Abstract
The second messenger molecule cAMP regulates the activation phase of the cAMP signaling pathway through high-affinity interactions with the cytosolic cAMP receptor, the protein kinase A regulatory subunit (PKAR). Phosphodiesterases (PDEs) are enzymes responsible for catalyzing hydrolysis of cAMP to 5′ AMP. It was recently shown that PDEs interact with PKAR to initiate the termination phase of the cAMP signaling pathway. While the steps in the activation phase are well understood, steps in the termination pathway are unknown. Specifically, the binding and allosteric networks that regulate the dynamic interplay between PKAR, PDE, and cAMP are unclear. In this study, PKAR and PDE from Dictyostelium discoideum (RD and RegA, respectively) were used as a model system to monitor complex formation in the presence and absence of cAMP. Amide hydrogen/deuterium exchange mass spectrometry was used to monitor slow conformational transitions in RD, using disordered regions as conformational probes. Our results reveal that RD regulates its interactions with cAMP and RegA at distinct loci by undergoing slow conformational transitions between two metastable states. In the presence of cAMP, RD and RegA form a stable ternary complex, while in the absence of cAMP they maintain transient interactions. RegA and cAMP each bind at orthogonal sites on RD with resultant contrasting effects on its dynamics through parallel allosteric relays at multiple important loci. RD thus serves as an integrative node in cAMP termination by coordinating multiple allosteric relays and governing the output signal response.
Introduction
Signaling pathways are exquisitely regulated by a complex interplay of reversible interactions with partner proteins, ligand cofactors, and posttranslational modifications. These multivalent interactions modulate the cell’s spatiotemporal recognition of and response to extracellular stimuli. Signaling pathways are also characterized by distinct activation and termination phases that govern the duration, intensity, and amplification of the signal as it is propagated through the cell (1). Signaling proteins are intrinsically dynamic and populate multiple conformational states in equilibrium and its ligands/partner proteins alter these conformational equilibria (2–4). Indeed, an overlay of protein dynamics is fundamental for bridging structure and function of signaling proteins and consequently for a molecular understanding of signal transduction (5–7). Reversible protein ligand and protein-protein interactions play a critical role in altering dynamic properties of signaling molecules. At a molecular level, signals mediated by specific ligands or partner proteins are propagated across the target protein from active sites to effector sites through allostery. This allosteric communication from one protein locus to another constitutes the basis of signaling proteins’ function (8,9). Consequently, signaling proteins have distinct loci for binding diverse ligands and partner proteins and these sites are allosterically coupled (10). An emerging challenge in protein chemistry lies in delineating binding interactions from long-range propagation of multivalent allosteric relays in signaling proteins.
Amide hydrogen/deuterium exchange mass spectrometry (HDXMS) has emerged as a powerful tool for mapping allosteric communication in proteins (11,12). The method relies on tracking the acid- and base-catalyzed abstraction of protein backbone amides and replacement by different protons. The rate of amide exchange is dependent on solvent accessibility as well as H-bond propensities and strengths and provides an overview of protein dynamics (13). In addition to mapping allosteric changes in proteins (14,15), HDXMS also has been useful for mapping dynamics of transient interactions in ternary complexes of multiple proteins with ligands and for monitoring progression of enzyme reactions in solution (16). In this study, we set out to apply HDXMS to characterize protein-ligand interactions and map associated allosteric networks in the second messenger cyclic AMP (cAMP)/protein kinase A (PKA) signaling pathway. In this pathway, a single protein (regulatory subunit) functions as a cAMP receptor and interacts with two important effector proteins: the kinase (catalytic subunit) and a phosphodiesterase (PDE) (17–19). In this study we describe how this protein functions as an integrative node in the signaling pathway by responding allosterically in myriad ways to cAMP and two antagonistic effector proteins to modulate the output response.
The second messenger 3′, 5′- cyclic adenosine monophosphate (cyclic AMP) transduces the effects of external hormonal stimulation and mediates a myriad of intracellular responses. In Dictyostelium discoideum, cAMP is both a chemoattractant and intracellular signaling molecule (20,21). Its most profound role is in initiating important physiological changes associated with transformation from a unicellular to a multicellular state in response to starvation (22,23). One of the main targets of cAMP is cyclic AMP-dependent protein kinase, also referred to as PKA, which consists of catalytic (C) and regulatory (R) subunits. In the absence of cAMP, PKA exists as an inactive complex of R- and C-subunits (referred to as the holoenzyme) (24). cAMP binding to the holoenzyme induces conformational changes and facilitates the release of the active C-subunit (17,25,26). This constitutes the activation phase of the cAMP signaling pathway. PDEs, enzymes responsible for catalyzing hydrolysis of cAMP to 5′AMP, initiate the termination phase of the cAMP signaling pathway by forming direct interactions with the cyclic-nucleotide-binding (CNB) pocket domains of PKA R-subunit and hydrolyzing the bound cAMP (Fig. 1A) (19,27). The R-subunit of D. discoideum (henceforth referred to as RD) differs from its mammalian homologs in being monomeric, and lacks an N-terminal dimerization domain, but contains two canonical cyclic AMP binding sites in two distinct domains, CNB domains A and B (denoted CNB:A and CNB:B) (Fig. 1B) (28–30). The CNB domains have a conserved fold with a cAMP binding site containing a characteristic motif for cAMP binding (Fig. 1B, inset). The binding site forms a buried pocket shielding cAMP from the action of phosphodiesterases (31). Previous studies have shown that in RD, CNB:A binds to cAMP with high affinity, while cAMP binding to CNB:B has not been detected and assumed to be a low-affinity site, despite having all the motifs typical of high-affinity cAMP binding domains across PKA R-subunits (28,30).
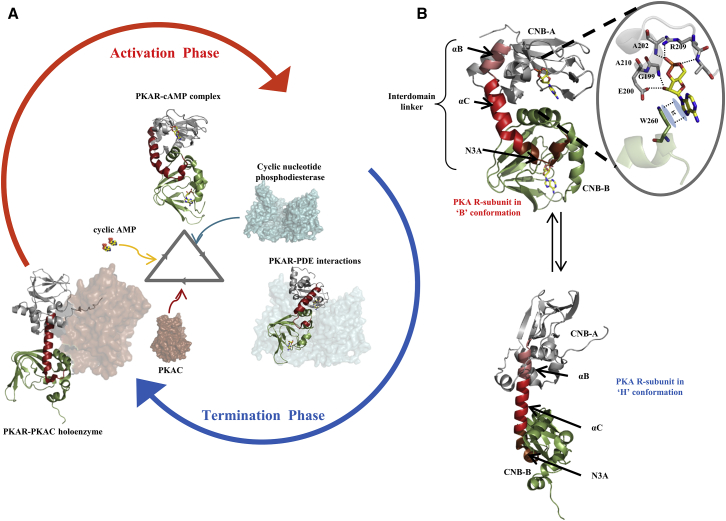
Figure 1.
Dynamic modulators of PKA regulatory subunit in cAMP signaling pathway. (A) The PKAR exhibits multiple end-point conformations in the course of the cAMP signaling pathway. Here PKAR is represented in cartoon form with (gray) CNB:A domain and (green) CNB:B domain, with the dynamic interdomain linker (B/C helix) (red) and cAMP (yellow sticks). In its inactive form, PKAR binds to PKA catalytic subunit (PKAC, brown surface representation) to form the inactive holoenzyme complex (H form) (PDB: 2QCS). In this form, PKAR is in an extended form. cAMP (yellow sphere representation) binds to PKAR, which in turn releases and activates PKAC. In the cAMP-bound form (B form), PKAR adopts a compact conformation (PDB: 1RGS). The transition from holoenzyme form to the cAMP-bound form constitutes the activation phase of the cAMP signaling pathway. In the termination phase, cyclic nucleotide PDEs form direct interactions with cAMP-bound PKAR (docking model (27)) and cause the release and dissociation of bound cAMP. Free PKAR can now bind PKAC and regenerate the holoenzyme, thereby terminating the cAMP pathway. PKAR is a dynamic protein and its conformations are modulated by PKAC, cAMP, and PDEs in different stages of the cAMP signaling pathway. (B) The interdomain linker is important in conformational transitions of PKAR from the cAMP-bound B form and the holoenzyme H form. The interdomain linker is composed of the α:B helix (in salmon), α:C helix (in red), and N3A helix (in brown). In the B form, the linker is kinked into three distinct segments, while in the H form, the linker adopts an extended helical conformation. Also depicted are the interactions that cAMP makes with residues in the cAMP binding pocket of CNB:A in PKAR (inset). Multiple hydrogen bonds (dotted lines) along with π stacking between W260 and the adenine ring of cAMP secure the molecule in the binding pocket.
RD is the primary receptor of cAMP and mediates multivalent interactions with the C-subunit and phosphodiesterases. The D. discoideum phosphodiesterase RegA, important in different stages of cell development, was shown to interact with PKA R-subunit and control PKA-mediated differentiation of prestalk and spore cells, and regulate encystation (22). We have earlier shown that the catalytic domain of RegA (RegAC) is sufficient for hydrolysis of cAMP tightly bound to the mammalian R-subunit (19). Distinct conformations of mammalian R-subunit have been captured by crystallography (32) and NMR (33–35) and the endpoint states have been denoted as B (cAMP-bound) and H (C-subunit or inactive) forms (36). These conformational transitions have been the focus of several studies to map the allosteric transitions in response to cAMP and C-subunit binding (33,37,38). There have been far fewer studies on how PDEs interact to hydrolyze cAMP bound to PKA R-subunit and initiate cAMP signal termination (19,27). Further, how signals in the cAMP signaling pathway converge and branch off from a target through alternate allosteric pathways is of enormous interest. RD is the primary cAMP receptor and a great model system for mapping interactions with ligand, cAMP, and partner proteins (RegAC). This has been achieved using HDXMS, which allows monitoring cAMP through the activation and termination phases of the pathway by following backbone amide hydrogens as reporters across RD. In the course of following cAMP, we also describe binding and allosteric networks associated with signal termination in cAMP signaling mediated via RD-RegAC-cAMP interactions.
Our results reveal how cAMP and the effector protein, RegAC, interact with RD at spatially distal sites, allosterically coupled to regulate the output response. We describe how PDEs access cAMP bound to the receptor, R-subunit, to mediate signal termination in cAMP signaling. We show that effects of cAMP binding and PDE interactions are transmitted through distinct, nonoverlapping allosteric relays. This underscores how the intracellular cAMP receptor, RD, functions as an integrative node by mediating multiple interactions with effector proteins, to regulate the output response.
Materials and Methods
Materials
A plasmid encoding full-length RD with codon optimization for Escherichia coli expression was obtained from DNA2.0 (Menlo Park, CA). Chemically competent E. coli BL21 (DE3) bacterial strains were from Life Technologies (Carlsbad, CA). TALON Cobalt affinity resin for His-tag purification was from Clontech (Mountain View, CA). LC/MS grade acetonitrile and water were from Fisher Scientific (Waltham, MA). Poroszyme immobilized pepsin cartridge was from Applied Biosystems (Foster City, CA); deuterium oxide (99.9%) was from Cambridge Isotope Laboratories (Tewksbury, MA). All other reagents were research grade from Sigma-Aldrich (St. Louis, MO).
Protein expression and purification
RD was first subcloned into pET28a plasmid and transformed into E. coli BL21 (DE3) for bacterial expression and purification. Bacterial cell pellet (15 g) was subject to sonication in lysis buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, and 5 mM β-mercaptoethanol) for 20 min. Lysate was centrifuged at 17,000 × g for 30 min and supernatant was then incubated with Cobalt metal affinity chromatography resin for 1 h. RD was eluted in lysis buffer containing 300 mM imidazole. Eluate was subsequently subjected to size-exclusion chromatography on a Superdex S-200 column (GE Healthcare Life Sciences, Marlborough, MA) as a finishing step. Purity of the purified RD was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. We factor that cAMP in E. coli would bind RD during expression and this dissociates during size-exclusion chromatography to generate apo RD. GST-tagged catalytic domain of RegA (RegAC) was purified as previously described in Moorthy et al. (19). Briefly, GST-tagged RegAC was overexpressed in E. coli BL21 (DE3) and purified using glutathione sepharose 4B resin-based (GE Healthcare Life Sciences) affinity chromatography followed by size-exclusion chromatography.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were carried out to monitor the binding and determine the dissociation constant of cAMP and RegAc to apo RD protein using a VP-ITC MicroCalorimeter (MicroCal, Northampton, MA). The cell reservoir was filled with 1.8 mL of 10 μM RD protein, reference cell with 1.8 mL of buffer (20 mM Tris-Cl pH 7.5, 100 mM NaCl) and syringe with final volume ∼400 μL of 200 μM cAMP prepared in the same buffer. The binding reaction was started with first injection volume of 2 μL followed by 39 sequential injections each of 4 μL cAMP at intervals of 240 s and set to stirring speed of 350 rpm. Continuous measurement of heat change inside the cell allowed determination of enthalpy change during the process ΔH (−53.7 kcal/mol ± 1.6 kcal/mol) and the equilibrium association constant KA (9.92 × 105 ± 1.02 × 104 M−1, dissociation constant (KD) ∼1 μM. Three independent titration experiments were carried out at 298 K with similar results and a representative ITC graph is depicted in Fig. S2 in the Supporting Material. Similar experiments were carried out for probing binding of RegAC to RD (data not shown).
Amide hydrogen/deuterium exchange mass spectrometry
Deuterium exchange experiments were first carried out on cAMP-free (apo) RD (4 μM final concentration). To test effects of cAMP on RD dynamics, cAMP at final reaction concentration of 300 μM, was added to apo RD. This high concentration of cAMP was used to fully saturate the CNB:A site and possibly the CNB:B site, which has been predicted to not bind at lower (<μM) concentrations of cAMP (28). To capture dynamics of apo RD in a binary complex with RegAC, RegAC (2 μM) was complexed with apo RD (6 μM) in a 1:3 molar ratio of RegAC to RD. A ternary complex of RD, cAMP, and RegAC was obtained by complexing RegAC (2 μM) with apo RD (6 μM) in the presence of 300 μM cAMP. In all conditions, complexation reaction was initiated simultaneously with the deuterium exchange reaction in the presence of deuterated buffer.
Buffer for deuterium exchange reaction was prepared by vacuum evaporation of aqueous buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl) until dry, which was subsequently reconstituted in 99.9% D2O. The deuterium exchange reaction was initiated by diluting the sample 10-fold in deuterated exchange buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl) resulting in a final deuterium concentration of 90% in the deuterium labeling reaction. The deuterium-oxide-exchanged buffer was maintained at a final pHread of 7.5. Deuterium exchange reactions were carried out for the following times: 30 s, and 1, 5, 10, 30, 60, and 100 min. The exchange reaction was quenched by lowering the pHread of the reaction to 2.5, using 0.1% trifluoro acetic acid. Samples were injected onto a nano-UPLC HDX sample manager (Waters, Milford, MA) as described in Wales et al. (39). Online immobilized pepsin digestion and reverse phase liquid chromatography were carried out as previously described in Krishnamurthy et al. (27). Peptides separated from previous liquid chromatography were subsequently sprayed onto a SYNAPT G2-Si mass spectrometer (Waters) acquiring in MSE mode. Continuous instrument calibration was carried out using Glu-fibrinogen peptide, according to manufacturer’s recommendations. Back-exchange factor for the system was found on average to be ∼30% (19), but in this study only the uncorrected raw values are reported.
Peptide identification and mass spectral data analysis
Peptides from MSE spectra of undeuterated samples were identified using PROTEIN LYNX GLOBAL SERVER, Ver. 3.0 (Waters). Deuterium exchange data was quantified by DYNAMX software (Ver. 2.0, Waters). The reported values are an average of three independent hydrogen/deuterium exchange experiments (Table S1 in the Supporting Material). Peptides showing EX1 kinetics of amide exchange were additionally analyzed using GRAPHPAD PRISM 6.0 (San Diego, CA). The bimodal deconvolutions were carried out by fitting the curves to a sum of Gaussian equation. Amplitudes and centroid values for each deconvoluted envelope are reported in Table S2.
Results
RD CNB:B domain exhibits distinct HDX bimodal spectra consistent with local unfolding
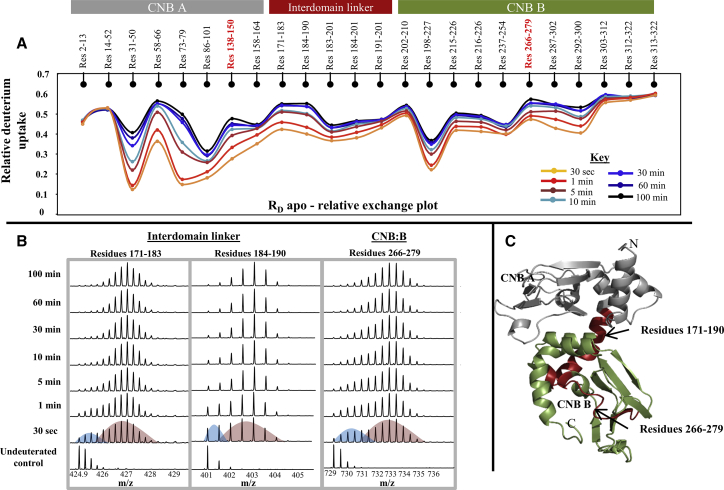
It has been previously shown that the mammalian PKA R-subunit is an intrinsically dynamic protein that forms high-affinity complexes with cAMP (KD ∼ nM), and cAMP-mediated allostery in the R-subunit has been explained by conformational selection (36). PKA is a highly conserved protein widely found in all eukaryotes (40,41), and we hypothesized that RD from D. discoideum with ∼51% sequence homology with mammalian PKA regulatory domain isoform Iα would exhibit similar dynamic properties to its mammalian homolog. To test this, we first set out to characterize protein dynamics of apo RD. HDXMS experiments of apo RD were carried out as described in the Materials and Methods and a total of 24 peptides was identified and analyzed, corresponding to ∼71% of the primary sequence of RD (Fig. S1). The overall dynamic profile of RD is provided in a relative exchange plot (Fig. 2A). A relative exchange plot is a plot of the ratio of average deuterons exchanged to the total number of exchangeable amides available for each peptide. Regions of the protein that are dynamic in nature have higher relative deuterium uptake values.
Figure 2.
Apo RD is a dynamic protein with regions that undergo slow structural transitions/local unfolding. (A) The relative deuterium uptake value (calculated as the ratio of average deuterium ions exchanging to the maximum exchangeable amides; y axis) for each pepsin digest fragment from the N- to C-terminus (x axis) of RD is plotted in a relative exchange plot. The relative exchange plot provides a snapshot of the overall dynamics of RD for each deuterium labeling time point as depicted in the key. Peptides spanning the cAMP binding pocket in CNB A and B are highlighted (red). Plots were generated using the software DYNAMX. (B) Stacked mass spectra of the three peptides exhibiting bimodal distributions are shown. Mass spectra are stacked in order of increasing deuterium labeling time (y axis) as shown. Colored curves are used to represent lower-exchanging (blue curve) and higher-exchanging (red curve) distributions in the 30-s labeling time of spectra. (C) Peptides showing bimodal characteristics are mapped (red) onto the modeled structure of RD. (Gray) CNB:A (residues 1–180) and (green) CNB:B (residues 181–327); the N- and C-termini of the protein and CNB pocket in A and B domains are labeled.
Closer examination of the mass spectral envelopes of all peptides revealed that three regions showed a bimodal distribution of mass spectra (Fig. 2B). Bimodal distributions are characterized by the presence of multiple spectral envelopes within the same mass spectrum. The most common reason for the presence of bimodal spectra in HDXMS experiments is attributed to the EX1 deuterium exchange regime (42,43). EX1 kinetics are rarely seen in soluble proteins at physiological conditions and are indicative of local unfolding events coupled to slow conformational changes (11,13,44). They also report on the dynamic interconversion between alternate population states of the protein (45). EX1 kinetics can serve as conformational probes and provide important insights into protein dynamics and function (44,46). Mass spectra from shorter deuterium exchange labeling time experiments (30 s to 5 min) exhibited bimodal characteristics, but as labeling time increased, the bimodality of the spectrum decreased and shifted toward a binomial spectrum. These phenomena are typical of EX1 kinetics in HDXMS experiments (47). The peptides exhibiting bimodal distribution were found to span loci important for RD function, mainly the putative C-helix (Residues 171–201) and the conserved arginine containing the CNB region in domain B (Residues 266–279) (48). Deconvolution of the bimodal spectral envelopes confirmed EX1 kinetics, and the interpretation of these results is described in a later section (see Fig. 6A). A peptide spanning residues 191–201 is binomial in nature (data not shown), thus by subtractive analysis we can localize the region showing bimodal distribution to the residues 171–190. To better visualize these results, the results were mapped onto the three-dimensional model of RD (Fig. 2C). A structural model for RD was generated using the structure of a close homolog, PKA RIα from Bos taurus (PDB: 1RGS) using SWISS-MODELER (http://swissmodel.expasy.org/).
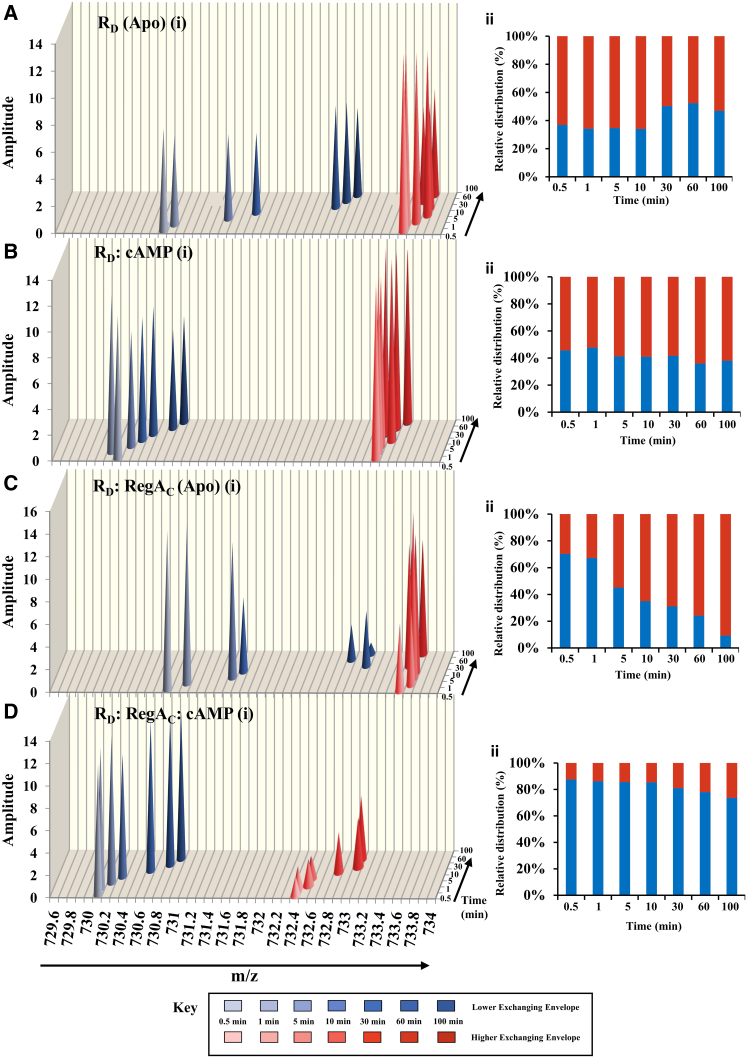
Figure 6.
Deconvolution of EX1 deuterium exchange kinetics for a CNB:B domain peptide (residues 266–279). Stacked three-dimensional graphs represent the distribution of the bimodal kinetics of the RD in its different conditions. (A) apo RD; (B) RD with excess of cAMP; (C) RD:RegAC binary complex in absence of ligand; and (D) RD:RegAC:cAMP ternary complex. Each peak depicts average amplitude value (y axis) for the respective centroid values (x axis) of the lower-exchanging (shades of blue) and higher-exchanging envelopes (shades of red) at different time points (Z axis). The intensities of the two populations were calculated using an equation for the sum of two Gaussians on GRAPHPAD PRISM 6.0. Relative envelope distributions (percentages) were calculated by normalizing the intensities of peaks from each envelope relative to sum total intensities. Y axes represent percentage of the normalized intensities and x axes show deuterium exchange (min). (Blue bars) Relative intensities of the distribution of lower-exchanging envelopes; (red bars) those for the higher-exchanging envelope. The corresponding intensities and deuterium exchange values are listed in Table S2.
HDX bimodal signatures as conformational probes to monitor perturbations in protein dynamics
Our results indicated at least three distinct loci showing bimodal kinetics of deuterium exchange and therefore highlighting these sites as undergoing slow conformational transitions in apo RD. We next examined the effects of cAMP on the overall dynamics of RD, including the above loci by HDXMS analysis of RD, in the presence of 300 μM cAMP. Like other regulatory subunits from homologous protein kinases, the RD CNB:A site has been shown to have a high affinity for cAMP with a dissociation constant in nanomolar range, KD = ∼3 nM (29,49). On the other hand, the binding affinity of cAMP at the CNB:B site, measured using isothermal titration calorimetry, was found to be KD ∼1 μM (Fig. S2). Knowledge of the binding constants guided concentrations of cAMP used (300 μM) in the HDXMS experiments to ensure ligand saturation of the cAMP binding pocket at both the high-affinity CNB:A and the low-affinity CNB:B domains in the HDXMS experiments.
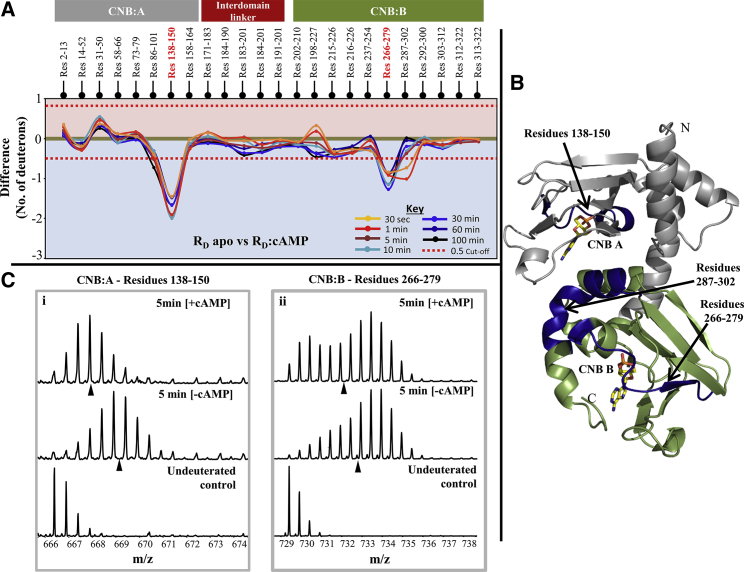
Results from cAMP-bound RD were compared with apo RD and revealed three regions showing significant differences in deuterium exchange. A difference plot depicting differences between apo RD and RD:cAMP is shown in Fig. 3A. This plot shows the average difference between the two states for each peptide and deuterium labeling time. The average error between replicate runs for each peptide was between 0.1 and 0.2 Da in our experimental setup, and a difference of 0.5 Da or more is considered significant (50). Two of the three regions showed significant decreases in deuterium exchange and are located within the cAMP binding pockets of CNB:A and CNB:B, respectively. A third region spanning residues 287–302 also showed significant differences at shorter deuterium labeling times, but at increased labeling times the difference between apo RD and RD:cAMP were negligible. This region is distal to both cAMP binding pockets and represents an allosteric effect upon ligand binding. We also observed increased deuterium exchange in a peptide spanning residues 31–50, which flanks the putative PKAC binding site. These results are mapped onto the modeled structure of RD in Fig. 3B. Representative mass spectra for peptides in CNB:A and CNB:B are shown in Fig. 3C.
Figure 3.
Effects of cAMP binding on RD dynamics. (A) Difference plot, plotting absolute difference in deuterons (y axis) between apo RD and RD:cAMP for each pepsin fragment peptide listed from the N- to C-terminus (x axis). Points in the negative scale (shaded blue) represent a decrease in deuterium exchange upon cAMP binding, while points in the positive scale (shaded red) represent increases in deuterium exchange upon cAMP binding. Peptides spanning the cAMP binding pocket in CNB:A and B are highlighted (in red). Each deuterium labeling time for every peptide is depicted and colored according to key. A difference of ±0.5 Da is considered significant (red dashed line). Plots were generated using the software DYNAMX. (B) Regions showing significant decreases in deuterium exchange are mapped (blue) onto the modeled structure of RD. cAMP molecules are represented in the structure (yellow sticks). (Gray) CNB:A (residue 1–180) domain and (green) CNB:B domain (residues 181–327); the N- and C-termini of the protein and CNB pockets in A and B domain are labeled. (C) Stacked mass spectra of peptides spanning residues (i) 138–150 in CNB-A and (ii) 266–279 in CNB-B are shown. Mass spectra compares the ligand free state to the ligand bound at 5-min deuterium labeling time. (Black triangles) Centroids for the spectra.
Of the three peptides showing bimodal kinetics of deuterium exchange in apo RD, only one of the peptides spanning residues 266–279 at CNB:B showed differences in the presence of cAMP. The spectrum exhibited sharper bimodal characteristics, resulting in a shift of the centroid to the left (Fig. 3C ii). This bimodal distribution is seen for the entire time course of the deuterium-labeling experiment. Interestingly, the centroid of the higher-exchanging population did not show any shifts, but the centroid of the lower-exchanging population shifted to the right with increasing labeling time (see Fig. 6B). The spectra for the lower-exchanging species are representative of EX2 deuterium exchange kinetics seen in stably folded regions (11). In comparison, apo RD showed bimodal distributions at shorter labeling times and binomial distribution after 10-min deuterium labeling times (Fig. S2). Interestingly, the interdomain linker with its characteristic bimodal spectra (residues 171–190) did not show any significant differences upon cAMP binding.
RD and RegAC form a stable ternary complex with cAMP
In a previous study, we monitored RegAC mediated cAMP dissociation in mammalian R-subunit prebound to a nonhydrolyzable cAMP analog (Sp-cAMPS) by HDXMS and observed a short-lived ternary complex of RD:RegAC:Sp-cAMPS (16). The main goal of this study was to characterize the dynamics of the ternary complex in greater detail to gain important insights into how the cAMP signal termination pathway is initiated. RD was added at a threefold molar excess to RegAC in the presence of 300 μM cAMP to prime formation of the termination complex. Simultaneous addition of the two proteins with cAMP in the deuterium labeling reaction allowed us to monitor the time-course dependent kinetics/dynamics as the reaction progresses, eventually leading to ternary complex formation. The rationale behind using excess RD was to channel all of the RegAC toward complex formation and to obviate any hydrolysis of cAMP by unbound RegAC in the experiment.
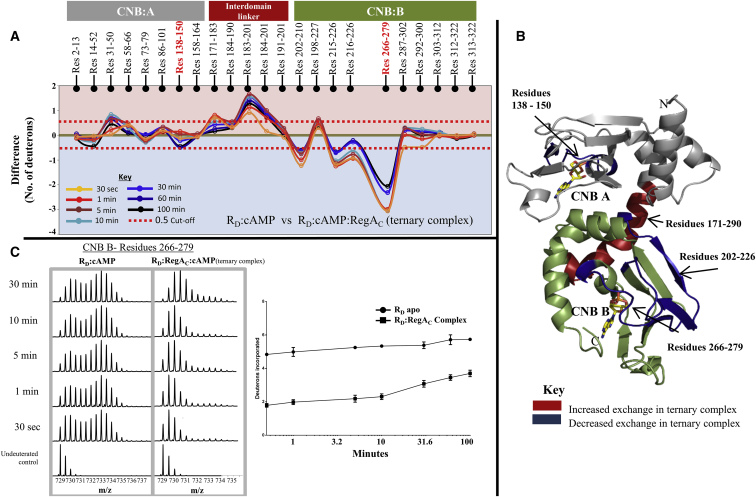
HDXMS results from the RD:RegAC complex in the presence of cAMP provide conclusive evidence for a stable ternary complex at the same loci. Importantly, regions in CNB:B of RD showed decreased exchange for the entire time course of the experiment in the ternary complex. HDXMS results, comparing the ternary complex with RD:cAMP, are summarized in Fig. 4A in a difference plot. It was also seen that the interdomain linker showed increased exchange. Interestingly, we also observed decreased deuterium exchange at cAMP binding pocket at domain A (residues 138–150) in the ternary complex, at longer labeling times.
Figure 4.
Mapping the RD:RegAC:cAMP ternary complex by HDXMS. (A) Difference plot, plotting absolute difference in deuterons (y axis) between RD:cAMP and the RD:cAMP:RegAC ternary complex for each pepsin fragment peptide listed from the N- to C-terminus (x axis). Points in the negative scale (shaded blue) represent a decrease in deuterium exchange in the ternary complex, while points in the positive scale (shaded red) represent increases in deuterium exchange. Peptides spanning the cAMP-binding pocket in CNB A and B are highlighted (red). Each deuterium labeling time for every peptide is depicted and colored according to key. A difference of ±0.5 Da is considered significant (red dashed line). Plots were generated using the software DYNAMX. (B) Regions showing significant differences in deuterium exchange in the ternary complex are mapped onto the modeled structure of RD according to key. (Yellow sticks) cAMP molecules. (Gray) The CNB A (residues 1–180) domain and (green) CNB B domain (residues 181–327); the N- and C-termini of the protein and CNB pocket in A and B domain are labeled. (C) (Left panel) Stacked mass spectra of peptides spanning residues 266–279 in CNB-B domain of RD. Bimodal spectra from RD:cAMP are compared with spectra from the ternary complex. Mass spectra are stacked in order of increasing deuterium labeling time as shown. (Right panel) Deuterium exchange plot comparing RD:cAMP and RD:cAMP:RegAC for the same peptide is depicted. Semilog deuterium exchange plots are generated with x axis in the log scale and y axis in linear scale; error bars are indicated. Plots are generated in GRAPHPAD PRISM 6 (San Diego, CA).
The results from the HDXMS experiment were mapped onto the modeled structure of RD in Fig. 4B. Close examination of the structure showed that the interaction interface can be localized to the cAMP binding pocket of CNB:B. Increased dynamics in the C-helix suggests a mode for allosteric communication between the two cAMP binding pockets. Further confirmation of the stable nature of the ternary complex came from observing the bimodal spectra at residues 266–279. Surprisingly, bimodality was almost negligible, with the entire population shifting to the lower-exchanging, ordered conformation (Figs. 4C and 6D). Deuterium exchange plots also depict the large differences in deuterium exchange between the RD:cAMP and ternary complex (Fig. 4C).
RegAC forms a transient complex with RD in the absence of cAMP
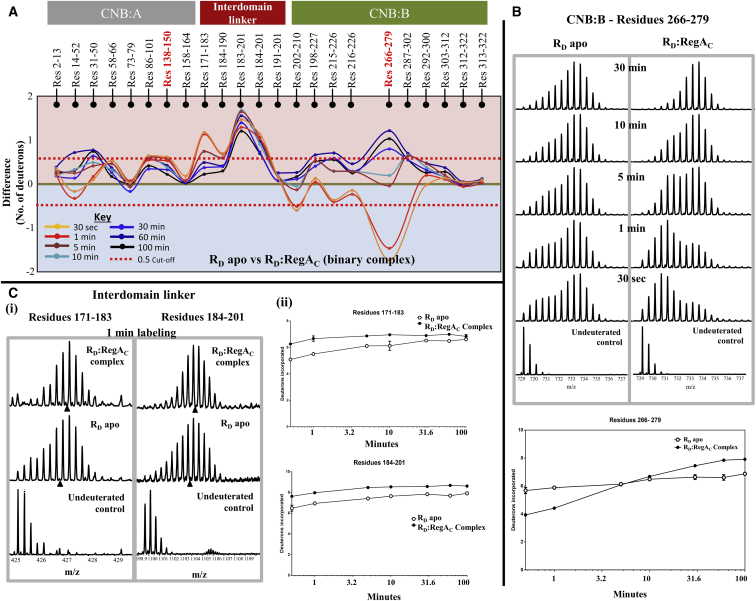
Previously we had shown that the mammalian PKA regulatory subunit (PKAR)-PDE interaction interface between the cAMP binding pocket of mammalian PKAR and the catalytic pocket of PDE is indicative of active site coupling (19,27). While these complexes represent stable endpoint complexes, we set out to monitor the dynamics of the RD-RegAC complex by HDXMS experiments with RD at a threefold molar excess to RegAC in the absence of excess cAMP. ITC experiments revealed no detectable binding between RegAC and apo RD at the experimental conditions, suggesting that the KD for the RD-RegAC complex to be weaker than 1 μM. Importantly, complex formation was initiated simultaneously with the deuterium exchange reaction to better follow steps in complex formation as well as dissociation. HDXMS results from the binary complex were compared with apo RD and are presented as a difference plot in Fig. 5A.
Figure 5.
Mapping transient RD:RegAC interactions by HDXMS. (A) Difference plot, plotting absolute difference in deuterons (y axis) between apo RD and RD:RegAC binary complex for each pepsin fragment peptide listed from the N- to C-terminus (x axis). Points in the negative scale (shaded blue) represent a decrease in deuterium exchange upon RegAC binding, while points in the positive scale (shaded red) represent increases in deuterium exchange upon RegAC binding. Peptides spanning the cAMP binding pocket in CNB:A and B are highlighted (red). Each deuterium labeling time for every peptide is depicted and colored according to key. A difference of ±0.5 Da is considered significant and is shown (red dashed line). Plots were generated using the software DYNAMX. (B) (Top panel) Stacked mass spectra of peptides spanning residues 266–279 in CNB-B domain of RD. Bimodal spectra from apo RD are compared with bimodal spectra from the binary complex. Mass spectra are stacked in order of increasing deuterium-labeling time as shown. (Bottom panel) Deuterium exchange plot comparing apo RD and RD:RegAC for the same peptide is depicted. (C) Representative mass spectra and deuterium exchange plots for two peptides spanning residues 171–183 and 184–201 are shown comparing RD apo state and RD:RegAC state. Semilog deuterium exchange plots are generated with x axis in the log scale and y axis in linear scale; error bars are indicated. All plots are generated in GRAPHPAD PRISM 6.
Most regions of RD showed increased deuterium exchange in the presence of RegAC, implying an overall increase in dynamics of RD in the binary complex. We observed decreased deuterium exchange for many regions in RD domain B for 30 s and 1-min labeling times, but interestingly, this trend is inversed as labeling time increases, with the same regions showing increased dynamics. These results are consistent with the formation of a RD-PDE complex with the interaction interface at RD CNB:B. With increasing labeling time, dissociation of the complex occurs, leading to deuterium exchange even at the interaction interface. While this should eventually result in no differences in deuterium exchange between the apo state and binary complex state, we see increased deuterium exchange at most loci of the CNB-B in the binary complex. Time-dependent differences in deuterium exchange were also observed in contiguous peptides spanning residues 14–52 and 31–50, which showed increased dynamics with increasing reaction time.
We can gain more detailed insights into complex formation and dissociation by using the bimodal spectra as conformational probes, to monitor the degree of disorder in residues 266–279. While ligand binding did not result in a significant ordering of structure in this region, RegAC binding results in significant ordering in early stages of the reaction. As the reaction progressed, the degree of disorder increased and by the end of the reaction (100 min labeling), the peptide is completely deuterated (relative deuterium uptake is ∼0.7), suggesting complete disorder in this region (Fig. 5B, top panel). This inversion is better visualized in deuterium uptake plots (Figs. 5B, bottom panel, and 6C).
We next probed conformational changes associated with complex formation at CNB:A. The bimodal spectra at the interdomain linker (residues 171–190) serve as conformational probes to monitor interconversion of various short-lived conformations of the interdomain linker. As described in the previous section, cAMP binding had no significant effects at the interdomain linker, but RegAC binding resulted in increased disorder in the linker. We can infer this increased disorder from mass spectral envelopes of peptides from this region (Fig. 5C i). When comparing mass spectra for apo RD with RD:RegAC binary complex, we see the apo spectra showed greater bimodality indicative of residual structure. Upon RegAC binding, this residual structure is eliminated and we observe a binomial distribution corresponding to increased dynamics. Deuterium exchange plots show that this increased dynamics is observed for the entire time course of the experiment (Fig. 5C ii).
These results indicate that RegAC binds to RD at CNB:B and causes ordering in RD domain B while simultaneously inducing long-range conformational changes in domain A as perceived by increased dynamics in domain A. Ordering in CNB:B is transient, and with increases in reaction time, we observed increased overall dynamics across most regions of RD, providing evidence for the transient nature of the RD:RegAC interaction in the absence of cAMP. While cAMP binding showed local effects in deuterium exchange at the cAMP binding pocket, RegAC binding has a proteinwide effect, providing a case for RegAC-mediated allosteric effects on RD.
Deconvolution and quantitation of EX1 kinetics at cAMP binding pocket of CNB:B
EX1 kinetics at the cAMP binding pocket of CNB:B were quantitated by deconvolution of the bimodal spectra to obtain centroid and amplitude values for the low-exchanging and high-exchanging envelopes. The key parameters in quantitating EX1 kinetics data are the amplitude ratios between the envelopes, centroids of each envelope, and the changes with respect to time. The quantitated data are summarized in Fig. 6.
In apo RD, the centroid of the lower-exchanging envelope shifts with time while the higher-exchanging envelope stays constant (Fig. 6A). Upon addition of cAMP, it is seen that the centroids of both the lower- and higher-exchanging populations stay constant with time (Fig. 6B). In the RD:RegAC binary complex, it is once again seen that the centroid of the lower-exchanging population shifts with time (Fig. 6C). In the binary complex, the amplitude ratio shows an inversion with time with lower-exchanging population predominating at earlier time points and the higher-exchanging population predominating at higher time points (Fig. 6C ii). RD:RegAC:cAMP ternary complex data shows that the lower-exchanging population predominates through the entire time course and the centroids stay constant with time (Fig. 6D). These results highlight the role of cAMP in stabilizing conformations and preventing interchange between the different conformations of RD.
Discussion
In this study we set out to monitor steps in the cAMP signal termination pathway by probing the dynamic interplay between phosphodiesterases and the regulatory subunit of PKA. We have used HDXMS as a tool to study complex formation and dissociation by monitoring protein dynamics as a function of reaction time. These results build on previous work mapping PDE-R-subunit interactions (27) with evidence for substrate channeling in the PKA R-subunit-PDE signaling complex. Importantly, these results also give key insights into PDE-mediated allosteric regulation in cAMP signal termination. While this work was carried out with proteins from a lower eukaryote, D. discoideum, the broad conservation of the components in the cAMP signaling pathway suggests that these results provide a universal model for the various steps in cAMP signal termination pathway.
Steps in the PDE-mediated cAMP signal termination pathway
Our previous work has proposed active site coupling and substrate channeling as a mode for signal termination, whereby PDEs interact with the active site of RD and hydrolyze cAMP bound to the receptor (27). Here we have deciphered how complex formation and dissociation occurs in the various stages of the cAMP signal termination pathway.
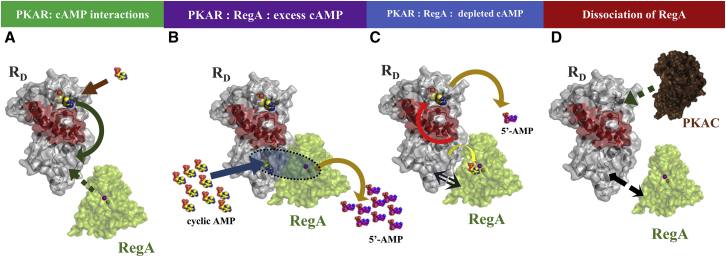
After cAMP-mediated activation of the PKA, the high local concentrations of cAMP would prevent reassociation of the PKA holoenzyme. Thus signal termination would occur only when cAMP is depleted by the action of phosphodiesterases. We have shown here that, in the presence of high concentrations of cAMP, RD and RegAC form a stable ternary complex at CNB:B of RD, which also causes increased exchange at important loci in CNB:A. This is further corroborated by deconvolution analysis of EX1 kinetics observed for a peptide spanning residues 266–279 in CNB:B domain (Fig. 6). The envelope distribution in this peptide is distinct for the ternary complex and is not merely a sum of profiles seen for the RD:RegAC and RD:cAMP states. This is inferred from the greatly diminished profile for the higher-exchanging envelope seen in this condition, compared to the RD:RegAC and RD:cAMP states. This provides important evidence for formation of a stable ternary complex in cAMP signal termination and warrants further characterization. Formation of the ternary complex lends itself to the substrate-channeling model for coordinated cAMP hydrolysis proposed earlier (Fig. 7) (27).
Figure 7.
Steps in PDE-mediated cAMP signal termination pathway. (A) cAMP binding at CNB:A of RD (in gray surface representation) results in an allosteric relay (green arrow) that stabilizes structural transitions in CNB:B. This presents an interaction interface for RegA (green surface representation) binding. (B) In the first step of the cAMP termination pathway, with high local cAMP (in yellow spheres) concentrations, RD and RegA form a stable catalytic complex. cAMP from the local environment binds at the RD CNB:B cAMP binding pocket and subsequently is channeled into the active site of RegA, as an example for substrate channeling in a signaling complex. (C) Once free cAMP is depleted by substrate channeling, RegA and RD form transient complexes (indicated by the black arrow). RegA binding also results in an allosteric relay (red arrow) that results in dissociation of cAMP from CNB:A. Thus RegA hydrolyzes all cAMP bound to the receptor and results in apo RD. (D) RegA dissociates from RD and subsequently primes RD to reassociate with PKAC resulting in termination of the cAMP signaling pathway. RegA, cAMP, and PKAC serve to regulate RD through unique allosteric pathways.
The dynamic loci spanning α:C helix and CNBs A and B are involved in allosteric regulation of RD, and suggest a role for RegAC in allosterically modulating RD function. In a cAMP-depleted state, the complex at CNB:B of RD is transient in nature, but CNB:A exhibits all the allosteric effects observed in the ternary complex. Importantly, increased dynamics is seen at the N-terminal PKAC binding motif of RD. The dynamics of the PKA binding motif has been shown to play a critical role in tuning PKAC interactions (37). NMR has provided the most detailed insights into the role of the dynamic linkers (51,52) that connect CNB:A and CNB:B domains and have been implicated in both cAMP cooperativity and PKA activation. The α:B-C helical segment serves as more than a passive covalent linker and functions to allosterically respond to cAMP binding to the PBC regions in CNBs A and B (53). Our results indicate that cAMP and RegAC (PDE) modulate the dynamics of RD via parallel allosteric pathways.
These results together suggest that in the presence of a large pool of cAMP, the RegAC and RD stay associated and function as a stable catalytic multiprotein complex, whereby cAMP that binds to RD is channeled to the PDE’s active site. We predict that this complex would stay stably bound until all cAMP is depleted, suggesting a role for substrate channeling in cAMP signal termination. After depletion of pools of cAMP, the only cAMP molecules left in the system are those associated with RD receptors. We hypothesize that, without cAMP to stabilize the complex, the PDE forms transient complexes with all RD molecules in the vicinity. In the process, the PDE dissociates and hydrolyzes all bound cAMP, thereby ensuring signal resetting. Allosteric effects in RD, particularly at the PKAC binding motif, suggest that the PDE is priming RD to reassociate with PKAC to terminate the cAMP signaling pathway (Fig. 7). This provides a model for how the cAMP signaling pathway can rapidly reset itself to respond to subsequent stimuli. This also provides a model for how the pathway responds to fluxes of cAMP rather than steady-state levels (54).
Allosteric networks in RD’s role as integrative node in cAMP signaling
HDXMS studies of cAMP-free RD showed that many regions of the proteins showed a bimodal distribution of mass spectra, specifically the cAMP-binding pocket at CNB:B showed significant bimodal characteristics. Bimodal distributions (EX1 kinetics) are indicative of local unfolding/slow structural transitions, while binomial distributions (EX2 kinetics) are representative of folded states (11). Local unfolding at the cAMP binding pocket provides a structural explanation for the proposed low cAMP affinity at the CNB:B cAMP binding pocket (41,49).
Ligand binding to dynamic proteins is known to shift the equilibrium of a protein from an inactive conformation to an active conformation (2). These effects are more prominent for proteins with disordered regions or in a state of constant structural transitions, with ligand binding causing large-scale ordering in protein structure (7). Thus it was interesting to observe that cAMP did not cause significant ordering at CNB-B, as evidenced from the bimodal nature of the mass spectra. By closely monitoring time-dependent changes in the bimodality of the spectra, it is possible to gain insights into the rate at which structural transitions occur. In apo RD, the transition from predominantly bimodal to predominantly binomial spectra occurs within 5 min of deuterium exchange reaction. In the cAMP-bound form, although bimodal spectra are observed, the relative ratio of the lower-exchanging and higher-exchanging populations do not change with labeling time (Fig. 6B). This implies that cAMP binding at CNB:A results in a major decrease in the rate of the structural transitions at CNB:B, without causing significant ordering at the binding pocket. The interdomain linker C-helix undergoes drastic conformational changes in mammalian RD between the cAMP-bound conformation (B-form) and the PKAC-bound conformation (H-form) (32). This helix serves as an important locus for allosteric communication between the two cAMP binding domains (33). In RD, the interdomain linker region also shows significant local unfolding, but cAMP binding does not have any effect at this loci. These results show cAMP-mediated effects localized mainly to the binding pockets with distinct allosteric communication relays connecting CNB:A to CNB:B.
PDE-mediated effects on RD are in contrast to cAMP, with RegAC influencing RD protein dynamics at many important loci. Importantly, RegAC causes stabilization of the cAMP binding pocket at CNB:B, as seen from reduced bimodality in spectra in the binary and ternary complex. Furthermore, RegAC also causes long-range conformational changes that lead to increased dynamics in the interdomain linker of RD.
Dynamics of RD play an important in the different stages of the cAMP signaling pathway. The R-subunit has been shown to allosterically respond to both C-subunit binding as well as cAMP (32,33,55–58). These highlight the importance of allostery in the R-subunit for coordinating the activation phase and proceed primarily through CNB:A. Our results reported here indicate the importance of CNB:B in binding PDEs and this interaction is then allosterically coupled to CNB:A through relays that are independent from that mediated by cAMP and the C-subunit. CNB:B domain of RD from D. discoideum is distinct as it has a very low affinity for cAMP compared to the mammalian homologs. Despite this low affinity, our results highlight its importance as a dynamic node for PDE action. This underscores the importance of R-subunits in serving as integrative nodes by coordinating multiple allosteric relays that ultimately govern the output response in both activation and termination phases of the cAMP signalosome (Fig. 7).
Author Contributions
S.K. and N.K.T. designed and performed the experiments; S.K., N.K.T., A.C., and G.S.A. analyzed and interpreted data; and all four authors wrote the article.
Acknowledgments
This work was supported by grants from the Singapore Ministry of Education Academic Research Fund Tier 3 (No. MOE2012-T3-1-008) and the National University of Singapore awarded to G.S.A.
Editor: Jason Kahn.
Footnotes
Srinath Krishnamurthy and Nikhil Kumar Tulsian contributed equally to this work.
Four figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00711-0.
Supporting Material
References
- 1.Gould C.M., Newton A.C. The life and death of protein kinase C. Curr. Drug Targets. 2008;9:614–625. doi: 10.2174/138945008785132411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussinov R., Tsai C.J. Unraveling structural mechanisms of allosteric drug action. Trends Pharmacol. Sci. 2014;35:256–264. doi: 10.1016/j.tips.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Nussinov R., Tsai C.J., Liu J. Principles of allosteric interactions in cell signaling. J. Am. Chem. Soc. 2014;136:17692–17701. doi: 10.1021/ja510028c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardino A.K., Villali J., Kern D. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henzler-Wildman K., Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 7.Smock R.G., Gierasch L.M. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moorthy B.S., Anand G.S. Multistate allostery in response regulators: phosphorylation and mutagenesis activate RegA via alternate modes. J. Mol. Biol. 2012;417:468–487. doi: 10.1016/j.jmb.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 9.Nussinov R., Ma B., Tsai C.J. Multiple conformational selection and induced fit events take place in allosteric propagation. Biophys. Chem. 2014;186:22–30. doi: 10.1016/j.bpc.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds K.A., McLaughlin R.N., Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoofnagle A.N., Resing K.A., Ahn N.G. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 12.Kaveti S., Engen J.R. Protein interactions probed with mass spectrometry. Methods Mol. Biol. 2006;316:179–197. doi: 10.1385/1-59259-964-8:179. [DOI] [PubMed] [Google Scholar]
- 13.Englander S.W., Kallenbach N.R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 14.Iacob R.E., Zhang J., Engen J.R. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS One. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sours K.M., Xiao Y., Ahn N.G. Extracellular-regulated kinase 2 is activated by the enhancement of hinge flexibility. J. Mol. Biol. 2014;426:1925–1935. doi: 10.1016/j.jmb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamurthy S., Moorthy B.S., Anand G.S. Dynamics of phosphodiesterase-induced cAMP dissociation from protein kinase A: capturing transient ternary complexes by HDXMS. Biochim. Biophys. Acta. 2013;1834:1215–1221. doi: 10.1016/j.bbapap.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Anand G.S., Krishnamurthy S., Johnson D.A. Cyclic AMP- and (Rp)-cAMPS-induced conformational changes in a complex of the catalytic and regulatory (RIα) subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Proteomics. 2010;9:2225–2237. doi: 10.1074/mcp.M900388-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moorthy B.S., Badireddy S., Anand G.S. Cooperativity and allostery in cAMP-dependent activation of protein kinase A: monitoring conformations of intermediates by amide hydrogen/deuterium exchange. Int. J. Mass Spectrom. 2011;302:157–166. [Google Scholar]
- 19.Moorthy B.S., Gao Y., Anand G.S. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002295. M110.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann S.K., Brown J.M., Firtel R.A. Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev. Biol. 1997;183:208–221. doi: 10.1006/dbio.1996.8499. [DOI] [PubMed] [Google Scholar]
- 21.Firtel R.A., Chapman A.L. A role for cAMP-dependent protein kinase A in early Dictyostelium development. Genes Dev. 1990;4:18–28. doi: 10.1101/gad.4.1.18. [DOI] [PubMed] [Google Scholar]
- 22.Shaulsky G., Fuller D., Loomis W.F. A cAMP-phosphodiesterase controls PKA-dependent differentiation. Development. 1998;125:691–699. doi: 10.1242/dev.125.4.691. [DOI] [PubMed] [Google Scholar]
- 23.Thomason P.A., Traynor D., Kay R.R. An intersection of the cAMP/PKA and two-component signal transduction systems in Dictyostelium. EMBO J. 1998;17:2838–2845. doi: 10.1093/emboj/17.10.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor S.S., Kim C., Anand G.S. Dynamics of signaling by PKA. Biochim. Biophys. Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 25.Anand G., Taylor S.S., Johnson D.A. Cyclic-AMP and pseudosubstrate effects on type-I A-kinase regulatory and catalytic subunit binding kinetics. Biochemistry. 2007;46:9283–9291. doi: 10.1021/bi700421h. [DOI] [PubMed] [Google Scholar]
- 26.Johnson D.A., Akamine P., Taylor S.S. Dynamics of cAMP-dependent protein kinase. Chem. Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 27.Krishnamurthy S., Moorthy B.S., Anand G.S. Active site coupling in PDE:PKA complexes promotes resetting of mammalian cAMP signaling. Biophys. J. 2014;107:1426–1440. doi: 10.1016/j.bpj.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mutzel R., Simon M.N., Véron M. Expression and properties of the regulatory subunit of Dictyostelium cAMP-dependent protein kinase encoded by λgt11 cDNA clones. Biochemistry. 1988;27:481–486. doi: 10.1021/bi00401a069. [DOI] [PubMed] [Google Scholar]
- 29.Veron M., Mutzel R., Wallet V. cAMP-dependent protein kinase from Dictyostelium discoideum. Dev. Genet. 1988;9:247–258. doi: 10.1002/dvg.1020090407. [DOI] [PubMed] [Google Scholar]
- 30.Canaves J.M., Taylor S.S. Classification and phylogenetic analysis of the cAMP-dependent protein kinase regulatory subunit family. J. Mol. Evol. 2002;54:17–29. doi: 10.1007/s00239-001-0013-1. [DOI] [PubMed] [Google Scholar]
- 31.Berman H.M., Ten Eyck L.F., Taylor S.S. The cAMP binding domain: an ancient signaling module. Proc. Natl. Acad. Sci. USA. 2005;102:45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim C., Cheng C.Y., Taylor S.S. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Das R., Esposito V., Melacini G. cAMP activation of PKA defines an ancient signaling mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:93–98. doi: 10.1073/pnas.0609033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das R., Melacini G. A model for agonism and antagonism in an ancient and ubiquitous cAMP-binding domain. J. Biol. Chem. 2007;282:581–593. doi: 10.1074/jbc.M607706200. [DOI] [PubMed] [Google Scholar]
- 35.McNicholl E.T., Das R., Melacini G. Communication between tandem cAMP binding domains in the regulatory subunit of protein kinase A-Iα as revealed by domain-silencing mutations. J. Biol. Chem. 2010;285:15523–15537. doi: 10.1074/jbc.M110.105783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badireddy S., Yunfeng G., Anand G.S. Cyclic AMP analog blocks kinase activation by stabilizing inactive conformation: conformational selection highlights a new concept in allosteric inhibitor design. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.004390. M110.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akimoto M., Selvaratnam R., Melacini G. Signaling through dynamic linkers as revealed by PKA. Proc. Natl. Acad. Sci. USA. 2013;110:14231–14236. doi: 10.1073/pnas.1312644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornev A.P., Taylor S.S., Ten Eyck L.F. A generalized allosteric mechanism for cis-regulated cyclic nucleotide binding domains. PLOS Comput. Biol. 2008;4:e1000056. doi: 10.1371/journal.pcbi.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wales T.E., Fadgen K.E., Engen J.R. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor S.S., Buechler J.A., Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 41.Mutzel R., Lacombe M.L., Veron M. Cloning and cDNA sequence of the regulatory subunit of cAMP-dependent protein kinase from Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1987;84:6–10. doi: 10.1073/pnas.84.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weis D.D., Wales T.E., Ten Eyck L.F. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectrom. 2006;17:1498–1509. doi: 10.1016/j.jasms.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Konermann L., Simmons D.A. Protein-folding kinetics and mechanisms studied by pulse-labeling and mass spectrometry. Mass Spectrom. Rev. 2003;22:1–26. doi: 10.1002/mas.10044. [DOI] [PubMed] [Google Scholar]
- 44.Wang L.C., Morgan L.K., Anand G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012;31:2648–2659. doi: 10.1038/emboj.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wildes D., Marqusee S. Hydrogen-exchange strategies applied to energetics of intermediate processes in protein folding. Methods Enzymol. 2004;380:328–349. doi: 10.1016/S0076-6879(04)80015-6. [DOI] [PubMed] [Google Scholar]
- 46.Engen J.R., Smithgall T.E., Smith D.L. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14384–14391. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 47.Fang J., Rand K.D., Engen J.R. False EX1 signatures caused by sample carryover during HX MS analyses. Int. J. Mass Spectrom. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Y., Dostmann W.R., Varughese K.I. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 49.Degunzburg J., Part D., Veron M. An unusual adenosine 3′,5′-phosphate dependent protein-kinase from Dictyostelium discoideum. Biochemistry. 1984;23:3805–3812. doi: 10.1021/bi00312a003. [DOI] [PubMed] [Google Scholar]
- 50.Houde D., Berkowitz S.A., Engen J.R. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J. Pharm. Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akimoto M., Moleschi K., Melacini G. Allosteric linkers in cAMP signalling. Biochem. Soc. Trans. 2014;42:139–144. doi: 10.1042/BST20130257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popovych N., Sun S., Kalodimos C.G. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulton S., Akimoto M., Melacini G. A tool set to map allosteric networks through the NMR chemical shift covariance analysis. Sci. Rep. 2014;4:7306. doi: 10.1038/srep07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leiser M., Fleischer N., Erlichman J. Enhanced activation of cAMP-dependent protein kinase by rapid synthesis and degradation of cAMP. J. Biol. Chem. 1986;261:15486–15490. [PubMed] [Google Scholar]
- 55.Abu-Abed M., Das R., Melacini G. Definition of an electrostatic relay switch critical for the cAMP-dependent activation of protein kinase A as revealed by the D170A mutant of RIα. Proteins. 2007;69:112–124. doi: 10.1002/prot.21446. [DOI] [PubMed] [Google Scholar]
- 56.Anand G.S., Hughes C.A., Komives E.A. Amide H/2H exchange reveals communication between the cAMP and catalytic subunit-binding sites in the R(I)α subunit of protein kinase A. J. Mol. Biol. 2002;323:377–386. doi: 10.1016/s0022-2836(02)00919-1. [DOI] [PubMed] [Google Scholar]
- 57.Das R., Abu-Abed M., Melacini G. Mapping allostery through equilibrium perturbation NMR spectroscopy. J. Am. Chem. Soc. 2006;128:8406–8407. doi: 10.1021/ja060046d. [DOI] [PubMed] [Google Scholar]
- 58.Kim C., Xuong N.H., Taylor S.S. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.