Abstract
AIM: To investigate the prognostic significance of c-Kit gene mutation and DNA ploidy in gastointestinal stromal tumors (GISTs).
METHODS: A total of 55 cases of GISTs were studied for the expression of c-Kit by immunohistochemistry, and the c-Kit gene mutations in exons 9, 11, 13, and 17 were detected by polymerase chain reaction-single strand confirmation polymarphism (PCR-SSCP) and denaturing high performance liquid chromatography (D-HPLC) techniques. DNA ploidy was determined by flow cytometry.
RESULTS: Of the 55 cases of GISTs, 53 cases (96.4%) expressed c-Kit protein. The c-Kit gene mutations of exons 11 and 9 were found in 30 (54.5%) and 7 cases (12.7%), respectively. No mutations were found in exons 13 and 17. DNA aneuploidy was seen in 10 cases (18.2%). The c-Kit mutation positive GISTs were larger in size than the negative GISTs. The aneuploidy tumors were statistically associated with large size, high mitotic counts, high risk groups, high cellularity and severe nuclear atypia, and epithelioid type. There was a tendency that c-Kit mutations were more frequently found in aneuploidy GISTs.
CONCLUSION: DNA aneuploidy and c-Kit mutations can be considered as prognostic factors in GISTs.
INTRODUCTION
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract. The term, GIST, was first introduced in the 1980s to include a group of nonlymphomatous, nonepithelial tumors of the gut[1]. With the advent of immunohistochemistry, CD117 (c-Kit) negative tumors, such as schawannomas, leiomyomas, and leiomyosarcomas, were excluded from the noncommittal term of “gastrointestinal stromal tumors”. Recently, based on immunophenotypic and ultrastructural similarities, it is widely accepted that the precursor cells in GISTs are the interstitial cells of Cajal (ICCs)[2]. Immunohistochemically, GISTs are typically positive for c-Kit and CD34, but negative for S-100 protein, desmin, and may express smooth muscle actin in 20% to 40% of cases[3].
Yet despite their recognition as a distinct pathologic entity, because GISTs are widely diverse in terms of clinical presentation, morphology, and biologic behavior, the prediction of malignancy on the basis of pathologic features is often difficult. Many studies have analyzed the prognostic relevance of a variety of parameters such as anatomical location, mucosal invasion, tumor necrosis, and high cellularity, rather often leading to conflicting conclusions[1,4-6]. However, two morphologic features have emerged as fairly reliable predictors of outcome: mitotic rate and tumor size[4,7-12]. A consensus conference held at National Institutes of Health in April 2001 provided both an evidence-based definition and a practical scheme for assessing the risk (very low risk, low risk, intermediate risk, and high risk) of aggressive clinical behavior[13]. Recently, a prognostic significance of c-Kit mutations was suggested by several clinicopathologic studies[14-17]. The largest series has shown that c-Kit mutation in exon 11 was more common in large tumors, and that the presence of this mutation was an adverse prognostic factor[14].
The conceptual evolution of GISTs and c-Kit mutation has been the subject of numerous previous articles. However, DNA ploidy studies in relation to c-Kit mutation are not well established. The purpose of this study was to examine the c-Kit mutations and DNA ploidy in GISTs to evaluate them as the prognostic factors.
MATERIALS AND METHODS
Histological review
The cases included in this study were retrieved from patients who were diagnosed as GISTs and treated at Korea University Hospitals between January 1997 and August 2003. The clinical history and postoperative courses were obtained from the review of clinical records. The tumor size in greatest dimension and the location were taken from the pathology reports. Histological slides were reviewed by two of the authors (Kim, I and Lee, JH) in all of the cases. The tumor cell type was determined by light microscopic study. The spindle cell type had a predominant fusiform morphology. Both cytoplasm and nuclei were elongated and aligned within the microscopic field. The epithelioid cell type had a predominantly spherical morphology. The nuclei were round and tended to be centrally placed within tumor cells. The number of mitosis was determined by counting the mitotic activity in 50 adjacent high-power fields (HPF) at a magnification of × 400. The cytologic atypia of the tumor cells was determined to be mild, moderate, or severe. The cytologic atypia was assessed within the most proliferative area. Mild atypia indicated that the tumor cells were histologically benign or mildly atypical. Moderate atypia indicated that the tumor cells had moderately enlarged hyperchromatic or vesicular nuclei with or without prominent nucleoli and some nuclear irregularity. Severe atypia indicated that the tumor cells had enlarged irregular, hyperchromatic, or vesicular pleomorphic nuclei with prominent nucleoli. The cellularity was scored as low if the cell count was below 1000/mm2, intermediate for a range from 1000 to 2000 cells per mm2, and high if exceeding 2000 cells/mm2. Tumor necrosis was determined to be present or absent. Tumor hemorrhage was determined to be present or absent. The growth pattern was determined to be expansive or invasive. GISTs were reclassified as very low, low, intermediate, and high risk groups for their estimated potential for aggressive clinical behavior as suggested by NIH consensus[13].
Immunohistochemistry
Representative sections from each lesion were subjected to immunohistochemical staining by using the avidin-biotin-peroxidase complex (ABC) method. The target differentiation antigens visualized by the monoclonal and polyclonal antibodies were c-Kit (1:400; Dako, Glostrup, Denmark), CD34 (1:200; Neomarker, CA, USA), smooth muscle actin (SMA, 1:200; Dako, Glostrup, Denmark), and S-100 protein (1:400; Neomarker, CA, USA). The tumors were designated as positive when more than 10% of the tumor cells showed a positive reaction for CD34, SMA and S-100 protein. Positive reactions were classified by using the following criteria for c-Kit: (-) negative, if < 10% cells were stained; (+), if 10%-50% cells were immunoreactive; (++), if 51%-100% cells were immunoreactive. The tumors which showed positive reaction for CD34 and/or c-Kit protein with or without SMA and S-100 protein were included in GISTs.
DNA extraction
DNA was extracted from the formaldehyde-fixed, paraffin-embedded tissue sections by using standard methods with proteinase K digestion and phenol/chloroform purification.
Polymerase - chain reaction - single strand confirmation polymorphism (PCR-SSCP)
PCR primers were designated to amplify exons 9, 11, 13, and 17 (Table 1). PCR was carried out with the following conditions: 50 μL total reaction volume, with 5 μL template, 5 μL of each oligonucleotide primer, 10 μL dNTP, 10 μL ddH2O, 2 μL Taq polymerase, 8 μL Mg2+ and 5 μL 10 × PCR buffer. Cycling conditions were as follows: an initial penetration at 95 °C for 4 min, 38 cycles each at 94 °C for 1 min, at 56 °C for 1 min, at 72 °C for 1 min, followed by one cycle at 72 °C for 10 min. PCR products were visualized by gel electrophoresis in 1.7 g/L agarose. Then the PCR products were subjected to 80 g/L non-denaturation polyacrylamide gel electrophoresis with 50 g/L glycerin and silver nitrate staining.
Table 1.
Primer sequence for c-Kit exons 9, 11, 13, and 17 and the corresponding annealing temperature (TA) and the size of expected PCR size products
| c-Kit exon No. | Primers | Primer sequence 5’->3’ | TA (°C) | Product size (bp) |
| 9 | hEx9-F | TTCCTAGAGTAAGCCAGGG | 53 | 298 |
| hEx9-R | AATCATGACTGATATGGT | |||
| 11 | hEx11-F | CAGGTAACCATTTATTTGT | 53 | 326 |
| hEx11-R | TCATTGTTTCAGGTGGAAC | |||
| 13 | hEx13-F | ATCAGTTTGCCAGTTGTGCT | 53 | 250 |
| hEx13-R | TTTATAATCTAGCATTGCC | |||
| 17 | hEx17-F | GTTTTCACTCTTTACAAGT | 53 | 277 |
| hEx17-R | TTACATTATGAAAGTCACAGGAAAC |
Denaturing high performance liquid chromatography (D-HPLC)
To enhance heteroduplex formation, the untreated PCR products (5-7 μL) were denaturated at 95 °C for 5 min followed by gradual reannealing to room temperature for over 30 min. Samples were analyzed in WAVE DHPLC. The gradient was formed by mixing buffer A (0.1 mmol/L TEAA) and buffer B (0.1 mol/L TEAA, 250 g/L acetonitrile). The analysis was carried out at a flow rate of 0.9 mL/min and buffer B gradient increase of 20 g/L per minute for 4 min. Oven temperature for optimal heteroduplex separation under partial DNA denaturation was determined for each amplified fragment by using WAVE Marker software. The most important criteria for assigning the presence of a sequence alteration in each DHPLC fragment were the numbers and the shape of elution peaks in comparison with a wild-type subject elution profile used as reference. To allow the detection of heteroduplex DNA molecules in homozygous patients, those amplicons were pre-mixed with sequence-confirmed wild-type PCR controls. The mixed samples were then denatured at 94 °C for 5 min and cooled for over 45 min at room temperature, before D-HPLC analysis.
DNA ploidy
Nuclear suspensions from the selected tissue blocks, previously used in the immunohistochemical study, were prepared in each case. Three 50-μm thick sections from the tissue block were dewaxed in xylene and rehydrated in a sequence of decreasing concentrations of alcohol. After being washed in Dulbecco’s phosphate buffered saline (DPBS), the tissue was incubated in 2 g/L pepsin (Sigma Chemical Company, St. Louis, USA) at 37 °C water bath for 2.5 h. Then, the samples were neutralized by citrate buffer and treated with trypsin at 37 °C water bath for 10 min. After RNase treatment at 37 °C water bath for 30 min, the samples were stained with propidium iodide at 4 °C for 30 min. By using a FACS analyzer, the DNA histograms were obtained.
Statistical analysis
The data were analyzed with Chi-square and t tests by using SPSS 10.0 program.
RESULTS
Clinicopathologic charateristics
There were 25 males and 30 females. The mean age of the patients at the time of diagnosis was 60 (range: 29-80) years. The locations of tumors were as follows: one in the esophagus, 43 in the stomach, 10 in the small intestine (including duodenum), and one in the mesentery. The symptoms of patients were as follows: 24 cases of abdominal pain, 11 cases of gastrointestinal bleeding, 4 cases of palpable mass and 12 cases of nonspecific. The mean diameter of tumors was 5.29 (range: 0.4-17.0) cm. Three patients showed liver metastasis at the time of surgery. Two patients received re-operation due to recurrence. One patient had a local gastric recurrence after 17 mo and the other had a local duodenal recurrence after 33 mo.
Patients included in this study were diagnosed to have a GIST by a variety of histologic criteria. Spindle cell lesions occurred with or without an epithelioid component, the most common tumor lesions showed interlacing fascicles of spindle cells. Some lesions showed a palisaded morphology resembling peripheral neurilemmomas, but storiform, fascicular, or mixed growth patterns were seen. Some of them were composed of round and epithelioid cells appearing with perinuclear shrinkage of cytoplasm. In some tumors or parts of tumors, there was a mixture of epithelioid cells and spindle cells. Fifteen (27.3%) tumors were epithelioid cell type, whereas the remaining 40 (72.7%) were spindle cell type (Figure 1). The mitotic index ranged from 0 to 100 (mean: 11.4). The degree of cytologic atypia was mild in 27 (49.1%) patients, moderate in 21 (38.2%), and severe in 7 (12.7%). The degree of cellularity was low in 11 (20.0%) patients, intermediate in 37 (67.3%), and high in 7 (12.7%). Necrosis was present in 11 (20.0%), and hemorrhage was present in 17 (30.9%) tumors. Expansive growth was found in 30 (54.5%), and invasive growth in 25 (45.5%) tumors.
Figure 1.
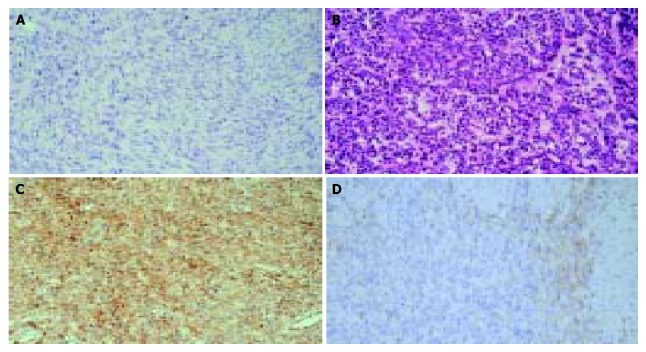
Histological features of various cell types of gastrointestinal stromal tumors and immunohistochemical staining for c-Kit. × 200. A: Spindle cell type; B: Epithelioid type; C: > 50% tumor cells are positive for c-Kit; D: 10%-50% tumor cells are positive for c-Kit.
Correlation of risk groups and clinicopatholgic parameters
Tumors were divided into very low (10/55, 18.2 %), low (26/55, 47.3 %), intermediate (4/55, 7.3%) and high risk (15/55, 27.3%) groups by NIH consensus[13]. Correlation of risk groups and other clinicopthologic parameters is shown in Table 2. Higher risk (intermediate and high risk) groups showed more frequent necrosis, more severe nuclear atypia and higher cellularity than lower risk (very low and low risk) groups which were statistically significant. The epithelioid cell type was more frequent in higher risk group.
Table 2.
Comparison of clinicopathologic parameters accord-ing to the risk groups of gastrointestinal stromal tumors
| Parameter\Risk group | Very low and low (n = 36) | Intermediate and high (n = 19) | P | |
| Cellularity | Low | 11 | 0 | |
| Intermediate | 22 | 15 | 0.019 | |
| High | 3 | 4 | ||
| Nuclear atypia | Mild | 22 | 5 | |
| Moderate | 12 | 9 | 0.019 | |
| Severe | 2 | 5 | ||
| Necrosis | Absent | 34 | 10 | 0.000 |
| Present | 2 | 9 | ||
| Hemorrhage | Absent | 28 | 10 | 0.055 |
| Present | 8 | 9 | ||
| Infiltrative | Absent | 22 | 8 | 0.178 |
| Growth pattern | Present | 14 | 11 | |
| Tumor cell type | Spindle | 29 | 11 | 0.001 |
| Epithelioid | 7 | 8 | ||
Results of immunohistochemistry
A positive reaction for c-Kit protein was obtained in 53 (96.4%), CD 34 in 53 (96.4%), SMA in 17 (30.9%), and S-100 protein in 3 (5.5%). Among 53 c-Kit positive cases, 10 cases (18.9%) were (+) and 43 cases (81.1%) were (++) (Figure 1).
Correlation of c-kit mutation with clinicopatholgic parameters
Thirty cases (54.5%) showed mutations of c-Kit in exon 11 (Figure 2). Among 30 cases, 16 cases showed mutations by both PCR-SSCP and D-HPLC methods, 9 cases by PCR-SSCP, and 5 cases by D-HPLC method. Seven cases (12.7%) showed mutations of c-Kit in exon 9. Among them, 3 cases showed mutations by both PCR-SSCP and D-HPLC methods, 3 cases by PCR-SSCP, and 1 case by D-HPLC method. Two cases showed mutations of both exons 9 and 11. No mutations were found in exons 13 and 17. The mutation-positive GISTs belonged to the large tumors (Table 3), but the association with the other parameters was not significant.
Figure 2.
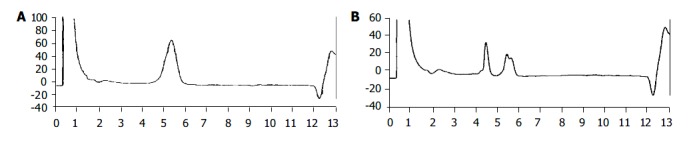
c-Kit mutations in exon 11 by denaturing high performance liquid chromatography. A: Normal; B: mutation.
Table 3.
Comparison of clinicopathologic parameters accord-ing to c-Kit mutations
| Parameter |
Mutations |
P | |
| Negative (n = 20) | Positive (n = 35) | ||
| Sex (M:F) | 11:9 | 14:21 | 0.283 |
| Age (mean, yr) | 58.2 | 61.0 | 0.360 |
| Mean tumor size (cm) | 3.8 | 6.2 | 0.020 |
| Mean mitoses counts | 7.8 | 13.4 | 0.360 |
| Risk group: Very low/low | 14 | 22 | 0.592 |
| Intermediate/high | 6 | 13 | |
| Cellularity: Low | 5 | 6 | 0.741 |
| Intermediate | 13 | 24 | |
| High | 2 | 5 | |
| Nuclear atypia Mild | 10 | 17 | 0.898 |
| Moderate | 7 | 14 | |
| Severe | 3 | 4 | |
| Necrosis Absent | 18 | 26 | 0.161 |
| Present | 2 | 9 | |
| Hemorrhage Absent | 17 | 21 | 0.054 |
| Present | 3 | 14 | |
| Infiltrative growth Absent | 12 | 18 | 0.539 |
| Present | 8 | 17 | |
| Tumor cell type Spindle | 13 | 27 | 0.330 |
| Epithelioid | 7 | 8 | |
Correlation of c-kit mutation with c-kit expression
Among 10 c-Kit (+) cases, 6 cases showed mutations of c-Kit in exon 9 or 11. Among 43 c-Kit (++) cases, 31 cases showed mutations of c-Kit in exon 9 or 11. The rate of c-Kit mutations was higher in c-Kit (++) group than that in c-Kit (+) group (P = 0.05).
Correlation of DNA aneuploidy with clinicopatholgic parameters
Ten cases (18.2%) were DNA aneuploidy (Figure 3). The tumors with aneuploidy were associated with the larger tumor size, high mitotic counts, higher risk groups, high cellularity, and severe nuclear atypia, and epithelioid cell type (Table 4).
Figure 3.
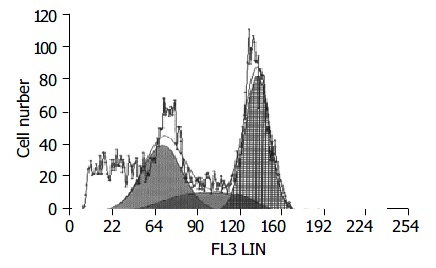
DNA aneuploidy in gastrointestinal stromal tumors.
Table 4.
Comparison of clinicopathologic parameters accord-ing to DNA ploidy
| Parameter | Diploid (n = 45) | Aneuploid (n = 10) | P |
| Sex (M:F) | 21:24 | 4:6 | 0.702 |
| Age (mean, yr) | 60.8 | 56.1 | 0.225 |
| Mean tumor size (cm) | 4.1 | 10.5 | 0.007 |
| Mean mitoses counts | 5.0 | 40.0 | 0.022 |
| Risk group: Very low/low | 34 | 2 | 0.001 |
| Intermediate/high | 11 | 8 | |
| Cellularity: Low | 11 | 0 | 0.008 |
| Intermediate | 31 | 6 | |
| High | 3 | 4 | |
| Nuclear atypia Mild | 24 | 3 | 0.016 |
| Moderate | 18 | 3 | |
| Severe | 3 | 4 | |
| Necrosis Absent | 38 | 6 | 0.080 |
| Present | 7 | 4 | |
| Hemorrhage Absent | 33 | 5 | 0.149 |
| Present | 12 | 5 | |
| Infiltrative growth Absent | 27 | 3 | 0.085 |
| Present | 18 | 7 | |
| Tumor cell type Spindle | 36 | 4 | 0.010 |
| Epithelioid | 9 | 6 |
Correlations of c-kit mutation and DNA aneuploidy with metastasis or recurrence
Three metastatic GISTs showed c-Kit mutation. Among them, two cases showed mutations in exon 11, and the other one showed mutation in exon 9. Two of these 3 tumors had DNA aneuploidy. Among two recurrent GISTs, only one case showed mutation in exon 11, and the two cases were DNA diploidy.
Correlation of c-kit mutation with DNA aneuploidy
Correlation of c-Kit mutation with DNA ploidy state was analyzed (Table 5). There was a tendency that c-Kit mutations were more frequent in aneuploid tumors than in diploid ones, but not statistically significant (P = 0.063).
Table 5.
Correlation of c-Kit mutation and DNA ploidy
| DNA ploidy1 |
c-Kit mutation |
|
| Negative | Positive | |
| Diploid | 19 | 26 |
| Aneuploid | 1 | 9 |
P = 0.063.
DISCUSSION
It has been reported that GISTs are strongly and nearly consistent c-Kit positive, and associated with the mutations of the c-Kit gene. Approximately 40% to 50 % of GISTs, mostly the malignant variants, had mutations in the juxtamembrane domain (exon 11) of the c-Kit gene[14-16], although some studies found mutations in only 15%[18] or the others in as many as 80%[19] of the analyzed cases. Lack of mutations in exon 11 in a significant portion of GISTs may suggest that mutations may occur in other domains of the c-Kit gene. However, no mutations were found in exon 17 (kinase domain) in a large group of GISTs studied[14], which was the area where c-Kit mutations occur in mastocytoma[20] and seminoma[21]. Recently, new mutational hotspots were identified as a result of comprehensive sequencing of c-Kit cDNA obtained from 13 GISTs, which were negative for exon 11 mutations. Mutations in exon 9 and exon 13 were detected in 6 (46%) and 2 (15%) of them, suggesting that such mutations may be relatively common in GISTs[19].
In this study, we analyzed 55 GISTs. Thirty cases (54.5%) showed mutations of c-Kit in exon 11 and 7 cases (12.7%) showed mutations in exon 9. No mutations were found in exon 13 and exon 17. The c-Kit mutation positive GISTs were large in size, and all three metastatic GISTs showed mutations, but the association with other clinicopathologic factors was not evident. The rate of c-Kit mutations was higher in c-Kit (++) group rather than in c-Kit (+) group.
The factors related to the tumor biology might usefully complement the clinical and morphological data. Thus, DNA aneuploidy has been claimed to allow a sensitive and specific discrimination between benign and malignant tumors of the gastrointestinal tract and to correlate with their prognosis[22-28]. However, in other reports, aneuploidy could not be used as a diagnostic criterion of malignancy, but was associated with a mere tendency to an adverse outcome[29,30]. In our study, 10 cases (18.2%) showed DNA aneuploidy. The aneuploid GISTs were associated with large tumor size, high mitotic counts, higher risk groups, high cellularity, severe nuclear atypia and epithelioid type. These results suggest that DNA aneuploidy represents high risk groups of GISTs suggested by NIH consensus[13].
In conclusion, there is a tendency that c-Kit mutations are more frequent in aneuploid GISTs. Therefore, DNA ploidy and c-Kit mutations might be closely related.
Footnotes
Edited by Wang XL and Xu CT Proofread by Pan BR and Xu FM
References
- 1.Appelman HD. Smooth muscle tumors of the gastrointestinal tract. What we know now that Stout didn't know. Am J Surg Pathol. 1986;10 Suppl 1:83–99. [PubMed] [Google Scholar]
- 2.Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol. 1998;152:1259–1269. [PMC free article] [PubMed] [Google Scholar]
- 3.Miettinen M, Sarlomo-Rikala M, Lasota J. Gastrointestinal stromal tumors: recent advances in understanding of their biology. Hum Pathol. 1999;30:1213–1220. doi: 10.1016/s0046-8177(99)90040-0. [DOI] [PubMed] [Google Scholar]
- 4.Appelman HD. Mesenchymal tumors of the gut: historical perspectives, new approaches, new results, and does it make any difference? Monogr Pathol. 1990:220–246. [PubMed] [Google Scholar]
- 5.Emory TS, Sobin LH, Lukes L, Lee DH, O'Leary TJ. Prognosis of gastrointestinal smooth-muscle (stromal) tumors: dependence on anatomic site. Am J Surg Pathol. 1999;23:82–87. doi: 10.1097/00000478-199901000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Miettinen M, Sarlomo-Rikala M, Sobin LH, Lasota J. Esophageal stromal tumors: a clinicopathologic, immunohistochemical, and molecular genetic study of 17 cases and comparison with esophageal leiomyomas and leiomyosarcomas. Am J Surg Pathol. 2000;24:211–222. doi: 10.1097/00000478-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 7.DeMatteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231:51–58. doi: 10.1097/00000658-200001000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans HL. Smooth muscle tumors of the gastrointestinal tract. A study of 56 cases followed for a minimum of 10 years. Cancer. 1985;56:2242–2250. doi: 10.1002/1097-0142(19851101)56:9<2242::aid-cncr2820560918>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Franquemont DW. Differentiation and risk assessment of gastrointestinal stromal tumors. Am J Clin Pathol. 1995;103:41–47. doi: 10.1093/ajcp/103.1.41. [DOI] [PubMed] [Google Scholar]
- 10.Miettinen M, Lasota J. Gastrointestinal stromal tumors--definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1–12. doi: 10.1007/s004280000338. [DOI] [PubMed] [Google Scholar]
- 11.Ranchod M, Kempson RL. Smooth muscle tumors of the gastrointestinal tract and retroperitoneum: a pathologic analysis of 100 cases. Cancer. 1977;39:255–262. doi: 10.1002/1097-0142(197701)39:1<255::aid-cncr2820390139>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Reith JD, Goldblum JR, Lyles RH, Weiss SW. Extragastrointestinal (soft tissue) stromal tumors: an analysis of 48 cases with emphasis on histologic predictors of outcome. Mod Pathol. 2000;13:577–585. doi: 10.1038/modpathol.3880099. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 15.Ernst SI, Hubbs AE, Przygodzki RM, Emory TS, Sobin LH, O'Leary TJ. KIT mutation portends poor prognosis in gastrointestinal stromal/smooth muscle tumors. Lab Invest. 1998;78:1633–1636. [PubMed] [Google Scholar]
- 16.Lasota J, Jasinski M, Sarlomo-Rikala M, Miettinen M. Mutations in exon 11 of c-Kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am J Pathol. 1999;154:53–60. doi: 10.1016/S0002-9440(10)65250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasota J, Wozniak A, Sarlomo-Rikala M, Rys J, Kordek R, Nassar A, Sobin LH, Miettinen M. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. A study of 200 cases. Am J Pathol. 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moskaluk CA, Tian Q, Marshall CR, Rumpel CA, Franquemont DW, Frierson HF. Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene. 1999;18:1897–1902. doi: 10.1038/sj.onc.1202496. [DOI] [PubMed] [Google Scholar]
- 19.Lux ML, Rubin BP, Biase TL, Chen CJ, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CD, Fletcher JA. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–795. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci U S A. 1995;92:10560–10564. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Q, Frierson HF, Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol. 1999;154:1643–1647. doi: 10.1016/S0002-9440(10)65419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federspiel BH, Sobin LH, Helwig EB, Mikel UV, Bahr GF. Morphometry and cytophotometric assessment of DNA in smooth-muscle tumors (leiomyomas and leiomyosarcomas) of the gastrointestinal tract. Anal Quant Cytol Histol. 1987;9:105–114. [PubMed] [Google Scholar]
- 23.Cooper PN, Quirke P, Hardy GJ, Dixon MF. A flow cytometric, clinical, and histological study of stromal neoplasms of the gastrointestinal tract. Am J Surg Pathol. 1992;16:163–170. doi: 10.1097/00000478-199202000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Kiyabu MT, Bishop PC, Parker JW, Turner RR, Fitzgibbons PL. Smooth muscle tumors of the gastrointestinal tract. Flow cytometric quantitation of DNA and nuclear antigen content and correlation with histologic grade. Am J Surg Pathol. 1988;12:954–960. [PubMed] [Google Scholar]
- 25.Suzuki H, Sugihira N. Prognostic value of DNA ploidy in primary gastric leiomyosarcoma. Br J Surg. 1993;80:1549–1550. doi: 10.1002/bjs.1800801219. [DOI] [PubMed] [Google Scholar]
- 26.Shimamoto T, Haruma K, Sumii K, Kajiyama G, Tahara E. Flow cytometric DNA analysis of gastric smooth muscle tumors. Cancer. 1992;70:2031–2034. doi: 10.1002/1097-0142(19921015)70:8<2031::aid-cncr2820700803>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham RE, Federspiel BH, McCarthy WF, Sobin LH, O'Leary TJ. Predicting prognosis of gastrointestinal smooth muscle tumors. Role of clinical and histologic evaluation, flow cytometry, and image cytometry. Am J Surg Pathol. 1993;17:588–594. doi: 10.1097/00000478-199306000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Tsushima K, Rainwater LM, Goellner JR, van Heerden JA, Lieber MM. Leiomyosarcomas and benign smooth muscle tumors of the stomach: nuclear DNA patterns studied by flow cytometry. Mayo Clin Proc. 1987;62:275–280. doi: 10.1016/s0025-6196(12)61904-1. [DOI] [PubMed] [Google Scholar]
- 29.Lerma E, Oliva E, Tugués D, Prat J. Stromal tumours of the gastrointestinal tract: a clinicopathological and ploidy analysis of 33 cases. Virchows Arch. 1994;424:19–24. doi: 10.1007/BF00197388. [DOI] [PubMed] [Google Scholar]
- 30.Lerma E, Lee SJ, Tugues D, Oliva E, Gich I, Prat J. Ploidy of 36 stromal tumors of the gastrointestinal tract. A comparative study with flow cytometry and image analysis. Anal Quant Cytol Histol. 1994;16:435–440. [PubMed] [Google Scholar]


