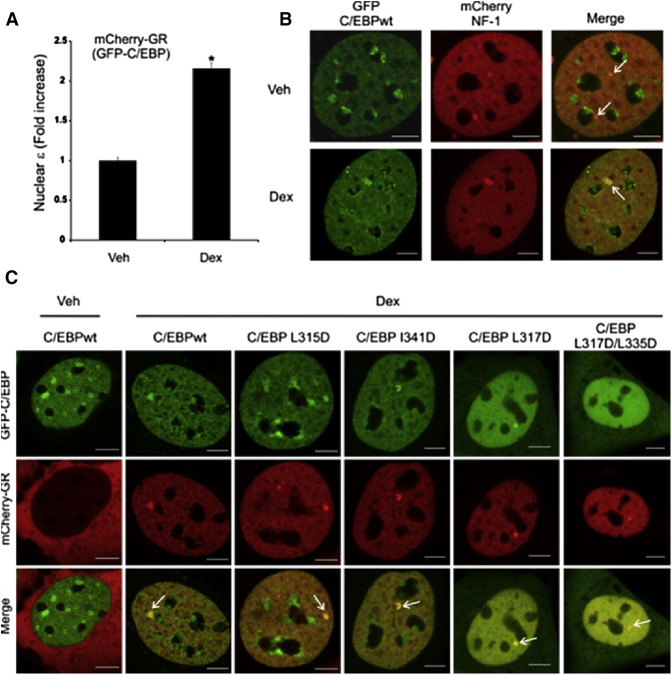
In silico analysis predicted that C/EBPα could interact with either the monomeric or the dimeric form of the GR (Fig. 7). To test which type of complexes form in vivo, we performed N&B analysis in living cells expressing fluorescently tagged GR and C/EBPα. This technique, based on moment analysis, provides the average number of moving fluorescent molecules and their brightness at every pixel in the images. With this information, the relative oligomerization state of a protein can be determined (58). Using this approach, it was shown previously that activated GR is mostly dimeric inside the nucleus (52,59). Thus, if C/EBP were to interact with GR as a monomer, we would expect a decrease in the dimeric population of GR when C/EBPα is coexpressed. On the other hand, if the GR-C/EBP complex contains GR dimers, then the GR population will remain dimeric independently of the presence of C/EBP. N&B analysis (Fig. 9 A) shows that GR nuclear brightness in the presence of C/EBP remains consistent with a fully dimeric receptor, suggesting that GR interacts with C/EBP as a dimer.
Figure 9.
In vivo analysis of GR-C/EBP interactions. (A) The 3617 cells (transfected with pEGFP-C/EBPα and pmCherry-GR) were incubated with ethanol vehicle (Veh) or 100 nM dexamethasone (Dex). For each cell (n = 67), the brightness (ε) was calculated as previously described (52). The figure shows the fold increase of GR nuclear brightness ε relative to the control (monomeric GR). A relative brightness of ∼2 indicates that most of the molecules are in dimeric form. ∗Mean ± SE, p < 0.05 (Student’s t-test). (B and C) Representative confocal images of 3617 cells transiently expressing both GFP-C/EBP and mCherry-NF-1 (B) or both mCherry-GR and GFP-C/EBP (wild-type (wt) or the indicated mutations) (C). Scale bar, 5 μm. To see this figure in color, go online.
Our in silico studies further predict that specific residues in C/EBP are involved in the GR-C/EBP interface and that mutation of these residues would disrupt GR-C/EBP complex formation (Table 4). To test this hypothesis in vivo, we analyzed the recruitment of C/EBP at a specific GR-DNA-binding locus inside living cells. The amplified array of a GR responsive promoter structure (the MMTV array) in 3617 cells contains 800–1200 GR binding sites (60) and DNA recognition sequences for the nuclear factor-1 (NF-1) protein (50). Thus, fluorescently tagged proteins binding to this region (e.g., GR or NF-1) can be directly visualized in living cells as a bright spot in the nucleoplasm. If GR interacts with C/EBP, then C/EBP should be recruited to the array in a manner dependent on GR activation by glucocorticoids.
First, we ruled out the possibility that C/EBP was able to bind the array in a glucocorticoid-independent manner. C/EBP is constitutively nuclear and presents a nonhomogenous intranuclear distribution, forming aggregates in heterochromatin regions (61) (Fig. 9B). As visual discrimination between the MMTV array and heterochromatin regions was not possible by examining only C/EBP distribution; we utilized mCherry-NF-1 to mark the array as it binds this structure independently of glucocorticoid (50). In the absence of glucocorticoids, C/EBP does not localize at the array (Fig. 9B, upper). Only after dexamethasone addition is a clear colocalization of C/EBP and NF-1 visualized at the array (Fig. 9B, lower). As GR is expressed endogenously by these cells, this finding suggests that C/EBP recruitment to the array is indeed GR-dependent.
In silico, mutations of residues in C/EBPα forming the interface with GR were predicted to disrupt the GR-C/EBP interaction. We therefore tested the effects of these mutations by introducing them into fluorescently tagged C/EBP and analyzing the subcellular distribution of these proteins after GR activation in living cells (Fig. 9C). Examination of 3617 cells transfected with C/EBP mutants revealed that L315D, I341D, and L317D mutants are able to bind to the MMTV array, suggesting that these mutations do not severely affect GR-C/EBP complex formation. Nevertheless, the L317D mutant appeared to lose the ability to bind to heterochromatin, and its nuclear distributions appeared more homogenous. Only after the introduction of both L317D and L335D mutations did we fail to detect the strong colocalization between C/EBP and GR at the array, suggesting a lack of interaction between these proteins (Fig. 9C, last column). The L317D+L335D mutant additionally presented a very homogenous intranuclear distribution, an absence of heterochromatin binding, and even a small population of molecules in the cytoplasmic compartment.
Discussion
Chromatin-remodeling complexes play an important role in transcriptional regulation by changing the access of proteins to DNA within the context of chromatin (62). Thus, it is essential to understand the mechanism of action of these complexes. Determining how individual subunits interact with one another contributes to knowledge of the mode of action of the SWI/SNF complex. Understanding how the structures interact within the SWI/SNF and how they interact with other transcription factors helps us to understand the steric limits imposed by structure, which in turn informs on the functional consequences of manipulations of the remodeling system. This computational work, as well as previous experimental results (17,20), supports the premise that multiple subunits of the SWI/SNF complex are involved in recruitment of the GR. GR lacking the LBD can interact with BAF60a and BAF57 but not with BGR1, BAF170, or BAF155. BAF250 also interacts with GR LBD in pull-down experiments via the BAF250 C-terminus (17), suggesting that there are at least three SWI/SNF components with which GR can associate. Our results suggest that the BAF250 C-terminus can indeed interact with GR through its LBD but not through its DBD. BAF57 conversely requires the DBD to bind to GR. We could not predict the interaction between the BAF60a N-terminus and GR DBD, since modeling did not produce a high-quality structure for the BAF60a N-terminus. The sequence identity was <30%. The accuracy of the homology model is limited to the sequence identity between target and templates and declines with decreasing sequence identity. Above 50% sequence identity, generally, reliable models are generated. On the other hand, a sequence identity <30% can produce misfolded protein models. Fortunately, we could model the BAF60a C-terminus and show that GR does not bind to the C-terminal region of BAF60a as suggested experimentally. The fact that GR can interact with multiple SWI/SNF subunits through its LBD or DBD suggests the possibility of redundancy and compensation. Thus, a mutation in one SWI/SNF component that abolished the interaction with GR would not bring about a total loss of interaction, since the other partners could compensate. Similarly, a mutation in the LBD or DBD of GR that affected the interaction with SWI/SNF subunits would not lead to complete loss of interaction. Multiple interactions may also enhance the stability of the complex. However, these interactions are not necessarily equally important. GR may interact with multiple SWI/SNF complex components, but which one or ones are used might depend on the context and conformation of the GR at a particular binding site. Thus, changes in one protein could be predicted to severely affect remodeling and gene expression if that protein is fairly unique to a location regarding a GR interaction. Studies suggest that the GR mutant R488Q, with impaired ability to bind the N-terminal BAF60a, exhibited reduced chromatin-remodeling activity. Similarly, the GR1-556 mutant, lacking the entire LBD, shows reduced activity of chromatin remodeling (20). Taken together, these results suggest that GR DBD interaction with the BAF60a N-terminus or GR LBD interaction with the BRG1 complex plays an important role in GR-mediated chromatin remodeling.
We also predicted the binary complexes of SWI/SNF subunits and constructed models for the multiprotein complex based on these binary interactions. The models presented here not only allow us to identify the interacting pairs but also help us understand how core SWI/SNF subunits interact with each other and with variant subunits to form the remodeling complex. Structural data enable us to predict how the presence or absence of a domain or a conformational change within it affects the complex. It allows us to identify the bridging molecules, like BAF60a, that are responsible for mediating interactions between different molecules. This is important because the absence of these molecules or a mutation at the interface will have profound effects on complex assembly and/or stability. However, we should keep in mind that the suggested models for the multiprotein complexes do not represent the entire complex. The vast majority of the SWI/SNF components do not have crystal structures in the PDB. Thus, we performed modeling to obtain the structural data. The quality and accuracy of a homology model are directly linked with the presence of a high-resolution experimental protein structure as a template and the sequence identity between the template and target protein (63,64). In this study, we only used the high-quality structural models in our predictions, but models may nonetheless miss some domains that could perhaps stabilize the complex through additional contacts or destabilize it due to steric hindrance.
In addition to the chromatin remodelers, GR has been shown to interact directly with other transcription factors and alter their function. C/EBP is one such factor (32,65). Predictions suggest that GR can interact with bZip domains of C/EBPα homodimer either as a monomer or as a dimer of the DBD. They further indicate that C/EBP can bind DNA while interacting with GR. Similarly, GR can bind DNA while interacting with C/EBP according to the first model. The first model also suggests that GR-C/EBP interaction might play a role in long-distance DNA looping between regions where GR and C/EBP bind separately. Similar cases have been suggested with other factors, such as CTCF, GATA, and nuclear hormone receptors (66–69). In the second model, the GR-C/EBP interface overlaps with the DNA-binding region of GR. Thus, GR cannot interact with DNA and C/EBP simultaneously in this case. The residues predicted to be structurally important for GR binding were confirmed by computational mutagenesis. In silico analysis showed that amino acid variations in C/EBP affect GR-C/EBP interaction. Our computational results show that single L315D and L317D and double L317D+L335D mutations have a significant impact on GR binding. As expected, these mutations affected the dimer interaction of C/EBP bZip domains, as they are located at the dimer interface. However, they did not disrupt the self-dimerization as they disrupted the interactions, they just weakened it. These results imply that these residues have important roles in GR binding. However, one should keep in mind that in silico tools are still limited and that their reliability seems to differ significantly between the various algorithms, so results often require experimental validation. Accordingly, studies using a variety of techniques support the predictions made for SWI/SNF remodeler subunits, and GR interaction with C/EBP has also been experimentally demonstrated (32,33). Notable methodological improvements in the provision of structural information for proteins with inherently disordered conformation-sensitive folding (70,71) offer promise for increased reliability of predictions.
In this study, GR was predicted to interact with C/EBP both as a monomeric and dimeric DBD, and our N&B data suggested that within the cell, GR took the dimeric form. It is not clear whether this preference is absolute or a cell-type-specific phenomenon since other cell types were not tested. However, in five cell lines tested to date using the N&B approach, activated nuclear GR has been dimeric independently of the presumed alternative transcription factor milieu expressed by these different cell types (52,59). Curiously, despite the strong structural predictions, when the effects of C/EBP mutations were tested in vivo, results showed that only the double C/EBP mutant L317D+L335D failed to be recruited to the MMTV array in a GR-dependent manner. These data suggest that only the double mutation severely impaired GR and C/EBP interactions. However, it is noteworthy that the in vivo experiment cannot technically discriminate between a loss of interaction between GR and C/EBP and the loss of dynamic assisted loading of C/EBP at a glucocorticoid responsive unit (29,72).
Overall, computational predictions can be extremely useful as a starting point to interrogate the functional constraints that structure imposes on biological molecules. In addition to computing the likely interfaces involved in protein-protein interactions, and building interactome maps (73), such models can also potentially inform on the downstream effects of pharmacological and experimental manipulations that produce conformational changes in proteins. Here, we used in silico tools to narrow down potential target residues for experimental confirmation of the predicted GR-C/EBP interface. As structural predictive algorithms continue to improve in line with advances in in vivo experimental technology, it is likely that computation will play an increasing role in the development and testing of biological hypotheses relating to the function of a variety of proteins. Such predictive power will be important for the development of novel therapeutic strategies that alter specific modalities of protein function to maximize clinical efficacy while minimizing side effects.
Author Contributions
S.M., J.R.P., D.M.P., L.G., O.K., and A.G. conceived and designed the study. S.M., O.K., and A.G. performed computational studies using PRISM. D.M.P. and J.R.P. performed N&B experiments. S.M., J.R.P., D.M.P., L.G., G.L.H., R.N., O.K., and A.G. prepared and wrote the manuscript. All authors edited and approved the manuscript.
Acknowledgments
A.G. and O.K. are members of the Science Academy, Turkey. O.K. was on sabbatical at the National Cancer Institute, National Institutes of Health (NIH).
This project was funded in whole or in part with federal funds from the National Cancer Institute, NIH, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Editor: H. Jane Dyson.
Footnotes
Diego M. Presman and John R. Pooley contributed equally to this work.
Four figures and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00627-X.
Contributor Information
Ozlem Keskin, Email: okeskin@ku.edu.tr.
Attila Gursoy, Email: agursoy@ku.edu.tr.
Supporting Material
References
- 1.Mangelsdorf D.J., Thummel C., Evans R.M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsiao P.W., Deroo B.J., Archer T.K. Chromatin remodeling and tissue-selective responses of nuclear hormone receptors. Biochem. Cell Biol. 2002;80:343–351. doi: 10.1139/o02-082. [DOI] [PubMed] [Google Scholar]
- 3.Kadmiel M., Cidlowski J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones K.A., Kadonaga J.T. Exploring the transcription-chromatin interface. Genes Dev. 2000;14:1992–1996. [PubMed] [Google Scholar]
- 5.Peterson C.L., Workman J.L. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 6.Becker P.B., Hörz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 7.Collingwood T.N., Urnov F.D., Wolffe A.P. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 1999;23:255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 8.Pazin M.J., Kadonaga J.T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 9.Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 11.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 12.Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 13.Yoshinaga S.K., Peterson C.L., Yamamoto K.R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 14.Peterson C.L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 15.Laurent B.C., Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- 16.Lemon B., Inouye C., Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 17.Nie Z., Xue Y., Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Z., Cui K., Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fryer C.J., Archer T.K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao P.W., Fryer C.J., Archer T.K. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebbar P.B., Archer T.K. Chromatin remodeling by nuclear receptors. Chromosoma. 2003;111:495–504. doi: 10.1007/s00412-003-0232-x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D.E., Zhang P., Tenen D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N.D., Finegold M.J., Darlington G.J. Impaired energy homeostasis in C/EBP α knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 24.Akai Y., Oitate T., Shiojiri N. Impaired hepatocyte maturation, abnormal expression of biliary transcription factors and liver fibrosis in C/EBPα(Cebpa)-knockout mice. Histol. Histopathol. 2014;29:107–125. doi: 10.14670/HH-29.107. [DOI] [PubMed] [Google Scholar]
- 25.Ellenberger T.E., Brandl C.J., Harrison S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 26.Ramji D.P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowenz-Leutz E., Leutz A. A C/EBP β isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen T.A., Kowenz-Leutz E., Nerlov C. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grøntved L., John S., Hager G.L. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32:1568–1583. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steger D.J., Grant G.R., Lazar M.A. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John S., Sabo P.J., Stamatoyannopoulos J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudiger J.J., Roth M., Block L.H. Interaction of C/EBPα and the glucocorticoid receptor in vivo and in nontransformed human cells. FASEB J. 2002;16:177–184. doi: 10.1096/fj.01-0226com. [DOI] [PubMed] [Google Scholar]
- 33.Johansson-Haque K., Palanichamy E., Okret S. Stimulation of MAPK-phosphatase 1 gene expression by glucocorticoids occurs through a tethering mechanism involving C/EBP. J. Mol. Endocrinol. 2008;41:239–249. doi: 10.1677/JME-08-0015. [DOI] [PubMed] [Google Scholar]
- 34.Tuncbag N., Gursoy A., Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat. Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baspinar A., Cukuroglu E., Gursoy A. PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes. Nucleic Acids Res. 2014;42:W285–W289. doi: 10.1093/nar/gku397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valencia A., Pazos F. Computational methods for the prediction of protein interactions. Curr. Opin. Struct. Biol. 2002;12:368–373. doi: 10.1016/s0959-440x(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 37.Planas-Iglesias J., Marin-Lopez M.A., Oliva B. iLoops: a protein-protein interaction prediction server based on structural features. Bioinformatics. 2013;29:2360–2362. doi: 10.1093/bioinformatics/btt401. [DOI] [PubMed] [Google Scholar]
- 38.Hamp T., Rost B. Evolutionary profiles improve protein-protein interaction prediction from sequence. Bioinformatics. 2015;31:1945–1950. doi: 10.1093/bioinformatics/btv077. [DOI] [PubMed] [Google Scholar]
- 39.Bhaskara R.M., Padhi A., Srinivasan N. Accurate prediction of interfacial residues in two-domain proteins using evolutionary information: implications for three-dimensional modeling. Proteins. 2014;82:1219–1234. doi: 10.1002/prot.24486. [DOI] [PubMed] [Google Scholar]
- 40.Radivojac P., Clark W.T., Friedberg I. A large-scale evaluation of computational protein function prediction. Nat. Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vreven T., Hwang H., Weng Z. Evaluating template-based and template-free protein-protein complex structure prediction. Brief. Bioinform. 2014;15:169–176. doi: 10.1093/bib/bbt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cukuroglu E., Gursoy A., Keskin O. HotRegion: a database of predicted hot spot clusters. Nucleic Acids Res. 2012;40:D829–D833. doi: 10.1093/nar/gkr929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Duarte J.M., Srebniak A., Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinformatics. 2012;13:334. doi: 10.1186/1471-2105-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu H., Domingues F.S., Lengauer T. NOXclass: prediction of protein-protein interaction types. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schymkowitz J., Borg J., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schymkowitz J.W., Rousseau F., Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl. Acad. Sci. USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker D., Htun H., Hager G.L. Using inducible vectors to study intracellular trafficking of GFP-tagged steroid/nuclear receptors in living cells. Methods. 1999;19:386–393. doi: 10.1006/meth.1999.0874. [DOI] [PubMed] [Google Scholar]
- 50.Stavreva D.A., Wiench M., Hager G.L. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat. Cell Biol. 2009;11:1093–1102. doi: 10.1038/ncb1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu Y., Zhao Y., Hager G.L. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol. Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 52.Presman D.M., Ogara M.F., Pecci A. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol. 2014;12:e1001813. doi: 10.1371/journal.pbio.1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kar G., Keskin O., Gursoy A. Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J. Proteome Res. 2012;11:1196–1207. doi: 10.1021/pr2009143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acuner Ozbabacan S.E., Keskin O., Gursoy A. Enriching the human apoptosis pathway by predicting the structures of protein-protein complexes. J. Struct. Biol. 2012;179:338–346. doi: 10.1016/j.jsb.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuzu G., Gursoy A., Keskin O. Exploiting conformational ensembles in modeling protein-protein interactions on the proteome scale. J. Proteome Res. 2013;12:2641–2653. doi: 10.1021/pr400006k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., Xue Y., Crabtree G.R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 57.Trotter K.W., Fan H.Y., Archer T.K. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Digman M.A., Dalal R., Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Presman D.M., Alvarez L.D., Pecci A. Insights on glucocorticoid receptor activity modulation through the binding of rigid steroids. PLoS One. 2010;5:e13279. doi: 10.1371/journal.pone.0013279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNally J.G., Müller W.G., Hager G.L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 61.Susperreguy S., Prendes L.P., Piwien-Pilipuk G. Visualization by BiFC of different C/EBPβ dimers and their interaction with HP1α reveals a differential subnuclear distribution of complexes in living cells. Exp. Cell Res. 2011;317:706–723. doi: 10.1016/j.yexcr.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vignali M., Hassan A.H., Workman J.L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker D., Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 64.Hillisch A., Pineda L.F., Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov. Today. 2004;9:659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arambasić J., Poznanović G., Grigorov I. Association of the glucocorticoid receptor with STAT3, C/EBPβ, and the hormone-responsive element within the rat haptoglobin gene promoter during the acute phase response. IUBMB Life. 2010;62:227–236. doi: 10.1002/iub.313. [DOI] [PubMed] [Google Scholar]
- 66.Rao S.S., Huntley M.H., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y., Bates D.L., Chen L. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Reports. 2012;2:1197–1206. doi: 10.1016/j.celrep.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Théveny B., Bailly A., Milgrom E. Association of DNA-bound progesterone receptors. Nature. 1987;329:79–81. doi: 10.1038/329079a0. [DOI] [PubMed] [Google Scholar]
- 69.Yasmin R., Yeung K.T., Noy N. DNA-looping by RXR tetramers permits transcriptional regulation “at a distance”. J. Mol. Biol. 2004;343:327–338. doi: 10.1016/j.jmb.2004.08.070. [DOI] [PubMed] [Google Scholar]
- 70.Goswami D., Devarakonda S., Griffin P.R. Time window expansion for HDX analysis of an intrinsically disordered protein. J. Am. Soc. Mass Spectrom. 2013;24:1584–1592. doi: 10.1007/s13361-013-0669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rochel N., Ciesielski F., Moras D. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 72.Voss T.C., Schiltz R.L., Hager G.L. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuzu G., Keskin O., Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr. Opin. Struct. Biol. 2012;22:367–377. doi: 10.1016/j.sbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.