Abstract
AIM: In recent years, studies have suggested that Epstein-Barr virus (EBV) is associated with HCC. The present study was to determine the prevalence of EBV in HCC patients, and whether EBV acted synergistically with hepatitis viruses in HCC carcinogenesis.
METHODS: Liver tissue 115 HCC patients and 26 non-carcinoma patients were studied. Polymerase chain reaction (PCR) was performed to detect EBV BamHI W DNA, EBV LMP1 DNA, HBV X DNA, and HBV S DNA. Reverse transcription PCR (RT-PCR) was performed to detect HCV RNA and HDV RNA. Immunohistochemistry was performed to detect LMP1, HBsAg, HBcAg and HCV. The positive ratios were compared between HCC group and control group by χ 2 test.
RESULTS: Totally, 78 HCC samples whose β -globulin DNA was positively detected by amplified PCR were selected. PCR was performed in all cases for EBV DNA and HBV DNA. RT-PCR was performed in 18 cases for HCV RNA and HDV RNA. EBV BamHI W and EBV LMP1 were positive in 18 and 6 cases, respectively. HBV X gene and HBV S gene were positive in 42 and 27 cases respectively. HCV was positive in one of the 18 cases, and none was positive for HDV. The positive rates were 28.2% (22 of 78) for EBV DNA (BamHI W and/or LMP1) and 56.4% (44 of 78) for HBV DNA (X gene and/or S gene) respectively. In addition, 12 cases were positive for both EBV DNA and HBV DNA. Among the 26 cases in the control group, 2 cases were positive for EBV BamHI W, 4 positive for HBV X gene and 3 positive for HBV S gene. The positive rates were 8.0% (2 of 26) and 23.1% (6 of 26), respectively, for EBV DNA and HBV DNA. The result of DNA sequencing of BamHI W was 100% homologous with the corresponding sequence of B95-8. There was significant difference in EBV infection rate between HCC patients and controls (χ 2 = 4.622, P < 0.05). The difference in HBV infection rate was also significant (χ 2 = 8.681, P < 0.05). However, there was no obvious correlation between HBV and EBV in HCC patients (χ 2 = 0.835, P > 0.05). LMP1, HBV (HBsAg, HBcAg) and HCV were detected positively in 25, 45 and 6 of 78 cases of HCC tissues respectively. In the 26 control cases, the corresponding positive cases were 2, 4 and 0. The difference in EBV infection rate between HCC patients and control cases was statistically significant (χ 2 = 6.02, P < 0.05). The difference in HBV infection rate was also statistically significant (χ 2 = 10.03, P < 0.05). In the 25 cases with positive LMP1 expression, 6 were in the nuclei of tumor cells, 9 in the cytoplasm of tumor cells and 10 in mesenchymal lymphocyte cytoplasm.
CONCLUSION: The existence of EBV infection in HCC tissues suggests that EBV may be involved in the hepatocellular carcinogenesis in China. HBV infection may be a major cause of HCC. There is no correlation between EBV and HBV in the development of HCC. The prevalence of HCV infection is low in our area, and HDV appears not to play a direct role in hepatocellular carcinogenesis.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most frequent malignant tumors. It possesses the characteristics of high malignancy, rapid progress and poor prognosis. In recent years, studies have suggested that Epstein-Barr virus (EBV) is associated with HCC although opposite results have been subsequently reported. The present study was to determine the prevalence of EBV in hepatocellular carcinoma (HCC) patients, and whether EBV acted synergistically with hepatitis viruses in HCC carcinogenesis.
MATERIALS AND METHODS
Patients and tissue samples
HCC samples from 115 patients, which were resected in the First Affiliated Hospital of Medical College, Shantou University and Shantou Central Hospital between October 1997 and October 2001, were fixed in 40 g/L neutral formaldehyde and embedded in paraffin. Of these, 78 samples satisfied the criteria that their β -globulin DNA presented positive. The patients consisted of 59 men and 19 women. Their ages ranged from 22 to 76 years (average 50.2 ± 2.4 years). According to the Edmondson (1956) HCC morphology standard, two, 34, 30 and 12 cases belonged to the levels I, II, III and IV, respectively. AFP was detected in 48 cases; the level was lower than 0.14 ng/mL in four cases, between 0.14 ng/mL and 5.6 ng/mL in 14 cases and higher than 5.6 ng/mL in 12 cases. Anti-hepatitis B virus (HBV) assay was performed by enzyme-linked immunosorbent assay (ELISA) in 62 cases, and 54 were positive. Anti- hepatitis C virus (HCV) assay was carried out in 58 cases and anti-EBV EA-IgA and VCA-IgA assays in 43 cases. The control group consisted of 20 cases with hepatolithiasis and lobectomy of liver, three cases with hepatic adenoma and three cases with hepatocyte focal nodular hyperplasia. HBV was positive in 10 cases. HCV and EBV were negative in all cases.
Reagents
Taq DNA polymerase, dNTP mixture, AMV reverse transcriptase XL, RNase inhibitor, random primer and DNA marker were purchased from TaKaRa® Biotechnology Co., Ltd (Dalian, China). The Trizol total RNA separating medium was purchased from Gibcobrl® Co., Ltd (Grand Island, NY, USA). LMP1 monoclonal antibody (Clone CS1-4), HBsAg monoclonal antibody (Clone ZMHB5), HBcAg polyclonal antibody, the immediately-used-type of the second generation immunohistochemistry Elivision™ plus broad spectrum test kit were from Maixin Co., Ltd (Fuzhou, China). Rat-anti-human HCV monoclonal antibody (Clone TORDJI-22) was from Zhongshan Biotechnology Co. Ltd (Beijing, China).
PCR, DNA sequencing and RT PCR
We downloaded the gene sequences of EBV, HBV, HCV and hepatitis D virus (HDV) from the GenBank. Each included four strains, and the homogeneity was compared among the strains by the Vector NTI suite7 software. The primers were designed by Oligo software (Table 1, LMP1[1] and β -globulin[2] primer sequences were adopted from literature) within the homogeneous gene regions, and synthesized in Shenggong Co., Ltd (Shanghai, China). The positive controls of EBV, HBV and HCV were, respectively, B95-8 cell line, PSDHBV-1 (HBV complete sequence plasmid, 7.8 k) and an HCV-positive serum. The HDV positive control could not be obtained. Fetal liver tissues served as the negative controls. Blank control was established. Primers flanking β -globulin gene were used as a positive control for DNA preservation. Full details of the sequences and genome coordinates of primers and amplification programs of PCR are given in Table 1. The PCR products were electrophoresed in 15 g/L agarose gel, stained with ethidium bromide (Et-Br), and photographed under a UV-transilluminator. After being retrieved and purified, PCR product was ligated with pMD 18-T vector, and then transfected E.coli competent cells. The clones were selected and identified. Sequencing of the positive clones was performed by Shenggong Co., Ltd. Bacteriophage M13 cloning vector was used to ligate the fragments. RNA extracts were prepared from paraffin sections. The tissue specimens were deparaffinized with xylene, then washed with ethanol. RNA was extracted using TRIZOL total RNA separating medium. Reverse transcription was performed using AMV reverse transcriptase XL with cDNA synthesis performed for 1 h at 42 °C. Then nested PCR was carried out using the resulting cDNA as template. The detail is shown in Table 1.
Table 1.
Primer sequences and amplification programs of PCR and RT-PCR detection
| Transcript | Product size | Genomic coordinates in GenBank | Primer sequence (5‘-3‘) | Amplification program |
| EBV | 337 bp | B95-8 | GAGTGTGTGCCAGTTAAGGT (168 056-16 8075 20 bp) | 94 °C 3 min (94 °C 30 s 55 °C |
| LMP1[1] | V01555 | CTAGCGACTCTGCTGGAAAT (168 373-16 8392 20 bp) | 40 s 72 °C 1 min) × 40 72°C 8 min | |
| EBV | 130 bp | B95-8 | CATCACCGTCGCTGACT (14 525-14 541 17 bp) | 95 °C 3 min (94 °C 30 s 52 °C |
| BamHI W | V01555 | GTTGGGCTTAGCAGAAA(14 638-14 654 17 bp) | 40 s 72 °C 45 s) × 40, 72°C 7 min | |
| HBV | 198 bp | HBV | AGGTATGTTGCCCGTTTGT (457-475 19 bp) | 95 °C 5 min (94 °C 30 s 52 °C |
| S gene | AB073856 | GAGGCCCACTCCCATAGG (637-654 18 bp) | 40 s 72 °C 50 s) × 35, 72°C 7 min | |
| HBV | 232 bp | HBV | TGTGCTGCCAACTGGATCCT (1 387-1 406 20 bp) | 95 °C 3 min (94 °C 30 s 55 °C |
| X gene | AB073856 | GTGGTCTCCATGCGACGTG (1600-1618 19 bp) | 40 s 72 °C 45 s) × 37 72°C 7 min | |
| HCV | 165 bp | HCV | GCGGAACCGGTGAGTACAC (138-156 19 bp) | 95 °C 5 min (94 °C 30 s 55 °C |
| cDNA | NC_001433 | GCACTCGCAAGCACCCTATC (283-302 20 bp) | 40 s 72 °C 50 s) × 40 72°C 7 min | |
| Outer | ||||
| HCV | 142 bp | HCV | GGAATTGCCAGGACGACC (158-175 18 bp) | 95 °C 5 min (94 °C 30 s 55 °C |
| cDNA | NC_001433 | CTCGCAAGCACCCTATCAG (281-299 19 bp) | 40 s 72 °C 50 s) × 40 72°C 7 min | |
| Inner | ||||
| HDV | 216 bp | HDV | GCGGGTCCGTCGTTCCAT (653-670 18 bp) | 95 °C 5 min (94 °C 30 s 55 °C |
| cDNA | AB037947 | GTCCGACCTGGGCATCCGT (850-868 19 bp) | 40 s 72 °C 50 s) × 35 72°C 7 min | |
| Outer | ||||
| HDV | 114 bp | HDV | TCGAGAATCGGCAAATG (752-768 17 bp) | 95 °C 5 min (94 °C 30 s 52 °C |
| cDNA | AB037947 | GACCTGGGCATCCGTAAG (847-864 18 bp) | 40 s 72 °C 50 s) × 35 72°C 7 min | |
| Inner | ||||
| β -globulin | 210 bp | Beta-globin | ACACAACTGTGTTCACTAGC (120-139 20 bp) | 94 °C 5 min (94 °C 30 s 55 °C |
| (PCO3-PCO5)[2] | human | GAAACCCAAGAGTCTTCTCT (310-329,20bp) | 40 s 72 °C 1 min) × 35 72°C 8 min | |
| DNA AY128650 |
Immunohistochemistry
We used a two-step method according to the instructions provided with the reagent kits. LMP1 located in nucleus or cytoplasm. HCV located in nucleus. HBsAg located in cytoplasm. HBcAg located in nucleus or cytoplasm. Positive cells were defined when there were clear small particles or granuloreticular reddening in cytoplasm or nucleus. The positive control was the positive tissue section provided by the company. Negative control was the sample of lobectomy of liver due to hepatolithiasis. Blank control was tissue section without addition of the first antibody.
Statistical analysis
The positive ratios were compared between HCC group and control group by means of χ 2 test. P < 0.05 was considered as statistically significant. The statistical software used was SPSS 10.0.
RESULTS
PCR and RT-PCR
Totally, 78 HCC samples whose β -globulin DNA was positively detected by amplified PCR were selected; PCR and RT-PCR were performed in all cases for EBV and HBV, and in 18 cases for HCV and HDV. EBV BamHI W and EBV LMP1 were positive in 18 and six cases, respectively. HBV X gene and HBV S gene were positive in 42 and 27 cases respectively. HCV was positive in 1 of the 18 cases, and none was positive for HDV. The positive rates were 28.2% (22 of 78) for EBV DNA (BamHI W and/or LMP1) and 56.4% (44 of 78) for HBV DNA (X gene and/or S gene) respectively. In addition, 12 cases were positive for both EBV DNA and HBV DNA. Among the 26 cases in the control group, 2 cases were positive for EBV BamHI W, four positive for HBV X gene and three positive for HBV S gene. The positive rates were 8.0% (2 of 26) and 23.1% (6 of 26), respectively, for EBV DNA and HBV DNA (Figure 1A-G). The result of DNA sequencing of BamHI W was 100% homologous with the corresponding sequence of B95-8 (Figure 2). There was significant difference in EBV infection rate between HCC patients and controls (χ 2 = 4.622, P < 0.05). The difference in HBV infection rate was also significant (χ 2 = 8.681, P < 0.05). However, there was no obvious correlation between HBV and EBV in HCC patients (χ 2 = 0.835).
Figure 1.
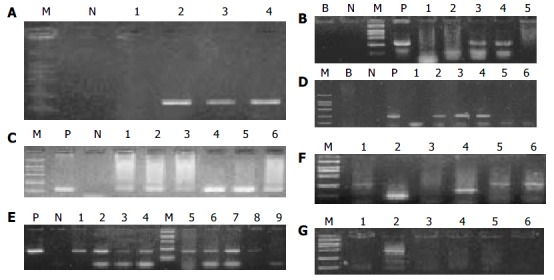
Results of PCR. P, positive control; N, negative control; B, blank control; M, marker of molecular mass. The sizes of the six bands of the marker are 2000 bp, 1000 bp, 750 bp, 500 bp, 250 bp, 100 bp, respectively. A: β -Globulin (Positive: Lanes 2-4, 210 bp), B: LMP 1 (Positive: Lanes 3 and 4, 337 bp), C: BamHI W (Positive: Lanes 1-6, 130 bp), D: X gene (Positive: Lanes 2-4, 232 bp), E: S gene (Positive: Lanes 1-8, 198 bp), F: HCV (Positive: Lane 4, other lanes are nonspecific, 142bp), G: HDV (All are negative, Lane 5 is nonspecific).
Figure 2.
Sequencing results.
Immunohistochemistry
The immunohistochemical detection was performed for LMP1, HBV (HBsAg, HBcAg) and HCV in tissues of 78 cases of HCC, with 25, 45 and six cases positive, respectively. In the 26 control cases, the corresponding positive cases were two, four and zero. The difference in EBV infection rate between HCC patients and control cases was statistically significant (χ 2 = 6.02, P < 0.05). The difference in HBV infection rate was also statistically significant (χ 2 = 10.03, P < 0.05). In the 25 cases with positive LMP1 expression, six presented in the nuclei of tumor cells, nine in the cytoplasm of tumor cells and 10 in mesenchymal lymphocyte cytoplasm (Figure 3A-H).
Figure 3.
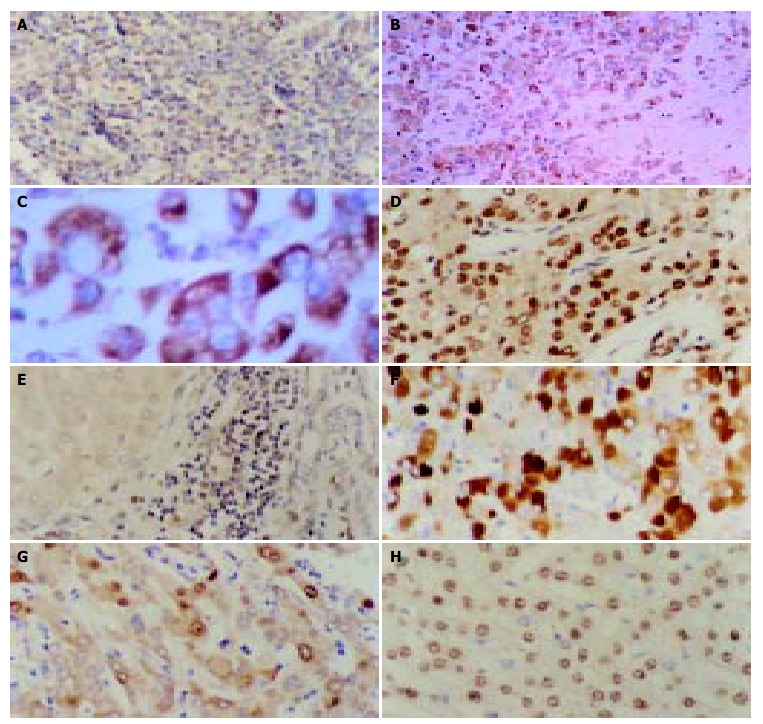
Results of immunohistochemistry. A: Positive-control of LMP1 from the tonsil tissue. The cytoplasm of lymphocytes showed positive signals. × 400, B: Immunohistochemical staining with anti-LMP1 antibody of an HCC specimen. The positive signals were localized in the cytoplasm and membrane of neoplasm cells. × 100, C: Immunohistochemical staining with anti-LMP1 antibody of an HCC specimen. The positive signals were localized in the cytoplasm and membrane of neoplasm cells. × 400, D: Immunohistochemical staining with anti-LMP1 antibody of an HCC specimen. The positive signals were localized in the nuclei of neoplasm cells. × 400, E: Immunohistochemical staining with anti-LMP1 antibody of an HCC specimen. The positive signals were localized in the interstitial lymphocytes but no clear positive signals were present in the adjacent neoplasm cells. × 400, F: Immunohistochemical staining with anti-HBsAg antibody of an HCC specimen. The positive signals were localized in the cytoplasm of neoplasm cells. × 400, G: Immunohistochemical staining with anti-HBcAg antibody of an HCC specimen. The positive signals were localized in the cytoplasm and nuclei of neoplasm cells. × 400, H: Immunohistochemical staining with anti-HCV antibody of an HCC specimen. The positive signals were localized in the nuclei of neoplasm cells. × 400.
DISCUSSION
EBV is considered as the first virus associated with human malignancies including Burkitt’s lymphoma[3], T cell lymphoma[4], parotid lymphoepithelioma[5,6], nasopharyngeal carcinoma[7], Hodgkin’s disease[8], gastric carcinoma[9] and breast carcinoma[10]. The virus proteins including EBNA1, 2, 3A, 3C, and LMP1 take part in the virus transformation and immortalization. Whether EBV has relation to HCC and whether it plays a role in the genesis of HCC has become the research focus of this field in recent years. However, the results so far are conflicting.
In Japan, Sugawara et al[11-13] found that the positive rate of EBNA1 in HCC was 37% and that cells infected by EBV could facilitate the replication of HCV. The positive rate of BamHI W in HCC was 33% detected by PCR. Among patients with positive BamHI W, anti-HCV positive rate was 40% and HBsAg positive rate was 14%, whereas the control subjects were all negative for anti-HCV and HBsAg. Moreover, EBV DNA load in HCC tissues was 1 000 times that in peripheral blood mononuclear cells of the healthy EBV-seropositive individuals. The authors concluded that EBV accelerates the onset of HCC and plays a role in oncogenesis of HCC, probably by promoting HCV replication, exacerbating inflammatory processes in liver tissue, promoting the proliferation of carcinoma cells or directly influencing tumorigenic potential. In China, Cheng et al[14] used EBV to transform the hepatitis C virus from an HCV positive patient to permanent lymphoblastoid cell lines. They found that HCV might exist and remain functional in a cultured cell line for a long period with the help of EBV. However, Chu et al[15] studied 41 cases of HCC from Los Angeles area in the United States, for evidence of EBV infection. Among these patients, one was positive for EBER1 by in situ hybridization, and none was positive for LMP1 and EBNA4 by PCR. The authors concluded that there is no association between EBV and genesis of HCC. In addition, later studies carried out in Germany, United Kingdom and The Netherlands[16-19] reported very low detection rates of EBV in HCC patients, further suggesting that EBV has little relation to hepatocellular carcinogenesis.
In our study, EBV DNA was detected in 28.2% of the HCC tissues by PCR while the positive rate of HBV DNA was 56.4%. HCV RNA was detected in 5.6% of the HCC tissues by RT-PCR while HDV RNA was not detected in all of 18 cases. The positive rate of LMP1 was 32.1% of the HCC tissues by immunohistochemistry. The positive signals of LMP1 were mainly localized in tumor cells. The infection rate of EBV was significantly higher in HCC patients than in control patients. These results are similar to those obtained by Sugawara et al Moreover, we observed LMP1 expression in cytoplasm of interstitial lymphocytes in a large proportion of HCC tissues. Experiments in vitro have demonstrated that EBV can infect human B-lymphocytes, combine with C3d receptor on the surface of B lymphocyte and enter the cells, escape from the immune surveillance of the host, and then transform the cells. We postulate that EBV, which infects lymphocytes, can promote the genesis and development of HCC in an unidentified way.
Why the results of Sugawara et al and ours are different from those of others? Previous studies produced controversial results on whether EBV played a role in the genesis and development of malignant tumors like breast cancer[20-23], esophageal cancer[24,25], gastric cancer and colorectal cancer. It has been reported that the correlation between EBV and tumor incidence is stronger in Japan and Taiwan than in the western countries. Therefore, we assume that the significance of EBV in HCC may vary because of regional and racial differences. Further studies are required to reveal the underlying mechanism by which EBV contributes to hepatocellular carcinogenesis.
It has been confirmed that HBV and HCV play a certain part in hepatocellular carcinogenesis. Our study showed that EBV and HBV had no synergistic action in HCC, which is consistent with the report by Yuan et al[26]. Because the infection rate of HCV in our region is much lower than that in Japan, the issue whether HCV interacts with EBV in hepatocellular carcinogenesis should be studied further. HDV is one kind of defective virus that can deteriorate the patient’s condition if co-infected with HBV. It has been suggested that HDV also plays a role in hepatocellular carcinogenesis to some extent. But our study did not support such a point of view.
The method using paraffin-imbedded samples for a retrospective molecular epidemiological study may be affected, to some extent, by sample processing and conservation. In this experiment, β -globulin gene was selected as an inner control in PCR amplification. Some cases in which DNA had been seriously degraded were excluded in order to reduce the false-negative rate. To increase the detectable rate, several indexes were used for each virus. BamHI W is regarded as a sensitive index as it repeats 7 to 12 times in EBV genome. LMP1, as a major virus transform protein, is detected in tumors that are related to EBV such as nasopharyngeal carcinoma, Burkitt’s lymphoma and other tumors. The primers and reaction conditions are also the important factors. We took advantage of the Internet and biology software to process sequence search, primer design and reaction condition simulation, and achieved a good performance.
The relationship between EBV and the malignant tumors including HCC is very complex and controversial. More studies with larger sample sizes are required to draw a scientific and precise conclusion. If the hypothesis that there is a relation between EBV and HCC comes into existence, then, research and development of EBV vaccines will become a more significant issue, and gene therapy aiming at EBV will be an important approach in the treatment of HCC.
ACKNOWLEDGEMENTS
The authors wish to acknowledge Professor Shu-Fang Tian from Heredity Laboratory, Institute for Viral Disease Prevention and Control, China CDC for kindly providing PSDHBV-1 plasmid. We also acknowledge Dr. Yi-Shu Yang, Dr. Shu-Hui Wang, and Dr. Hai-Jun Du from Oncogenic Virus Laboratory, Institute for Viral Disease Prevention and Control, China CDC for excellent assistance in the study.
Footnotes
Supported by the Technology Program of Guangdong Province, China, No. 2KM04504s
Edited by Xia HHX and Zhu LH Proofread by Xu FM
References
- 1.Huang YP, Guo XC, He ZG, Zhao J, Zeng Y. Epstein-Barr virus induced thymus malignant T cell lymphoma. Bindu Xuebao. 2001;17:289–294. [Google Scholar]
- 2.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 3.Bell A, Rickinson AB. Epstein-Barr virus, the TCL-1 oncogene and Burkitt's lymphoma. Trends Microbiol. 2003;11:495–497. doi: 10.1016/j.tim.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Yachie A, Kanegane H, Kasahara Y. Epstein-Barr virus-associated T-/natural killer cell lymphoproliferative diseases. Semin Hematol. 2003;40:124–132. doi: 10.1053/shem.2003.50012. [DOI] [PubMed] [Google Scholar]
- 5.Kuo T, Hsueh C. Lymphoepithelioma-like salivary gland carcinoma in Taiwan: a clinicopathological study of nine cases demonstrating a strong association with Epstein-Barr virus. Histopathology. 1997;31:75–82. doi: 10.1046/j.1365-2559.1997.5830814.x. [DOI] [PubMed] [Google Scholar]
- 6.Mok MY, Shek WH, Wong RW. Lymphoepithelioma-like carcinoma of the parotid gland in a patient with rheumatoid arthritis. Clin Exp Rheumatol. 2002;20:848–850. [PubMed] [Google Scholar]
- 7.Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 8.Gandhi MK, Tellam JT, Khanna R. Epstein-Barr virus-associated Hodgkin's lymphoma. Br J Haematol. 2004;125:267–281. doi: 10.1111/j.1365-2141.2004.04902.x. [DOI] [PubMed] [Google Scholar]
- 9.Mladenova I, Pellicano R. Infectious agents and gastric tumours. An increasing role for Epstein-Barr virus. Panminerva Med. 2003;45:183–188. [PubMed] [Google Scholar]
- 10.Wakiguchi H. Overview of Epstein-Barr virus-associated diseases in Japan. Crit Rev Oncol Hematol. 2002;44:193–202. doi: 10.1016/s1040-8428(02)00111-7. [DOI] [PubMed] [Google Scholar]
- 11.Sugawara Y, Mizugaki Y, Uchida T, Torii T, Imai S, Makuuchi M, Takada K. Detection of Epstein-Barr virus (EBV) in hepatocellular carcinoma tissue: a novel EBV latency characterized by the absence of EBV-encoded small RNA expression. Virology. 1999;256:196–202. doi: 10.1006/viro.1999.9619. [DOI] [PubMed] [Google Scholar]
- 12.Sugawara Y, Makuuchi M, Kato N, Shimotohno K, Takada K. Enhancement of hepatitis C virus replication by Epstein-Barr virus-encoded nuclear antigen 1. EMBO J. 1999;18:5755–5760. doi: 10.1093/emboj/18.20.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugawara Y, Makuuchi M, Takada K. Detection of Epstein-Barr virus DNA in hepatocellular carcinoma tissues from hepatitis C-positive patients. Scand J Gastroenterol. 2000;35:981–984. doi: 10.1080/003655200750023075. [DOI] [PubMed] [Google Scholar]
- 14.Cheng JL, Liu BL, Zhang Y, Tong WB, Yan Z, Feng BF. Hepatitis C virus in human B lymphocytes transformed by Epstein-Barr virus in vitro by in situ reverse transcriptase-polymerase chain reaction. World J Gastroenterol. 2001;7:370–375. doi: 10.3748/wjg.v7.i3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu PG, Chen YY, Chen W, Weiss LM. No direct role for Epstein-Barr virus in American hepatocellular carcinoma. Am J Pathol. 2001;159:1287–1292. doi: 10.1016/s0002-9440(10)62515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhter S, Liu H, Prabhu R, DeLucca C, Bastian F, Garry RF, Schwartz M, Thung SN, Dash S. Epstein-Barr virus and human hepatocellular carcinoma. Cancer Lett. 2003;192:49–57. doi: 10.1016/s0304-3835(02)00695-x. [DOI] [PubMed] [Google Scholar]
- 17.Junying J, Herrmann K, Davies G, Lissauer D, Bell A, Timms J, Reynolds GM, Hubscher SG, Young LS, Niedobitek G, et al. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology. 2003;306:236–243. doi: 10.1016/s0042-6822(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 18.zur Hausen A, van Beek J, Bloemena E, ten Kate FJ, Meijer CJ, van den Brule AJ. No role for Epstein-Barr virus in Dutch hepatocellular carcinoma: a study at the DNA, RNA and protein levels. J Gen Virol. 2003;84:1863–1869. doi: 10.1099/vir.0.19217-0. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann K, Niedobitek G. Epstein-Barr virus-associated carcinomas: facts and fiction. J Pathol. 2003;199:140–145. doi: 10.1002/path.1296. [DOI] [PubMed] [Google Scholar]
- 20.Bonnet M, Guinebretiere JM, Kremmer E, Grunewald V, Benhamou E, Contesso G, Joab I. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–1381. doi: 10.1093/jnci/91.16.1376. [DOI] [PubMed] [Google Scholar]
- 21.Chu PG, Chang KL, Chen YY, Chen WG, Weiss LM. No significant association of Epstein-Barr virus infection with invasive breast carcinoma. Am J Pathol. 2001;159:571–578. doi: 10.1016/S0002-9440(10)61728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Touitou R, Bonnet-Duquenoy M, Joab I. Association of Epstein-Barr virus with human mammary carcinoma. Pros and cons. Dis Markers. 2001;17:163–165. doi: 10.1155/2001/649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preciado MV. Lack of evidence for an association of Epstein-Barr virus infection with breast carcinoma-another point of view. Breast Cancer Res 2003; 5: E6. doi: 10.1186/bcr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LS, Chow KC, Wu YC, Li WY, Huang MH. Detection of Epstein-Barr virus in esophageal squamous cell carcinoma in Taiwan. Am J Gastroenterol. 1999;94:2834–2839. doi: 10.1111/j.1572-0241.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- 25.Kijima Y, Hokita S, Takao S, Baba M, Natsugoe S, Yoshinaka H, Aridome K, Otsuji T, Itoh T, Tokunaga M, et al. Epstein-Barr virus involvement is mainly restricted to lymphoepithelial type of gastric carcinoma among various epithelial neoplasms. J Med Virol. 2001;64:513–518. doi: 10.1002/jmv.1079. [DOI] [PubMed] [Google Scholar]
- 26.Yuan FP, Huang PS, Wang Y, Gong HS. Relationship between EBV infection in fujian HCC and HBV and P53 protein expression. Shijie Huaren Xiaohua Zazhi. 1999;7:491–493. [Google Scholar]


