Summary
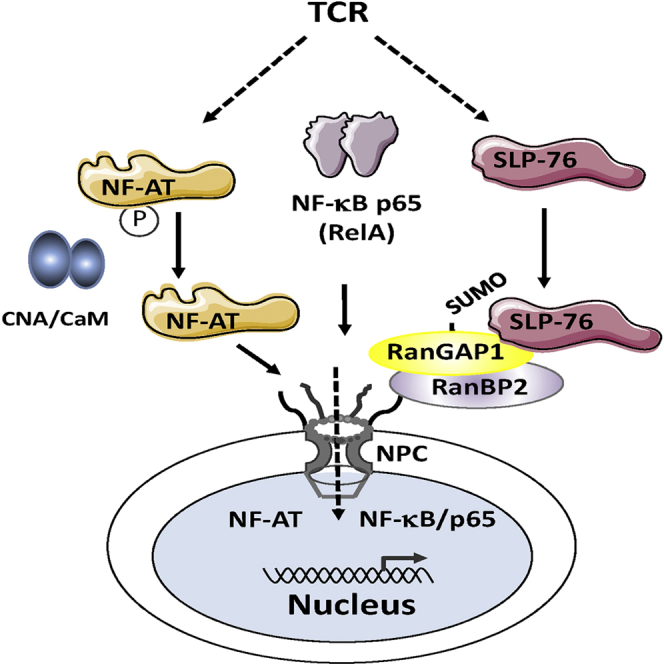
While immune cell adaptors regulate proximal T cell signaling, direct regulation of the nuclear pore complex (NPC) has not been reported. NPC has cytoplasmic filaments composed of RanGAP1 and RanBP2 with the potential to interact with cytoplasmic mediators. Here, we show that the immune cell adaptor SLP-76 binds directly to SUMO-RanGAP1 of cytoplasmic fibrils of the NPC, and that this interaction is needed for optimal NFATc1 and NF-κB p65 nuclear entry in T cells. Transmission electron microscopy showed anti-SLP-76 cytoplasmic labeling of the majority of NPCs in anti-CD3 activated T cells. Further, SUMO-RanGAP1 bound to the N-terminal lysine 56 of SLP-76 where the interaction was needed for optimal RanGAP1-NPC localization and GAP exchange activity. While the SLP-76-RanGAP1 (K56E) mutant had no effect on proximal signaling, it impaired NF-ATc1 and p65/RelA nuclear entry and in vivo responses to OVA peptide. Overall, we have identified SLP-76 as a direct regulator of nuclear pore function in T cells.
Graphical Abstract
Highlights
-
•
Immune adaptor SLP-76 binds to SUMO-RanGAP1 of cytoplasmic fibrils of the NPC
-
•
SLP-76 K-56 binding needed for optimal RanGAP1 localization and exchange activity
-
•
SLP-76 K56E mutant impaired NF-ATc1 and NFκB p65 (RelA) nuclear entry
-
•
Immune adaptors directly regulate nuclear entry of transcription factors in T cells
Liu et al. show that the direct binding of the immune cell adaptor SLP-76 to SUMO-RanGAP1 of cytoplasmic fibrils of the nuclear pore complex (NPC) is needed for the optimal NFAT and NFkB nuclear entry in T cells.
Introduction
T cells express protein-tyrosine kinases and adaptors that integrate signals for T cell activation (Rudd, 1999; Rudd et al., 2010; Samelson, 2002; Smith-Garvin et al., 2009). Adaptors possess binding sites and discrete modular domains that integrate signals. Immune cell adaptors include SH2 domain containing leukocyte protein of 76 kDa (SLP-76) (Jackman et al., 1995; Smith-Garvin et al., 2009), linker for the activation of T cells (LAT) (Zhang et al., 1998), and adhesion- and degranulation-promoting adaptor protein (ADAP) (da Silva et al., 1997; Liu et al., 1998; Musci et al., 1997). SLP-76 has a N-terminal sterile-α motif (SAM), tyrosine motifs and a SH2 domain and is needed for T cell differentiation and function (Jackman et al., 1995; Jordan et al., 2003; Pivniouk et al., 1998). SLP-76-deficient T cells show an impaired phospholipase Cγ1 (PLCγ1) activation and calcium mobilization (Yablonski et al., 1998), while N-terminal residues are phosphorylated by ZAP-70 (Bubeck Wardenburg et al., 1996; Raab et al., 1997). Y-113 and Y-128 bind exchange factor Vav1 and adaptor Nck (Bubeck Wardenburg et al., 1998; Jackman et al., 1995; Wu et al., 1996), resting lymphocyte kinase (Rlk) (Schneider et al., 2000), and inducible tyrosine kinase (Itk) (Bunnell et al., 2000). SLP-76 binds to the SH3 domain of PLCγ1 (Grasis et al., 2010; Yablonski et al., 2001), while GADs SH2 domain forms a complex with LAT (Zhang et al., 1998). SLP-76 also forms microclusters (Bunnell et al., 2002; Yokosuka et al., 2005), exerts feedback control on ZAP-70 (Liu et al., 2010), and interacts with subsynaptic LAT clusters (Purbhoo et al., 2010; Williamson et al., 2011). The SLP-76 SH2 domain binds to ADAP (da Silva et al., 1997; Musci et al., 1997) and hematopoietic progenitor kinase-1 (HPK-1) (Di Bartolo et al., 2007; Shui et al., 2007). In turn, ADAP binds to adaptor SKAP1 (SKAP-55) for integrin adhesion (Raab et al., 2010, 2011; Wang and Rudd, 2008).
SLP-76 is also needed downstream to activate transcription factors NFAT (nuclear factor for the activation of T cells) and NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) (Yablonski et al., 1998). NFAT possesses two basic nuclear localization sequences (NLSs) for nuclear import dependent on dephosphorylation by calcineurin (Müller and Rao, 2010; Wu et al., 2007). Dephosphorylation unmasks nuclear-location signals (Shibasaki et al., 1996). Similarly, NF-κB plays roles in inflammation, cell activation, and differentiation (Ghosh and Karin, 2002; Sen, 2011). Coreceptor CD28 and innate receptors activate NF-κB transcription via different pathways in T cells (Marinari et al., 2002; Thaker et al., 2015).
Nuclear transport is mediated by the nuclear pore complex (NPC) (Chatel and Fahrenkrog, 2012; Hoelz et al., 2011). The NPC is composed of more than 30 nucleoporins (Nups) needed for anchorage and the formation of a central mesh in the channel (Allen et al., 2008; D’Angelo and Hetzer, 2008). Intriguingly, eight filaments extend into the cytoplasm comprised of RanBP2 (Nup358) and RanGAP1, the latter having GTPase activity for GTP-Ran (Bischoff et al., 1994). This interaction requires the ATP-dependent posttranslational conjugation of RanGAP1 with SUMO-1 (for small ubiquitin-related modifier) (Lee et al., 1998; Mahajan et al., 1997). Ran binding to GTP causes importins to release protein in the nucleus, while nonhydrolysable GTP accumulates Ran-GTP at the filaments (Melchior et al., 1995). RanBP2/RanGAP1 and associated SUMO1/Ubc9 form a multisubunit SUMO E3 ligase (Pichler et al., 2002; Werner et al., 2012).
SLP-76 microclusters at the cell surface translocate to the perinuclear region of T cells (Bunnell et al., 2002). While adaptors mediate TCR proximal signaling, direct regulation of the NPC has not been reported. Here, we show that direct SLP-76 binding to the SUMO-RanGAP1 of cytoplasmic filaments of the NPC is required for the regulation of transcription factor entry into the nucleus of T cells. Our findings identify a surprising direct mechanism of NPC regulation by an immune adaptor in T cells.
Results
SLP-76 Localizes to the Nuclear Pore and Binds to SUMO-RanGAP1
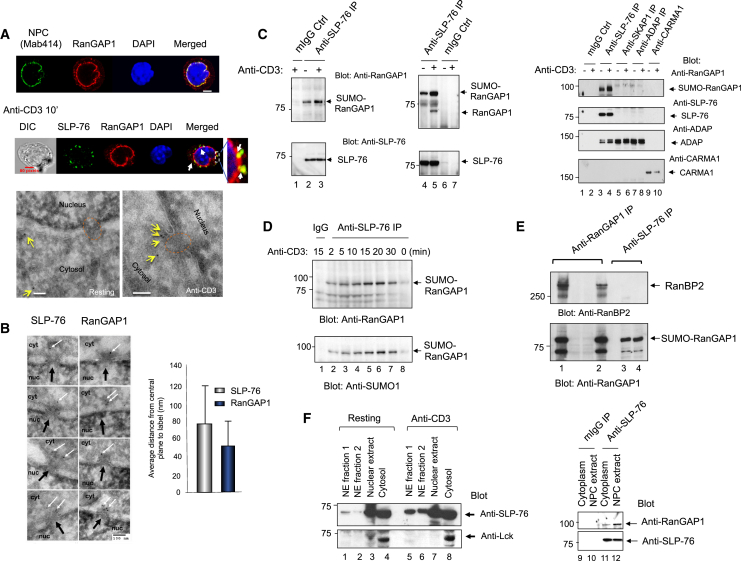
NPC fibrils could potentially interact with cytoplasmic signaling proteins in T cells. Mouse DC27.10 T cells were anti-CD3 ligated for 10 min and imaged by confocal fluorescence microscopy (Figure 1A). Monoclonal antibody to NPC proteins (Mab414) and anti-RanGAP1 stained the nuclear envelop around the DAPI-stained nucleus (upper and middle panels). Anti-CD3-induced endogenous SLP-76 microclusters overlapped with RanGAP1 as detected by antibody staining (middle panel; right expanded image). Immune-gold transmission electron microscopy (TEM) using anti-SLP-76 showed labeling of the cytoplasmic site of the NPC in response to anti-CD3 ligation (lower left versus right panel). Quantitation showed that anti-SLP-76 stained 13% of randomly selected NPCs with gold particles in resting cells (n = 44) (Figure 1B). Remarkably, 76% of NPCs had at least one gold particle within a distance of 130 nm from the cytoplasmic ring in response to anti-CD3 ligation, (i.e., an average of 0.84 particles per NPC of 100 randomly selected NPCs with zero to three labels each). A total of 77% of labels were cytoplasmic (n = 109), with an average distance of ∼35 nm from the cytoplasmic ring, a distance comparable to RanGAP staining (histogram).
Figure 1.
SLP-76 Localizes at the Cytoplasmic Face of the Nuclear Pore Complex and Interacts with SUMO-RanGAP1
(A) Confocal images of mouse DC27.10 T cells stained with Mab414-Alexa Fluor 488, anti-RanGAP1-Alexa Fluor 633, anti-SLP-76-Alexa Fluor 488 and DAPI (upper and middle panel) (scale bar, 5 μm). TEM images of gold-labeled anti-SLP-76 (yellow arrows) on the cytoplasmic face of the NPC (red dashed circle) (n = 3).
(B) TEM images of gold-labeled anti-SLP-76 and anti-RanGAP1 (white arrows) relative to the NPC (black arrows) upon anti-CD3 stimulation (left). Histogram shows distance of SLP-76 and RanGAP1 from central plane of NPC (right).
(C) Anti-SLP-76 coprecipitated RanGAP1. Resting or anti-CD3 activated DC27.10 T cells (left) or human peripheral blood lymphocytes (middle). Precipitation followed by blotting with anti-RanGAP1 (upper) or anti-SLP-76 (lower) (n = 4). (Right panel) RanGAP1 does not interact with immune adaptors SKAP1, ADAP, or CARMA1 (n = 3).
(D) Time course of RanGAP1 binding to SLP-76 upon anti-CD3 ligation. Blotted with anti-RanGAP1 (upper panel) or anti-SUMO1 (lower) (n = 3).
(E) Anti-SLP-76 does not coprecipitate RanBP2 from anti-CD3-stimulated Jurkat T cells. Blotted with anti-RanBP2 (upper panel) or anti-RanGAP1 (lower). Also see Figure S1.
(F) Anti-CD3 increases SLP-76 recruitment to the NPC. Mouse DC27.10 T cells stimulated with anti-CD3 followed by subcellular fractionation and blotted with anti-SLP-76 (left upper) or anti-lck (left lower). Anti-SLP-76 coprecipitates from NE fraction. Blotted with anti-RanGAP1 (right upper) and anti-SLP-76 (right lower) (n = 4).
NPC cytoplasmic fibrils are comprised of RanBP2 and RanGAP1 (Hutten et al., 2008). RanGAP1 exists as a nonsumoylated 70 Kd and a 90 Kd sumoylated form that associates with that NPC (Bernier-Villamor et al., 2002; Matunis et al., 1996). Anti-SLP-76 coprecipitated a 90 Kd protein from mouse DC27.10 T cells and preactivated human primary T cells as recognized by anti-RanGAP1 blotting (Figure 1C, left panel, lanes 2 and 3, and 4 and 5, respectively). Anti-CD3 ligation increased coprecipitated RanGAP1 (upper panel, lane 3 versus lane 2 and lane 5 versus lane 4). Occasionally, a smaller amount of nonsumoylated RanGAP1 was apparent in the anti-SLP-76 precipitates (3/16 experiments). Specificity was shown by the failure of anti-SKAP1 (right panel, lanes 5 and 6), anti-ADAP (lanes 7 and 8), or anti-CARMA1 (lanes 9 and 10) to coprecipitate RanGAP1. A time course showed that the optimal coprecipitation of RanGAP1 was a relatively late event with maximal levels at 10–20 min following anti-CD3 ligation (Figure 1D). Anti-SUMO blotting confirmed that SUMO-RanGAP1 at 90 KD was coprecipitated with anti-SLP-76 over the time course (lower panel). SUMO-RanGAP1 selectively associates with the fibrils of the NPC (Mahajan et al., 1997).
By contrast, anti-SLP-76 failed to precipitate RanBP2 (Nup358) from anti-CD3 activated Jurkat T cells (Figure 1E, upper panel, lanes 3 and 4), while it coprecipitated SUMO-RanGAP1 (lower panel, lanes 3 and 4). Anti-RanGAP1 precipitated itself (lower panel, lanes 1 and 2) and RanBP2 (upper panel, lanes 1 and 2). Similarly, anti-RanGAP1 coprecipitated SLP-76 from 293 T cells transfected with the adaptor (see Figure S1A available online, upper panel, lane 2 versus lane 1). By contrast, anti-RanBP2 failed to coprecipitate SLP-76 from 293 T cells expressing RanBP2 plus SLP-76 (lower panel, lane 4). Likewise, anti-NFATc1 failed to precipitate SLP-76 or RanGAP1 from resting or activated Jurkat cells (Figure S1B, lanes 3 and 4). Anti-SLP-76 coprecipitated RanGAP1, but no NFATc1 (lanes 5 and 6). These observations indicated that SLP-76 bound to RanGAP1, rather than RanBP2 or NFATc1.
Anti-SLP-76 coprecipitation of RanGAP1 was confirmed by MALDI-MS/MS (Figure S1C). Colloidal blue staining identified proteins that were coprecipitated by anti-SLP-76 (upper panel) that included GADs, ADAP-130 (Fyb-130) and 14-3-3. Six distinct peptides corresponding to RanGAP1 were detected (middle and lower boxes).
We also observed SLP-76 and the SLP-76/RanGAP complex in biochemically purified nuclear fractions (Figure 1F). Anti-CD3 ligation markedly increased SLP-76 in the nuclear envelope extract containing the NPC (left panel, lanes 5 and 6 versus lanes 1 and 2). p56lck was enriched in the cytosolic but not nuclear fraction (lower panel, lanes 4 and 8). Further, anti-SLP-76 coprecipitated RanGAP1 from the NPC extract of anti-CD3 ligated cells (right panel, lane 12). These data confirmed that anti-CD3 increased the presence of SUMO-RanGAP1 in the NPC.
SLP-76 Binding Promotes RanGAP1 Binding and Function at the NPC
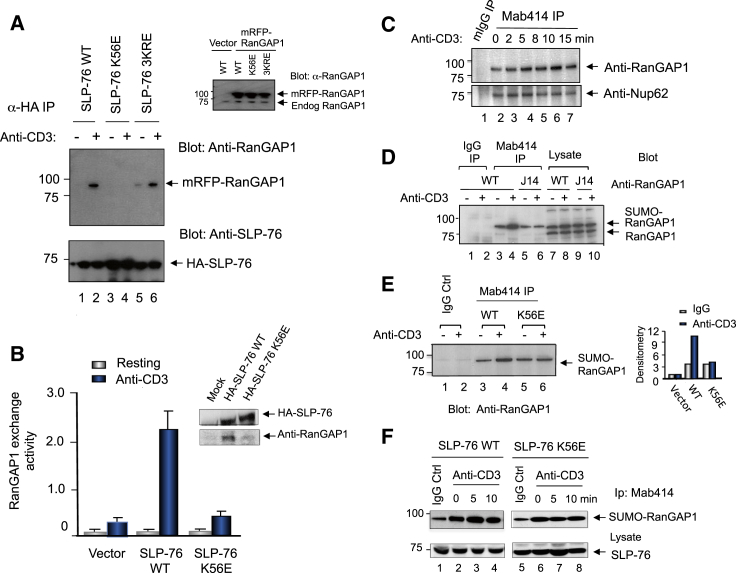
We next mapped RanGAP1 binding to the N terminus of SLP-76 (Figure 2A). RanGAP1 was coexpressed with HA-tagged SLP-76 wild-type (WT) or mutants in SLP-76-deficient J14 T cells followed by anti-HA precipitation and anti-RanGAP1 blotting. Anti-CD3 ligation induced the complex formation as shown by anti-SLP-76 coprecipitation of SUMO-RanGAP1 (lane 2 versus lane 1). Random mutagenesis showed that mutation of lysine-56 to glutamic acid (termed K56E) disrupted the interaction (lanes 3 and 4), while mutation of residues 74/181/491 (i.e., 3KRE) had no effect (lanes 5 and 6). Blotting of lysates showed similar RanGAP1 expression in transfected cells (upper inset). HA-tagged SLP-76 WT and mutant expression was confirmed by anti-SLP-76 blotting (lower panel). Further, anti-CD3 ligation increased anti-SLP-76-associated RanGAP1 exchange activity by 7-fold, an effect lost with mutation of the K56 residue (Figure 2B). Blotting confirmed the concurrent loss of RanGAP1 binding with K56E (upper inset).
Figure 2.
RanGAP1 Binds to SLP-76 Lysine 56 and Regulates RanGAP1 Binding to NPC
(A) RanGAP1 binds to lysine 56 in the SLP-76 N-terminal domain. SLP-76-deficient J14 T cells cotransfected with HA-tagged SLP-76 or mutants and mRFP-RanGAP1 (n = 3).
(B) Anti-CD3 increases RanGAP1 exchange activity that is lost with K56E. (Upper inset) Blot for HA-SLP-76 wild-type and K56E expression. Data are represented as mean ± SE of a representative experiment (n = 3). Also see Figure S2.
(C) Anti-CD3 ligation increases SUMO-RanGAP1 coprecipitated with anti-NPC Mab414. Cell lysates from DC27.10 T cells were stimulated with anti-CD3 for 15 min, followed by Mab414 precipitation and blotting with anti-RanGAP1 (upper) or Nup62 (lower) (n = 3).
(D) Anti-CD3-induced increase in SUMO-RanGAP1 association with the NPC is impaired in SLP-76-deficient T cells (n = 3).
(E) Anti-CD3-induced increase in SUMO-RanGAP1 association with the NPC is impaired in K56E-expressing cells. Right panel shows histogram of densitometric readings of SUMO-RanGAP1.
(F) Time course of anti-CD3-induced increase in SUMO-RanGAP1 coprecipitated by Mab414 from J14 cells expressing SLP-76 WT or K56E mutant. Anti-RanGAP1 blot (upper). Anti-SLP-76 blot of cell lysates (lower) (n = 4).
We also assessed whether SLP-76 and binding to RanGAP1 influenced RanGAP1 binding to the NPC. Anti-CD3 ligation increased SUMO-RanGAP1 coprecipitated by Mab414 (anti-NPC) over 2–15 min (Figure 2C, upper panel, lanes 3–7 versus lane 2). Nup62 coprecipitation served as a control (lower panel). The increase in coprecipitated SUMO-RanGAP1 from SLP-76-expressing cells at 15 min postligation (Figure 2D, lane 4 versus lane 3) was not evident in SLP-76-deficient J14 cells (lane 6 versus lane 5). Similarly, the anti-CD3-induced increase in SUMO-RanGAP1 in Mab414 precipitates from J14 cells expressing SLP-76 was not observed from cells with K56E (Figure 2E, lane 4 versus lane 3 and lane 6 versus lane 5; histogram). A time course also showed an increase in RanGAP1 that was not evident in K56E-expressing cells (Figure 2F, lanes 3 and 4 versus lane 2 and lanes 7 and 8 versus lane 6). SLP-76 expression and SLP-76-RanGAP1 binding is needed for the optimal anti-CD3-induced association of SUMO-RanGAP1 with the NPC.
We also observed that the specific activity for RanGAP1 for Ran-GTP was higher in anti-SLP-76 than anti-RanGAP1 precipitates lacking SLP-76 (Figure S2A, upper and lower panels). Specific activities were obtained by normalizing the activity relative to the amount of RanGAP1 in the precipitates as detected by blotting. Anti-RanGAP1 precipitates also showed lower exchange activity from anti-CD3-activated cells expressing K56E compared to wild-type SLP-76 (Figure S2B). These observations suggested that, in addition to its role in promoting RanGAP1 binding to the NPC, SLP-76 binding to RanGAP1 increased its GAP activity for Ran-GTP.
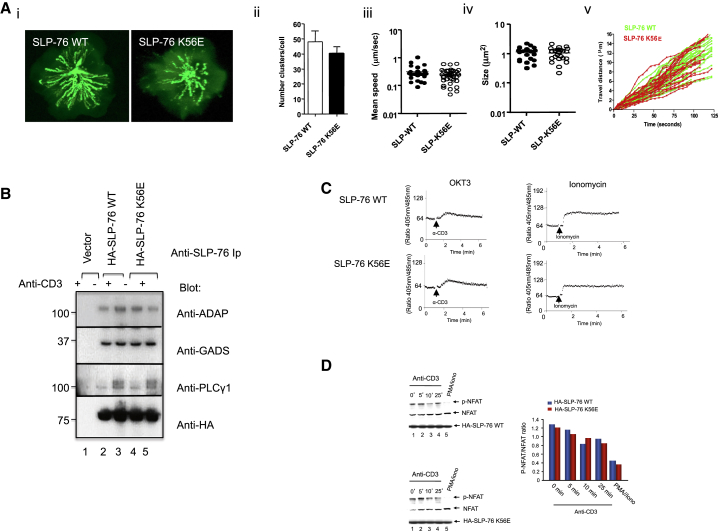
SLP-76-RanGAP1 Binding Fails to Affect Proximal Signaling
It was next important to assess whether the mutation of K56 affected TCR proximal signaling (Figure 3). Microcluster formation is an early event needed for the stimulation of T cells (Bunnell et al., 2002; Yokosuka et al., 2005). SLP-76-EYFP and K56E-EYFP J14 cells were imaged on immobilized anti-CD3 on slides. Maximum overtime (MOT) images showed the formation and movement of clusters to central contact area (Figure 3Ai). No difference was detected between WT and K56E in terms of the number (ii), speed (iii), size (iv), or migration of clusters (v) (Movie S1). K56E and wild-type SLP-76 also bound to similar amounts of ADAP, GADS, and PLCγ1 (Figure 3B). In both cases, PLCγ1 shifted in migration in response to anti-CD3, consistent with increased phosphorylation. Anti-CD3-induced increase in intracellular calcium was also the same in J14 cells expressing K56E and WT SLP-76 (Figures 3C and S3A). The ionomycin-induced increase was also the same in SLP-76 WT and K56E-transfected cells (right panels).
Figure 3.
SLP-76 K56E Does Not Interfere with Early TCR Signaling Events
(A) K56E does not interfere with clustering. EYFP-SLP-76 WT and EYFP-SLP-76 K56E mutant cluster formation in Jurkat J14 cells on anti-CD3 coated coverslides (0–125 s). (i) Maximum overtime images; (ii) average number of microclusters; (iii) mean speed of EYFP-SLP-76 WT (black dots) and EYFP-SLP-76 K56E (white dots) clusters; (iv) size of microclusters; (v) trafficking analysis (n = 3).
(B) SLP-76 K56E associates normally with ADAP, GADS, and PLCγ1. J14 cells transfected with HA-SLP-76 WT or HA-SLP-76 K56E were stimulated by anti-CD3 followed by precipitation with anti-SLP-76 and blotting (n = 3).
(C) K56E supports calcium mobilization. Intracellular calcium levels were measured in J14 T cells transfected with HA-SLP-76 WT or HA-SLP-76 K56E mutant upon anti-CD3 stimulation (left) or ionomycin treatment (right) (n = 4). Also related to Figure S3A.
(D) SLP-76 WT- and SLP-76 K56E-expressing cells show the same levels of NFATc1 dephosphorylation. Transfected as above and stimulated with PMA/ionomycin, or anti-CD3 for various times followed by blotting with anti-NFAT1c1 (left) (n = 3). Histogram shows the similar decreasing phospho-NFATc1/NFATc1 ratio (right).
NFATc1 dephosphorylation by calcineurin is a prerequisite for its nuclear translocation (Crabtree and Olson, 2002; Jain et al., 1992). J14 cells expressing K56E and WT SLP-76 showed a similar decrease in phosphorylated versus dephosphorylated NFAT in response to anti-CD3 ligation (Figure 3D). Further, a measurement of calcineurin phosphatase activity showed a similar increase in K56E and WT SLP-76-expressing cells (data not shown). These data indicated that K56E had no obvious effect on key aspects of proximal signaling by SLP-76.
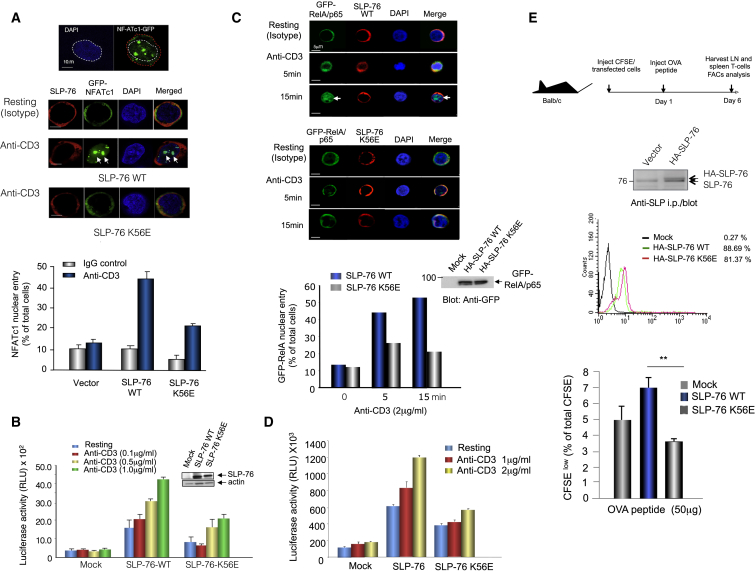
SLP-76-RanGAP1 Binding Is Needed for Optimal NFATc1 and Rel/p65 Nuclear Entry
Despite having no effect on proximal signaling, the K56E mutant impaired NFATc1 entry into the nucleus. GFP-tagged NFATc1 nuclear translocation was imaged in J14 T cells expressing wild-type and K56E SLP-76 (Figure 4A). Transfected cells were incubated with anti-CD3 or isotype control (resting) for 5 min followed by anti-HA-Alexa Fluor 633 staining. Cells were scored positive when >70% of total GFP-NFATc1 overlapped with DAPI nuclear staining as quantified by ImageJ (Figure 4A, upper images; Figure S3B). Anti-HA staining was used as an internal control for WT or K56E HA-SLP-76 expression. While 45% of SLP-76-expressing cells showed nuclear NFATc1-GFP in response to anti-CD3, only 18% of cells expressing the K56E showed nuclear entry (Figure 4A, lower histogram). These data showed that SLP-76–RanGAP1 is needed for optimal anti-CD3-induced NFATc1 nuclear entry. K56E also impaired anti-CD3-induced transcription of an NFAT-driven IL-2 reporter construct by 50%–60% at different anti-CD3 concentrations (Figure 4B). Coexpression of SLP-76 and RanGAP1 also synergised to enhance anti-CD3-induced NFATc1 transcription, an effect impaired by K56E (Figure S4A). Anti-SLP-76 blotting confirmed similar transfected SLP-76 wild-type and K56E expression, comparable to endogenous SLP-76 in Jurkat cells (upper left inset). Anti-HA-blotting showed similar RanGAP1 expression in transfected cells including the higher MW SUMO-RanGAP1 (upper right panel). As a control, mRFP-RanGAP1 transfection showed exclusive localization to the cytoplasm (Figure S4B). K56E expression also impaired NFAT transcription induced by ionomycin/phorbol ester (PMA), indicating that the SLP-76-RanGAP1 was needed for NFAT transcription induced by receptor-independent activation (Figure S4C). Intracellular staining for IL-2 also showed its reduced presence in SLP-76 K56E-expressing cells (Figure S4D).
Figure 4.
RanGAP1-SLP-76 Is Needed for Optimal NFATc1 and p65/RelA Nuclear Entry and In Vivo Responses to Antigen
(A) SLP-76 K56E shows impaired anti-CD3-induced-NFATc1 nuclear translocation. Images of cytoplasmic and nuclear GFP-NFATc in J14 T cells expressing HA-SLP-76 WT or HA-SLP-76 K56E. (Upper panel) Example of lines that delineate DAPI stained nucleus (white dotted line) from total cell (red dotted line). Also see Figure S3B. Histogram showing percentage of cells with nuclear NFATc1 (lower). Data are represented as a means of ± SD from a representative experiment (n = 3).
(B) SLP-76 K56E impairs anti-CD3-induced NFAT transcription activity. Anti-CD3 activation of Jurkat J14 cells with SLP-76 WT or SLP-76 K56E and NFAT (3x) luciferase reporter. (Inset) Expression levels of transfected SLP-76WT and mutant K56E. Data are means of ± SD from a representative experiment (n = 4).
(C) SLP-76 K56E impairs NFκB p65 (RelA) nuclear entry. Anti-CD3 stimulated J14 Jurkat cells expressing GFP-RelA/p65 and HA-SLP-76 or HA-K56E. Image of cells with SLP-76 (upper) or K56E (middle). Histogram showing percentage GFP-p65/RelA entry into nucleus (lower). (Upper inset) Blotting with anti-GFP show equal GFP-RelA/p65 in SLP-76 WT and K56E cells.
(D) Luciferase NF-kB reporter levels impaired in K56E-expressing J14 cells. Cells were transfected with SLP-76 or K56E plus a luciferase NF-kB reporter construct of interleukin 2 promoter. Data are means of ± SD from a representative experiment (n = 2). Also see Figure S4.
(E) RanGAP1-SLP-76 binding is needed for optimal in vivo responses of DO11.10 T cells to OVA peptide. CFSE-labeled DO11.10 CD4+ T cells were i.v. injected into the tail of Balb/c mice, followed by I.V. injection of 50 ug OVA- peptide 24 hr later. Experimental design (upper). Intracellular staining of transfected cells for SLP-76 wild-type and K56E (middle). (Inset) Anti-SLP-76 blotting of cell lysate showed that the level of expressed HA-SLP-76 is similar to endogenous SLP-76. Proliferation as measured by FACs analysis of CFSE intensity at day 6 (percentage CFSE low relative to total CFSE) (lower). Data are means of ± SD from representative experiments. Differences between means were tested using two-tailed unpaired Student’s t test; ∗p < 0.05 was considered significant.
SLP-76 K56E also inhibited the translocation of transport of NFκB p65/RelA into the nucleus (Figure 4C). GFP-RelA/p65 and HA-SLP-76 were cotransfected into J14 Jurkat cells, stimulated with anti-CD3, and imaged by confocal microscopy. SLP-76 K56E showed a reduction of 45% of GFP-RelA/p65 nuclear entry at 5 min and 65% at 15 min relative to WT SLP-76 (upper panels and lower histogram). Similar levels of GFP-RelA/p65 were expressed in cells with HA-SLP-76 WT and K56E (inset). Consistently, the anti-CD3-induced NF-κB driven luciferase reporter activity was 50%–60% lower in cells expressing K56E SLP-76 compared to WT SLP-76 (Figure 4D).
SLP-76-RanGAP1 Binding Is Needed for Optimal T Cell In Vivo Responses to Antigen
Given this, we next assessed whether the SLP-76-RanGAP1 interaction affected T cell responses in vivo (Figure 4E). DO11.10 transgenic T cells were transfected with either SLP-76-K56E or WT SLP-76, labeled with CFSE, and injected i.v. into Balb/c mice. Twenty-four hours later, 50 μg OVA peptide was then injected i.v. into mice (Greenwald et al., 2001). On day 6, spleens were extracted for FACs analysis (upper panel). Intracellular staining confirmed similar SLP-76 and K56E expression (middle panel). HA-SLP-76 was expressed similar to endogenous SLP-76 (inset). We found that while HA-SLP-76 WT supported OVA peptide-induced T cell proliferation, K56E-expressing cells were markedly impaired in their in vivo response to peptide (lower panel). These data confirmed that the SLP-76-RanGAP1 interaction plays a significant role in in vivo responses of T cells to antigen.
Discussion
Although the NPC controls the entry of transcription factors into nucleus in response to antigen receptor ligation on T cells, it has been unclear whether cytoplasmic adaptors directly modulate the NPC. The cytoplasmic localization of NPC filaments makes them ideal targets. Our findings now identify immune cell adaptor SLP-76 binding to SUMO-RanGAP1 of the NPC as a mechanism to ensure the optimal transport of NFATc1 and RelA/p65 in the nucleus in response to antigen-receptor ligation. We found that this surprising interaction accounts for at least 40%–50% of NFATc1 and NF-κB p65 entry into nucleus of T cells. The interaction represents a second checkpoint for NFATc1 nuclear entry, and may reflect the immense pressure on T cells to induce a rapid response to infection.
SLP-76 bound to RanGAP1, an event increased by TCR antigen receptor ligation. The principle form was SUMO-RanGAP1 that is exclusively associated with NPC filaments. SUMO1 is added to residue K526 of RanGAP1 and regulates the Ran-GDP-GTP gradient for importin α/β-dependent nuclear import (Mahajan et al., 1998). Specificity was shown by the failure of antibodies to ADAP, SKAP1, and CARMA1 to coprecipitate RanGAP1. SLP-76-SUMO-RanGAP1 was seen in mouse and human cell lines and primary cells, and appeared to occur maximally as a later event following TCR ligation. Its presence was confirmed by mass spectrometry after a late activation period, but is less evident in precipitates from T cells activated over shorter periods (Roncagalli et al., 2014). Remarkably, TEM showed that 76% of NPCs had at least one gold particle of SLP-76 with an average distance of 35 nm from the cytoplasmic face of the NPC, a distance comparable to RanGAP1. We found that binding depended on lysine 56 in SLP-76, where its mutation caused a loss of RanGAP1 binding and associated GTP/GDP exchange activity. Whether posttranslational modifications to K56 such as SUMOylation facilitate the interaction is unclear. Preliminary results with MaxQuant matching and Q-exactive failed to show SUMOylation (data not shown). SLP-76 therefore interfaces with the NPC complex by binding to SUMO-RanGAP1 of the cytoplasmic filaments of the pore complex.
Mechanistically, anti-CD3 ligation increased SLP-76 binding to RanGAP1 as well as GAP activity and complex binding to the NPC, in accord with the TEM studies. Antigen receptor modulation of RanGAP1 activity has not been previously reported. Further, SLP-76 affected RanGAP1 by acting on its binding to the NPC and GTP-GDP exchange activity. J14 cells lacking SLP-76, or transfected with K56E, had reduced coprecipitated RanGAP1 with MAb414, and lower RanGAP1 activity when normalized to levels of coprecipitated protein. It was the anti-CD3 stimulated increase (rather than basal activity) that was affected by SLP-76 deficiency, or the K56E mutant. The enhanced presence and activity of RanGAP1 at the NPC are in keeping with its role in regulating the Ran-GDP/GTP gradient (Quimby and Dasso, 2003).
Importantly, we showed that the SLP-76-RanGAP1 regulated the nuclear entry of two key transcription factors, NFATc1 and NF-κB p65. K56E impaired entry of each to the same degree. Further, SLP-76 and RanGAP1 synergized to greatly enhance anti-CD3-induced NFAT-driven transcription, an effect lost with the K56E mutant. Importantly, K56E had no effect on proximal signaling events such as SLP-76 microcluster formation, calcium mobilization, or NFATc1 dephosphorylation. Despite this, the K56E mutant impaired NFATc1-GFP and GFP-RelA/p65 nuclear entry and transcription by 40%–50%. An even more pronounced effect was observed in the in vivo responses of DO11.10 T cells to OVA peptide. NFAT entry therefore involves two steps, canonical dephosphorylation followed by direct SLP-76 binding to RanGAP1 and regulation at the NPC. This second tier of regulation may reflect the need for rapid transcription to respond to infections by viruses and other pathogens. The full range of cargo and the different receptors (i.e., innate versus adaptive) involved in the pathway remains to be determined. However, a broad effect on signaling was suggested by K56E inhibition of receptor-independent stimulation by PMA/ionomycin.
The SLP-76-SUMO-RanGAP1 interaction is unique to immune cells and has evolved to complement generic NPC regulation by RanBP2 that helps to localize SUMO1-RanGAP1 and UBC9 at the NPC. SLP-76 bound to RanGAP1, but not RanBP2, suggesting that SLP-76 is likely to complement RanBP2. Further, K56E preferentially affected the TCR/CD3 increase in the RanGAP1-NPC association in accord with a specific connection to TCR signaling. Whether SLP-76 homologs with a similar function exist in other cell types such as neuron cells remains to be determined.
Whether SLP-76 is derived from the cell surface or a distinct pool is unclear. SLP-76 microclusters accumulate in a perinuclear structure (Bunnell et al., 2002). It is also a later event, peaking at 15–20 min postligation, consistent with time needed to move from the cell surface to the nucleus. We postulate that SLP-76 surface clusters may eventually migrate and regulate the NPC. Alternatively, a separate pool of compartmentalised SLP-76 could also interact. Future studies will determine the range of cargo regulated by the SLP-76-RanGAP1 complex and whether it is needed for nuclear entry induced by other receptors such as Toll-like receptors (TLRs).
Experimental Procedures
Cell Culture and Reagents
Jurkat, SLP-76-deficient J14, T8.1, and DC27.10 cells were cultured as previously described (Liu et al., 2010; Raab et al., 1999; Schneider et al., 2008). Antibodies included anti-human CD3 (OKT3), hamster anti-mouse CD3 145- 2C11 (ATCC), anti-SLP-76 (BioXcell, West Lebanon, NH), anti-RanGAP1 (Santa Cruz Biotechnology, USA), Mab414 (Covance), anti-SUMO1 (gift from R. Hay, Dundee, UK), anti-Carma1 (Cell Signaling USA), anti-HA (Covance), anti-Lck, anti-ADAP, anti-SKAP1, and anti-GADS (Upstate Biotechnology, USA), anti-NF-ATc1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-RanBP2 (Thermo Scientific), and anti-actin (Sigma-Aldrich, USA). Carboxyfluorescein diacetate succinimidyl ester (CFSE) was purchased from Sigma (Poole, UK). HA-tagged SLP-76 and RanGAP1, SLP-76-YFP, and the K56E SLP-76 mutant were constructed as described in the Supplemental Experimental Procedures.
Transfection and Stimulation
Mouse and human T cells were stimulated with 2–5 μg/ml of 145-2C11 or OKT3 as described (Raab et al., 1999). Jurkat and J14 T cells were transfected by microporation (Digital Bio Technology) using a single pulse of 30 ms at 1410 V, while DC27.10 cells received 2 pulses of 20 ms at 1,400 V. Human primary T cells were transfected using a single pulse of 20 ms at 2,259 V. Primary naive mouse cells were transfected with various vectors using the Amaxa Nucleofector Kit (Lonza, Germany).
RanGAP1 GAP Activity Assays
RanGAP1 activity was quantified using either a Phos-Free Phosphate Assay Kit (Cytoskeleton Inc., Denver, CO) or a radioactive assay as described in Bischoff et al. (1994) and in the Supplemental Experimental Procedures. RanGAP1 GAP activity measured by isotope labeling (Figures 2B and S2A), or using a nonradioactive protocol (Figure S2B), ∗∗p < 0.01, ∗∗∗p < 0.001 versus SLP-76 WT. The average results were shown from three independent experiments.
Immune Precipitation and Nuclear Pore Fraction
Precipitation and blotting were performed as described (Raab et al., 2011). Nuclear pore fractions were separated from cytoplasmic fractions of DC27.10 T cells by using the Thermo NE-PER Nuclear and Cytoplasmic Extraction Kit (Fischer Scientific).
Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
Bands from anti-SLP-76 precipitation from DC27.10 or T8.1 T cells were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (conducted by Cambridge Centre for Proteomics, Cambridge University) as described in the Supplemental Experimental Procedures. Database searches of the mass fingerprint data were performed using Mascot (http://www.matrixscience.com). For tandem ms/ms analysis, desalted peptides were delivered to a ThermoFinnigan LCQ Classic ion-trap mass spectrometer using a static nanospray needle (Thermo Proxeon). Fragment ions were matched to possible sequence interpretations using MS-Product and/or MS-Tag (http://prospector.ucsf.edu/).
Immunofluorescence and Confocal Imaging
Immunofluorescence and confocal imaging were conducted as previously described (Liu et al., 2010; Purbhoo et al., 2010) and as described in the Supplemental Experimental Procedures. J14 cells were cotransfected with GFP-NFATc1 or GFP-RelA/p65 together with either HA-SLP-76 WT or HA-SLP76 K56E.
In Vivo Adoptive T Cell Transfer Assay
Adoptive T cell transfer assay were conducted as described in the Supplemental Experimental Procedures. DO11.10 T cells were transfected with HA-SLP-76 or HA-SLP-76 K53E and labeled with CFSE (5 μM). Proliferation of CFSE labeled T cells was assessed 6 days after the OVA-peptide injections by FACs analysis.
Transmission Electron Microscopy
TEM was conducted as described (Zuleger et al., 2011). Images were taken on a Hitachi H7600 electron microscope. In order to measure the distances, since the cytoplasmic ring could not be directly visualized, first the distance from the central plane to the particle was measured, then the known distance (40 nm) from the central plane to the top of the cytoplasmic ring (Maimon et al., 2012) was subtracted to give the ring-label distance.
Measurement of Intracellular Calcium
SLP-76-deficient J14 cells transfected with pcDNA-SLP-76 WT or K56E mutant were subjected to calcium influx measurement with Indo-1 (Invitrogen) (Parry et al., 2000). For statistical comparisons, the calcium response was normalized and expressed as the ratio of the maximum peak poststimulation to baseline. Kinetic plots were generated on ligated (Indo-1-labeled) cells using FlowJo software.
Promoter Reporter Assays
Promoter Reporter Assays were conducted as previously described (Liu et al., 2010; Raab et al., 1999; Veale et al., 1999) and as described in the Supplemental Experimental Procedures. Luciferase activity was measured using a luminometer (MicroLumat, EG&G Berthold).
Author Contributions
C.E.R. conceived of the original basis of the project and coordinated the activities of coauthors; H.L. and H.S. carried out the biochemical, cellular and imaging studies; A.R. carried out GAP assays and imaging studies; H.L., C.R., and M.W.G. conducted the transmission electron microscopy; C.E.R and H.L. wrote the manuscript with contributions from H.S., A.R., C.R., and M.W.G.
Acknowledgments
The work was supported by the program grant from Wellcome Trust 092627/Z/10/Z (C.E.R.).
Published: August 27, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes four figures, one movie, and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.molcel.2015.07.015.
Supplemental Information
References
- Allen T.D., Rutherford S.A., Murray S., Drummond S.P., Goldberg M.W., Kiseleva E. Scanning electron microscopy of nuclear structure. Methods Cell Biol. 2008;88:389–409. doi: 10.1016/S0091-679X(08)00420-2. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Klebe C., Kretschmer J., Wittinghofer A., Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck Wardenburg J., Fu C., Jackman J.K., Flotow H., Wilkinson S.E., Williams D.H., Johnson R., Kong G., Chan A.C., Findell P.R. Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J., Pappu R., Bu J.Y., Mayer B., Chernoff J., Straus D., Chan A.C. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Diehn M., Yaffe M.B., Findell P.R., Cantley L.C., Berg L.J. Biochemical interactions integrating Itk with the T cell receptor-initiated signaling cascade. J. Biol. Chem. 2000;275:2219–2230. doi: 10.1074/jbc.275.3.2219. [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Hong D.I., Kardon J.R., Yamazaki T., McGlade C.J., Barr V.A., Samelson L.E. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 2002;158:1263–1275. doi: 10.1083/jcb.200203043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel G., Fahrenkrog B. Dynamics and diverse functions of nuclear pore complex proteins. Nucleus. 2012;3:162–171. doi: 10.4161/nucl.19674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree G.R., Olson E.N. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- D’Angelo M.A., Hetzer M.W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva A.J., Li Z., de Vera C., Canto E., Findell P., Rudd C.E. Cloning of a novel T-cell protein FYB that binds FYN and SH2-domain-containing leukocyte protein 76 and modulates interleukin 2 production. Proc. Natl. Acad. Sci. USA. 1997;94:7493–7498. doi: 10.1073/pnas.94.14.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolo V., Montagne B., Salek M., Jungwirth B., Carrette F., Fourtane J., Sol-Foulon N., Michel F., Schwartz O., Lehmann W.D., Acuto O. A novel pathway down-modulating T cell activation involves HPK-1-dependent recruitment of 14-3-3 proteins on SLP-76. J. Exp. Med. 2007;204:681–691. doi: 10.1084/jem.20062066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Grasis J.A., Guimond D.M., Cam N.R., Herman K., Magotti P., Lambris J.D., Tsoukas C.D. In vivo significance of ITK-SLP-76 interaction in cytokine production. Mol. Cell. Biol. 2010;30:3596–3609. doi: 10.1128/MCB.01657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald R.J., Boussiotis V.A., Lorsbach R.B., Abbas A.K., Sharpe A.H. CTLA-4 regulates induction of anergy in vivo. Immunity. 2001;14:145–155. doi: 10.1016/s1074-7613(01)00097-8. [DOI] [PubMed] [Google Scholar]
- Hoelz A., Debler E.W., Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- Hutten S., Flotho A., Melchior F., Kehlenbach R.H. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol. Biol. Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J.K., Motto D.G., Sun Q., Tanemoto M., Turck C.W., Peltz G.A., Koretzky G.A., Findell P.R. Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem. 1995;270:7029–7032. doi: 10.1074/jbc.270.13.7029. [DOI] [PubMed] [Google Scholar]
- Jain J., McCaffrey P.G., Valge-Archer V.E., Rao A. Nuclear factor of activated T cells contains Fos and Jun. Nature. 1992;356:801–804. doi: 10.1038/356801a0. [DOI] [PubMed] [Google Scholar]
- Jordan M.S., Singer A.L., Koretzky G.A. Adaptors as central mediators of signal transduction in immune cells. Nat. Immunol. 2003;4:110–116. doi: 10.1038/ni0203-110. [DOI] [PubMed] [Google Scholar]
- Lee G.W., Melchior F., Matunis M.J., Mahajan R., Tian Q., Anderson P. Modification of Ran GTPase-activating protein by the small ubiquitin-related modifier SUMO-1 requires Ubc9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- Liu J., Kang H., Raab M., da Silva A.J., Kraeft S.K., Rudd C.E. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA. 1998;95:8779–8784. doi: 10.1073/pnas.95.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Purbhoo M.A., Davis D.M., Rudd C.E. SH2 domain containing leukocyte phosphoprotein of 76-kDa (SLP-76) feedback regulation of ZAP-70 microclustering. Proc. Natl. Acad. Sci. USA. 2010;107:10166–10171. doi: 10.1073/pnas.0909112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mahajan R., Gerace L., Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T., Elad N., Dahan I., Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Marinari B., Costanzo A., Viola A., Michel F., Mangino G., Acuto O., Levrero M., Piccolella E., Tuosto L. Vav cooperates with CD28 to induce NF-kappaB activation via a pathway involving Rac-1 and mitogen-activated kinase kinase 1. Eur. J. Immunol. 2002;32:447–456. doi: 10.1002/1521-4141(200202)32:2<447::AID-IMMU447>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F., Guan T., Yokoyama N., Nishimoto T., Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J. Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M.R., Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat. Rev. Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- Musci M.A., Hendricks-Taylor L.R., Motto D.G., Paskind M., Kamens J., Turck C.W., Koretzky G.A. Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J. Biol. Chem. 1997;272:11674–11677. doi: 10.1074/jbc.272.18.11674. [DOI] [PubMed] [Google Scholar]
- Parry C.M., Simas J.P., Smith V.P., Stewart C.A., Minson A.C., Efstathiou S., Alcami A. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J. Exp. Med. 2000;191:573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Pivniouk V., Tsitsikov E., Swinton P., Rathbun G., Alt F.W., Geha R.S. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- Purbhoo M.A., Liu H., Oddos S., Owen D.M., Neil M.A., Pageon S.V., French P.M., Rudd C.E., Davis D.M. Dynamics of subsynaptic vesicles and surface microclusters at the immunological synapse. Sci. Signal. 2010;3:ra36. doi: 10.1126/scisignal.2000645. [DOI] [PubMed] [Google Scholar]
- Quimby B.B., Dasso M. The small GTPase Ran: interpreting the signs. Curr. Opin. Cell Biol. 2003;15:338–344. doi: 10.1016/s0955-0674(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Raab M., da Silva A.J., Findell P.R., Rudd C.E. Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR zeta/CD3 induction of interleukin-2. Immunity. 1997;6:155–164. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- Raab M., Kang H., da Silva A., Zhu X., Rudd C.E. FYN-T-FYB-SLP-76 interactions define a T-cell receptor zeta/CD3-mediated tyrosine phosphorylation pathway that up-regulates interleukin 2 transcription in T-cells. J. Biol. Chem. 1999;274:21170–21179. doi: 10.1074/jbc.274.30.21170. [DOI] [PubMed] [Google Scholar]
- Raab M., Wang H., Lu Y., Smith X., Wu Z., Strebhardt K., Ladbury J.E., Rudd C.E. T cell receptor “inside-out” pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity. 2010;32:541–556. doi: 10.1016/j.immuni.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Smith X., Matthes Y., Strebhardt K., Rudd C.E. SKAP1 PH domain determines RAPL membrane localization and Rap1 complex formation for TCR activation of LFA-1. J. Biol. Chem. 2011;286:29663–29670. doi: 10.1074/jbc.M111.222661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncagalli R., Hauri S., Fiore F., Liang Y., Chen Z., Sansoni A., Kanduri K., Joly R., Malzac A., Lähdesmäki H. Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor-independent TCR signaling hub. Nat. Immunol. 2014;15:384–392. doi: 10.1038/ni.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd C.E. Adaptors and molecular scaffolds in immune cell signaling. Cell. 1999;96:5–8. doi: 10.1016/s0092-8674(00)80953-8. [DOI] [PubMed] [Google Scholar]
- Rudd C.E., Trevillyan J.M., Dasgupta J.D., Wong L.L., Schlossman S.F. Pillars article: the CD4 receptor is complexed in detergent lysates to a protein-tyrosine kinase (pp58) from human T lymphocytes. 1988. J. Immunol. 2010;185:2645–2649. [PMC free article] [PubMed] [Google Scholar]
- Samelson L.E. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- Schneider H., Guerette B., Guntermann C., Rudd C.E. Resting lymphocyte kinase (Rlk/Txk) targets lymphoid adaptor SLP-76 in the cooperative activation of interleukin-2 transcription in T-cells. J. Biol. Chem. 2000;275:3835–3840. doi: 10.1074/jbc.275.6.3835. [DOI] [PubMed] [Google Scholar]
- Schneider H., Smith X., Liu H., Bismuth G., Rudd C.E. CTLA-4 disrupts ZAP70 microcluster formation with reduced T cell/APC dwell times and calcium mobilization. Eur. J. Immunol. 2008;38:40–47. doi: 10.1002/eji.200737423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R. The origins of NF-κB. Nat. Immunol. 2011;12:686–688. doi: 10.1038/ni.2071. [DOI] [PubMed] [Google Scholar]
- Shibasaki F., Price E.R., Milan D., McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- Shui J.W., Boomer J.S., Han J., Xu J., Dement G.A., Zhou G., Tan T.H. Hematopoietic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell-mediated immune responses. Nat. Immunol. 2007;8:84–91. doi: 10.1038/ni1416. [DOI] [PubMed] [Google Scholar]
- Smith-Garvin J.E., Koretzky G.A., Jordan M.S. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker Y.R., Schneider H., Rudd C.E. TCR and CD28 activate the transcription factor NF-κB in T-cells via distinct adaptor signaling complexes. Immunol. Lett. 2015;163:113–119. doi: 10.1016/j.imlet.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veale M., Raab M., Li Z., da Silva A.J., Kraeft S.K., Weremowicz S., Morton C.C., Rudd C.E. Novel isoform of lymphoid adaptor FYN-T-binding protein (FYB-130) interacts with SLP-76 and up-regulates interleukin 2 production. J. Biol. Chem. 1999;274:28427–28435. doi: 10.1074/jbc.274.40.28427. [DOI] [PubMed] [Google Scholar]
- Wang H., Rudd C.E. SKAP-55, SKAP-55-related and ADAP adaptors modulate integrin-mediated immune-cell adhesion. Trends Cell Biol. 2008;18:486–493. doi: 10.1016/j.tcb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Flotho A., Melchior F. The RanBP2/RanGAP1∗SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase. Mol. Cell. 2012;46:287–298. doi: 10.1016/j.molcel.2012.02.017. [DOI] [PubMed] [Google Scholar]
- Williamson D.J., Owen D.M., Rossy J., Magenau A., Wehrmann M., Gooding J.J., Gaus K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- Wu J., Motto D.G., Koretzky G.A., Weiss A. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- Wu H., Peisley A., Graef I.A., Crabtree G.R. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007;17:251–260. doi: 10.1016/j.tcb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Yablonski D., Kuhne M.R., Kadlecek T., Weiss A. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science. 1998;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- Yablonski D., Kadlecek T., Weiss A. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol. Cell. Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A., Tokunaga M., Dustin M.L., Saito T. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- Zhang W., Sloan-Lancaster J., Kitchen J., Trible R.P., Samelson L.E. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- Zuleger N., Kelly D.A., Richardson A.C., Kerr A.R., Goldberg M.W., Goryachev A.B., Schirmer E.C. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J. Cell Biol. 2011;193:109–123. doi: 10.1083/jcb.201009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.