Graphical abstract
Keywords: Apicomplexa, FIKK kinase, Kinase, Cryptosporidium
Highlights
-
•
We studied FIKK kinases from Plasmodium falciparum and Cryptosporidium parvum.
-
•
Soluble and active samples of PfFIKK8 and CpFIKK contain a N-terminal extension.
-
•
Both FIKK samples preferentially phosphorylated serines with flanking arginines.
Abstract
FIKKs are protein kinases with distinctive sequence motifs found exclusively in Apicomplexa. Here, we report on the biochemical characterization of Plasmodium falciparum FIKK8 (PfFIKK8) and its Cryptosporidium parvum orthologue (CpFIKK) – the only member of the family predicted to be cytosolic and conserved amongst non-Plasmodium parasites. Recombinant protein samples of both were catalytically active. We characterized their phosphorylation ability using an enzymatic assay and substrate specificities using an arrayed positional scanning peptide library. Our results show that FIKK8 targets serine, preferably with arginine in the +3 and −3 positions. Furthermore, the soluble and active FIKK constructs in our experiments contained an N-terminal extension (NTE) conserved in FIKK8 orthologues from other apicomplexan species. Based on our results, we propose that this NTE is an integral feature of the FIKK subfamily.
1. Introduction
Transmission of malaria, perennially one of the deadliest infectious diseases in the world, is mediated by transmission of Plasmodium parasites between a vertebrate host and a mosquito vector. Such a complex life cycle is regulated by a signal transduction system of ∼90 protein kinases (PK) [1], a majority of which having human orthologues. The Plasmodium PKs absent from mammalian genomes are likely involved in parasite-specific functions and potential targets of selective drugs. One such group, known as FIKK kinases, is exclusive to the phylum of Apicomplexa and thus, far the subject of few studies.
The defining architecture of the FIKK subfamily includes a highly variable N-terminal region followed by a conserved C-terminal kinase domain (KD) where the eponymous quartet of residues, Phe–Ile–Lys–Lys, is located. Two species, namely Plasmodium falciparum and Plasmodium reichenowi [2–4] have been reported to have 21 and 6 FIKK members, respectively. Most of these members contain a PEXEL motif – designating export Plasmodium proteins [4–7] – as well as a signal peptide in the N-terminus. The sole exception is a single member, orthologous to FIKK8 (PfFIKK8) which is also the longest member. It is also the only member conserved in all Plasmodium species, as well as in the genomes of Toxoplasma, Cryptosporidium, Neospora and Eimeria [4].
FIKK kinases are expressed mostly during the blood stage of the parasitic life cycle [8]. PfFIKK4.1, PfFIKK9.3, PfFIKK9.6 and PfFIKK12 have been found to be localized in the Maurer’s cleft [8,9]. Furthermore, erythrocytic membrane rigidity has been found to be moderated by gene disruption of either PfFIKK7.1 or PfFIKK12 [10], as well as by dematin, a cytoskeletal protein that is potentially a substrate of PfFIKK4.1 [11]. An attempt to disrupt the FIKK8 orthologue in mouse malaria-causing parasite Plasmodium berghei (PbFIKK8) was not successful, suggesting that FIKK8 might play an essential role in cell cycle regulation of Plasmodium parasites [12].
Most of the catalytically important residues of typical eukaryotic protein kinases are conserved in FIKK kinases [3], with the exception of the glycine triad, resulting in speculation on their catalytically activity. To date, recombinant samples of FIKK4.1 [11] and FIKK4.2 [9] have been reported to be active. Furthermore, samples of PfFIKK12 and PfFIKK4.1 immuno-precipitated from cell lysates have been found to be capable of phosphorylating myelin basic protein (MBP) [8,11]. In our study, we expressed and purified recombinant protein samples of P. falciparum FIKK8 (PfFIKK8, gene ID PF3D7_0805700 at www.plasmodb.org) and its C. parvum orthologue CpFIKK (cgd5_4390 at www.cryptodb.org), and studied their enzymatic properties.
2. Results
We cloned multiple constructs of PfFIKK8 and CpFIKK (Table S1; method and materials in Supplementary materials). Using an Escherichia coli system previously proven for parasite PKs [13], only constructs including an extension of 38 or more residues at the N-terminus of the predicted KD yielded soluble and stable protein samples (Table S1, Fig. S1). This N-terminal extension (NTE) contains both polar and non-polar residues as well as multiple potential phospho-acceptor residues (S/Y) (Fig. S1B). Using a previously described method [14], we purified two constructs of PfFIKK8 and one of CpFIKK containing the NTE (PfFIKK8l, PfFIKK8o and CpFIKKd in Table S1). We then proceeded to assess their auto-phosphorylation behavior and determine their ability to phosphorylate a set of common peptide substrates.
To assess their auto-phosphorylation behavior, the purified PfFIKK8l and CpFIKKd samples were incubated with ATP and MgCl2. The samples were then trypsinized and analyzed using LC–MS–MS. We used the maps of the resulting peptides (Fig. S2) to identify multiple phospho-serines (pS) on the Plasmodium sample, both phospho-serines and phospho-threonines (pT) on the Crytposporidium sample and one phospho-tyrosine (pY) on each. One of the phosphorylated serines (pS1320) on PfFIKK8l is located in the predicted activation loop. Unfortunately, the corresponding serine on CpFIKKd was not part of any of the detectable peptides. Furthermore, some of the phosphorylated residues were found in the NTE, including a phospho-serine that is conserved on both samples.
We also found PfFIKK8l and CpFIKKd to be active against a set of standard kinase substrates – bovine casein, bovine MBP and Syntide-2 (PLARTLSVAGLPGKK), with MBP producing the highest level of activity.
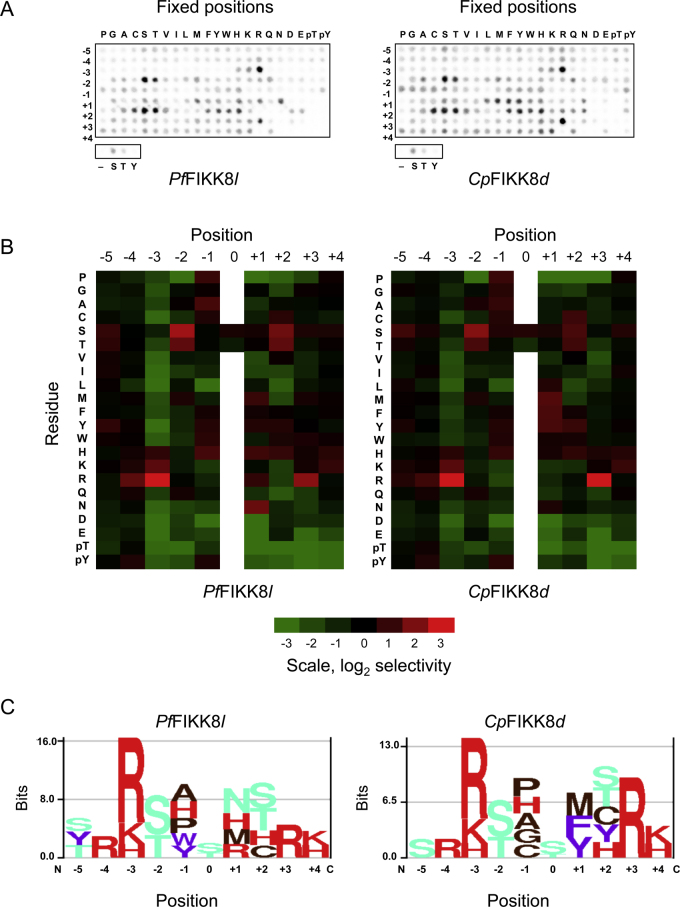
To systematically determine the sequence preferences of PfFIKK8l and CpFIKKd in an unbiased manner, we assayed the proteins using a positional-scanning peptide array [15]. Both demonstrated a strong preference for basic residues (Figs. 1 and S3 ), primarily Arg at positions −3 and +3 relative to the phosphorylation site. Arginine was also favored by both in the −4 position, albeit not as strongly. In addition, both selected Ser over Thr as the phosphate acceptor by about 2–3-fold (Tables S3 and S4).
Fig. 1.
The substrate preferences of PfFIKK8l and CpFIKKd were assessed using a positional peptide scanning array. (A) A matrix of biotinylated peptides with the indicated residue at the indicated position relative to Ser or Thr was allowed to be phosphorylated by either sample. The reaction mixtures were spotted onto a streptavidin membrane and exposed to a phosphor screen, resulting in a spot image array. (B) The heat maps correspond to normalized signal values in the arrays (average of two runs). Cleary, both samples preferred serine over threonine. (C) Sequence logos illustrate Arg was favored at the –3 and +3 positions by both proteins (preference for Arg at +3 is much stronger for the Cryptosporidium sample).
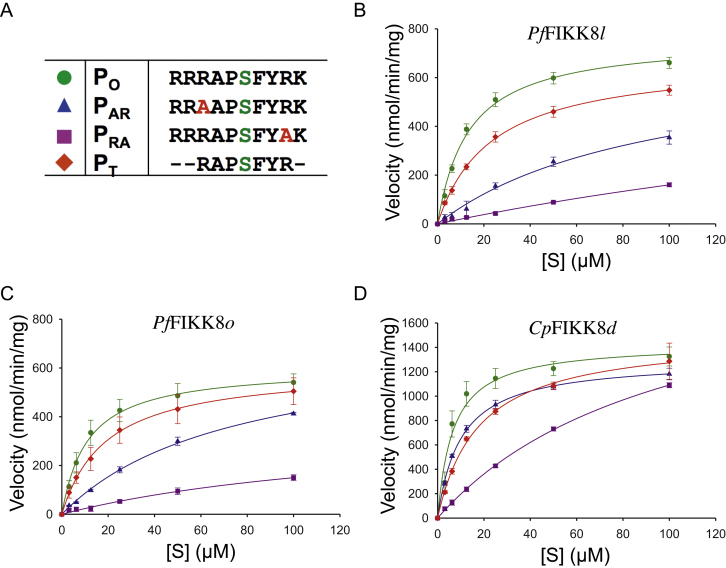
To evaluate the contributions of the arginines and other flanking residues to phosphorylation efficiency, we designed an optimized peptide substrate (PO) with the sequence RRRAPSFYRK and three variants. The variants featured mutation of Arg to Ala at the +3 (PAR) and −3 (PRA), as well as PT, a truncated version of PO. Using an LDH–PK coupled kinase assay [16], we compared the phosphorylation kinetics of these substrates with PfFIKK8l, PfFIKK8o and CpFIKKd respectively as catalysts. The two PfFIKK8 constructs behaved very similarly to each other with almost all the peptides (Fig. 2). CpFIKKd appeared more active than its P. falciparum orthologues; however, all the kinetic parameters are within the same order of magnitude. Similar Michaelis constants (Km) and catalytic efficiency values (kcat) were obtained using PO and PT for all three FIKK samples, with all indicating at least average substrate binding. On the other hand, both mutants (PAR and PRA) resulted in higher Km and lower kcat values, thus, emphasizing the importance of the flanking arginines.
Fig. 2.
(A) We tested the activity of both PfFIKK8l and CpFIKKd against four substrates. PO featured Arg at −3 and +3 positions. In PAR and PRA, one flanking Arg was replaced in order to assess their importance. PT was a truncated form of PO. (B) PfFIKK8l is most active against PO and PT (the truncated version of PO). Replacing Arg in the −3 or +3 position (PAR and PRA) substantially reduced activity for the Plasmodium sample. (C) While the shorter PfFIKK8o construct was also less active against PAR and PRA, the reduction of activity against PT was minimal. (D) A similar trend was seen with the Cryptosporidium sample but reduction in activity against the PAR, PRA and PT was not as pronounced.
3. Discussion
Many protein kinases, including PKA, PKC and rhoptry kinases (found primarily in Toxoplasma [17,18]), have N-terminal and/or C-terminal extensions flanking the canonical bilobed structure. Our results suggest the active domains of PfFIKK8 and CpFIKK have an NTE of at least 38 residues in length. We hypothesize that this extension is an integral component of the kinase domain of both, as neither can be expressed as soluble recombinant proteins in its absence.
In addition to the NTE, the FIKK motif and the absence of a C-terminal extension, FIKK kinases are also defined by a number of conserved divergences in some common kinase motifs. First, Phe and Gly in the DFG triad are often replaced by other hydrophobic residues. Second, in many FIKK kinases, the activation loop features proline in place of the more common alanine in the APE. Furthermore, the HRD motif in subdomain VI features a leucine in place of arginine. The absence of arginine in this position typically signifies that a kinase does not need auto-phosphorylation of the activation loop to become active. Interestingly, we found S1320 in PfFIKK8l, which is located in the activation loop and conserved in all FIKK kinases, to be auto-phosphorylated (Fig. S2). To investigate the relevance of this phosphoserine to FIKK8 activation, we attempted to express a mutant form of PfFIKK8l with S1320 mutated to alanine; however, the mutated protein did not express. Therefore, a possible regulation mechanism for FIKK kinases mediated by phosphorylation of the activation loop remains to be confirmed.
The inclusion of the NTE as an integral component of PfFIKK8l and CpFIKKd is corroborated by the kinetic parameters we obtained, all of which are in the range of active protein kinases with average to above average binding affinities for ATP and the optimized substrates used (PO and PT). Furthermore, sequence alignment (not shown) indicates that the NTE is conserved among available FIKK8 orthologues from apicomplexan parasites and, to a lesser degree, the other FIKK paralogues found in P. falciparum and P. reichenowi (Fig. S4). Given that PfFIKK8l and PfFIKK8o behaved nearly indistinguishably in the kinetics study, we propose that PfFIKK8o specifically defines the boundaries of the active FIKK8 domain and that M1049 is the start of the NTE. Identifying the functional significance of this NTE is left for future research; however, the evidence of auto-phosphorylation discussed above suggests the possibility of a regulatory role.
In addition to phosphoserines in the NTE, our auto-phosphorylation experiment also revealed one phosphorylated site in the N-lobe and 2 more in the C-lobe of PfFIKK8l. The N-lobe site, namely S1099, has previously been reported in a phospho-proteomics study of P. falciparum [19]. Furthermore, this phosphoserine is conserved in our autophosphorylated CpFIKKd sample (Fig. S2) and, significantly, located on the glycine-rich loop of both kinases – implicated in positioning of γ–phosphate in ATP hydrolysis. Previously, a phosphoserine in the same region of yeast ATG1 protein kinase was found to be inhibitory [20].
The peptide array study revealed a preference for Arg at the −3 and +3 positions for PfFIKK8l and CpFIKKd (Fig. 1) with both enzymes showing the strongest selection for arginine at the +3 position when analyzed using consensus peptide substrates. Notably, the consensus peptides included basic residues at multiple positions upstream of the phosphorylation site (−5, −4 and −3). Therefore, it is possible that the more modest effect of replacing the −3 Arg residue is due to compensation by nearby basic residues. Both PfFIKK8l and CpFIKKd largely preserved their catalytic efficiencies (based on kcat/Km values in Table S2) when the shortened substrate PT was used in place of PO, suggesting that this short substrate may be as an ideal tool for assaying FIKK8 activity and screening for small molecule inhibitors.
In conclusion, our recombinant samples of PfFIKK8 and CpFIKK are orthologous and catalytically active protein kinases, both of which featuring an approximately 40-residue long integral N-terminal extension. Future research to determine the function of this extension may reveal the mechanism of FIKK kinases. It is also possible that, in vivo, regions of the proteins not included in our active constructs may play catalytic, regulatory and localization roles.
Acknowledgements
The authors would like to thank Drs. Abdellah, Guillermo Senisterra and Majida El Bakkouri at the SGC for helpful advice on assays. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada through the Ontario Genomics Institute [OGI-055], GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z]. This work was also supported by U.S. NIH grant R01 GM104047 to B.E.T.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.molbiopara.2015.06.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Doerig C., Billker O., Haystead T., Sharma P., Tobin A.B., Waters N.C. Protein kinases of malaria parasites: an update. Trends Parasitol. 2008;24:570–577. doi: 10.1016/j.pt.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Doerig C., Billker O., Pratt D., Endicott J. Protein kinases as targets for antimalarial intervention: Kinomics, structure-based design, transmission-blockade, and targeting host cell enzymes. Biochim. Biophys. Acta. 2005;1754:132–150. doi: 10.1016/j.bbapap.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Ward P., Equinet L., Packer J., Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kooij T.W., Carlton J.M., Bidwell S.L., Hall N., Ramesar J., Janse C.J. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2005;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiller N.L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 6.Marti M., Good R.T., Rug M., Knuepfer E., Cowman A.F. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 7.Schneider A.G., Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes M.C., Goldring J.P., Doerig C., Scherf A. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Mol. Microbiol. 2007;63:391–403. doi: 10.1111/j.1365-2958.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- 9.Kats L.M., Fernandez K.M., Glenister F.K., Herrmann S., Buckingham D.W., Siddiqui G. An exported kinase (FIKK4. 2) that mediates virulence-associated changes in Plasmodium falciparum-infected red blood cells. Int. J. Parasitol. 2014;44:319–328. doi: 10.1016/j.ijpara.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Nunes M.C., Okada M., Scheidig-Benatar C., Cooke B.M., Scherf A. Plasmodium falciparum FIKK kinase members target distinct components of the erythrocyte membrane. PloS One. 2010;5:e11747. doi: 10.1371/journal.pone.0011747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt G.S., Bailey S. Dematin, a human erythrocyte cytoskeletal protein, is a substrate for a recombinant FIKK kinase from Plasmodium falciparum. Mol. Biochem. Parasitol. 2013;191:20–23. doi: 10.1016/j.molbiopara.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Tewari R., Straschil U., Bateman A., Bohme U., Cherevach I., Gong P. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wernimont A.K., Artz J.D., Finerty P., Jr., Lin Y.H., Amani M., Allali-Hassani A. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Mol. Biol. 2010;17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Mok J., Kim P.M., Lam H.Y., Piccirillo S., Zhou X., Jeschke G.R. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiianitsa K., Solinger J.A., Heyer W.D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 17.Qiu W., Wernimont A., Tang K., Taylor S., Lunin V., Schapira M. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 2009;28:969–979. doi: 10.1038/emboj.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim D., Gold D.A., Julien L., Rosowski E.E., Niedelman W., Yaffe M.B. Structure of the Toxoplasma gondii ROP18 kinase domain reveals a second ligand binding pocket required for acute virulence. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.523266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solyakov L., Halbert J., Alam M.M., Semblat J.P., Dorin-Semblat D., Reininger L. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Na. Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 20.Yeh Y.Y., Shah K.H., Chou C.C., Hsiao H.H., Wrasman K.M., Stephan J.S. The identification and analysis of phosphorylation sites on the Atg1 protein kinase. Autophagy. 2011;7:716–726. doi: 10.4161/auto.7.7.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.