Abstract
AIM: Immune escape mutations of HBV often occur in the dominant epitope, the second-loop of the a determinant of hepatitis B surface antigen (HBsAg). To let the hosts respond to the subdominant epitopes in HBsAg may be an effective way to decrease the prevalence of immune escape mutants. For this reason, a man-made clone of HBV S gene with the second-loop deletion was constructed. Its antigenicity was evaluated by yeast expression analysis and DNA immunization in mice.
METHODS: HBV S gene with deleted second-loop, amino acids from 139 to 145, was generated using splicing by overlap extension. HBV deleted S gene was then cloned into the yeast expression vector pPIC9 and the mammalian expression vector pcDNA3 to generate pHB-SDY and pHB-SD, respectively. The complete S gene was cloned into the same vectors as controls. The deleted recombinant HBsAg expressed in yeasts was detected using Abbott IMx HBsAg test kits, enzyme-linked immunoadsorbent assay (ELISA) and immune dot blotting to evaluate its antigenicity in vitro. The anti-HBs responses to DNA immunization in BALB/c mice were detected using Abbott IMx AUSAB test kits to evaluate the antigenicity of that recombinant protein in vivo.
RESULTS: Both deleted and complete HBsAg were successfully expressed in yeasts. They were intracellular expressions. The deleted HBsAg could not be detected by ELISA, in which the monoclonal anti-HBs against the α determinant was used, but could be detected by Abbott IMx and immune dot blotting, in which multiple monoclonal anti-HBs and polyclonal anti-HBs were used, respectively. The activity of the deleted HBsAg detected by Abbott IMx was much lower than that of complete HBsAg (the ratio of sample value/cut off value, 106 ± 26.7 vs 1814.4 ± 776.3, P < 0.01, t = 5.02). The anti-HBs response of pHB-SD to DNA immunization was lower than that of complete HBV S gene vector pHB (the positive rate 2/10 vs 6/10, 4.56 ± 3.52 mIU/mL vs 27.60 ± 17.3 mIU/mL, P = 0.02, t = 2.7).
CONCLUSIONS: HBsAg with deleted second-loop of the α determinant still has antigenicity, and can also raise weak anti-HBs response in mice to DNA immunization, suggesting that it is possible to develop a subdominant vaccine for preventing infections of immune escape mutants of HBV.
INTRODUCTION
The prevalence of hepatitis B virus (HBV) is still high in some areas of the world[1,2]. Universal inoculation of hepatitis B vaccine helps to sharply decrease the prevalence of HBsAg carriers from about 10% to 2% among the urban children in China[3]. However, there are about 5%-10% of healthy individuals demonstrating no or inadequate responses following a standard vaccination schedule[4,5]. A portion of these non-responders may be with a breakthrough infection of immune escape mutants[6,7]. The escape mutant infections were also frequently occurred in liver transplant recipients under hepatitis B immunoglobulin prophylaxis[8,9]. The prevalence of the mutants, usually as occult HBV infections, will progressively increase in the future since certain mutants are stable enough to be horizontally transmittable[10,11] and have a potential to be transmitted through blood transfusion because of escaping the routine screening assays. The second-loop from aa139 to aa147 of the α determinant of HBsAg is the dominant epitope. The escape mutants usually have mutations in this region, including K141E, P142S, D144E and G145R [8,9,12-16], though there are mutations in the rest part of HBsAg[17,18], and induce an altered immunity against the second-loop so that the mutants can escape vaccine-raised antibodies or rabbit polyclonal antibodies to some extent[19,20]. Fortunately, recent researches suggested that weak epitopes, such as subdominant epitopes, might not be escaped[21,22]. To let hosts respond to subdominant epitopes or to both dominant and subdominant epitopes may be an effective way for the prevention of escape mutant infections. For these reasons, a man-made clone of HBV S gene with the second-loop deletion of the α determinant region was constructed in order to destroy the dominant epitope and let the subdominant epitopes be responded by the hosts. The antigenicity of that deleted S gene was evaluated by means of yeast expression analysis and DNA immunization in mice.
MATERIALS AND METHODS
Reagents
pTZ19U-HBV containing double copies of HBV DNA (adw) was presented from professor Zhi-Min Huang, Zhongshan University, Guangzhou, China. pcDNA3 and pPIC9 were purchased from Invitrogen Company (the United States of America). T4 DNA ligase and pfu DNA polymerase were purchased from Promega Company (the United States of America). DNA gel extraction kits and plasmid isolation kits were purchased from Qiagen Company (Germany). Primers shown in Table 1 were synthesized by Bioasia Biological Engineering Company (Shanghai, China). ELISA HBsAg kit was purchased from Zhongshan Biological Engineering Company (Guangdong, China). Abbott IMx HBsAg and AUSAB test kits were purchased from Abbott Laboratory (the United States of America). Sheep polyclonal anti-HBs and labeled streptavidin biotin detecting kit were purchased from DAKO Company (the United States).
Table 1.
Primers used in construction of eukaryotic expression vectors
| Name | Sequences (5’→3’) |
| HBS-SD1 | TCC AAG CTT ATG GGA TCC GAG AAC ATC ACA TCA GGA TTC |
| HBS-SD2 | GCA ACA TGA GGG AAA CAT AG |
| HBS-SD3 | TCT ATG TTT CCC TCA TGT TGC AAT TGC ACC TGT ATT CCC ATC |
| HBS-SD4 | TCC GAA TTC TTT TGT TAG GGT TTA AAT GTA TAC C |
| HBPIC9-1 | CCG GAA TTC GAC GAT GAC GAT AAG GAG AAC ATC ACA TCA GGA TTC |
| HBPIC9-2 | CAA CGC GGC CGC TTA AAT GTA TAC CCA GAG AC |
Animal
Eight to twelve week-old inbred BALB/c female mice were obtained from Guangzhou Traditional Chinese Medicine University.
Construction of HB-SD and HB-SDY fragments
HB-SD was the fragment of HBV S gene with deletion of seven amino acid residues from 139 to 145 of the second-loop of the α determinant. In order to construct HB-SD, Two fragments of HBV S gene from codon 1 to codon 138 and from codon 146 to codon 226 were obtained by polymerase chain reaction (PCR) using HBS-SD1/HBS-SD2 and HBS-SD3/HBS-SD4 as primers, respectively, and pTZ19U-HBV as template. The fragments were then connected using splicing by overlap extension after the PCR products were run on 20 g/L agarose gel and a given band was extracted using DNA gel extraction kit[23]. HB-SD was obtained at last by purifying the spliced products. HB-SDY was obtained by PCR using HB-SD as template and HBSPIC9-1/HBSPIC9-2 as primers. The control fragments of complete HBV S gene were obtained using pTZ19U-HBV as template, and HBS-SD1/HBS-SD4 or HBSPIC9-1/HBSPIC9-2 as primers.
Construction of recombinant vector pHB-SDY and pHB-SD
For construction of pHB-SDY, the fragments of HB-SD and pPIC9 were digested by restriction endonucleases EcoRI and Not I, respectively. For construction of pHB-SD, the fragment of HB-SD was digested by restriction endonucleases Hind III and Eco RI, respectively as well as pcDNA3. Digested DNA fragment and vector DNA were ligated using T4 DNA ligase after purification. Plasmid DNA was obtained after Escherichia coli was transformed by ligated products. Candidate recombinant plasmids were selecteds by restriction fragment length polymorphism (RFLP) analysis and automatic DNA sequencing by Bioasia Biological Engineering Company. Control vectors pHB and pHBY were generated using the control fragments of complete HBV S gene in the same way.
Expression of pHB-SDY in yeast
The recombinant vector DNA of pHB-SDY and pHBY was prepared from transformed bacteria using Qiagen’s Max-Prep kits. Yeast cells from a single colony of Pichia pastoris GS115 strain were cultivated using YPD culture. Vector DNA was transformed into yeast cells by lithium chloride transformation method. Transformants were grown on minimal dextrose and minimal methanol plates to screen Mut+ and MutS phenotypes. PCR using AOX1 primers was utilized to screen integrants. MutS strain was cultivated in BMGY culture and induced using 5 mL/L methanol. Protein expression of the supernatants and cell pellets was analyzed by Coomassie-stained SDS-PAGE, ELISA, Abbott IMx and immune dot blotting assay. ELISA and IMx tests were carried out as the manufacturer’s protocol. Immune dot blotting was carried out using sheep polyclonal anti-HBs as first antibody and LSAB kit to demonstrate the results.
DNA immunization analysis of pHB-SD
Large scale plasmid DNA of recombinant plasmids pHB-SD and pHB was prepared using Qiagen’s Max-Prep kits. Plasmid DNA of pcDNA3 was also prepared to be used as control. Plasmid DNA was adjusted to 1 µg/µL in normal saline. Thirty BLBA/c mice were randomly divided into 4 groups. Each mouse was injected 100 µL of plasmid DNA which was distributed over five different sites into the anterior tibialis muscle 5 d after the injection of an equal volume of 2 g/L bupivacaine. Boost injection was carried out 3 times every 3 wk with equal amount of plasmid DNA. Four weeks after the last boost injection, all mice were put to death for serum. Anti-HBs in serum was detected using Abbott AUSAB kits.
Statistical analysis
For anti-HBs level, geometric mean titer (GMT) for each group was calculated at first. Then Student-Newman-Keuls-q was used for statistical analysis. For positive rate, Fisher exact probability analysis was used. SPSS 10.0 for Windows was used for all statistical analysis. P < 0.05 was considered statistically significant.
RESULTS
Construction of HB-SD and HB-SDY
Two fragments for constructing HB-SD fragment (Figure 1) were successfully obtained with base pair number as expected. The base pair number of spliced HB-SD fragment (Figure 1) was just a little smaller than that of HB fragment (Figure 1), which was the PCR fragment of complete HBV S gene with 711 bp in length. That was conformed to the fact that HB-SD fragment was only 21 bp smaller than HB fragment. HBY and HB-SDY fragments are shown in Figure 1. Their base pair numbers were the same as designed.
Figure 1.
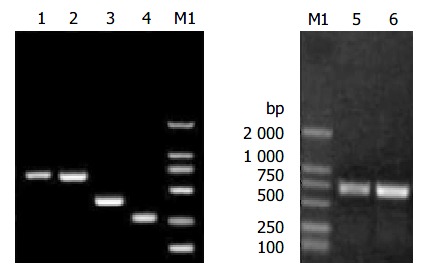
Electrophoresis of the fragments of HBV S gene. Lane 1: HB fragment of complete HBV S gene; Lane 2: HB-SD fragment with the second-loop deletion of the α determinant; Lane 3: DNA fragment of HBV S gene from codon 1 to codon 138; Lane 4: DNA fragment of HBV S gene from codon 146 to codon 226; Lane 5: HBY fragments; Lane 6: HB-SDY fragments; Lane M1: DNA marker.
Construction of pHB-SD and pHB-SDY
HB-SD fragment was inserted into the vector of pcDNA3 between restriction endonucleases Hind III and EcoRI. When recombinant plasmids were digested with the two restriction endonucleases, the molecular weight of the small restriction fragment was the same as that of HB-SD (Figure 2). After its sequence was confirmed by DNA sequencing, the recombinant plasmid was denominated as pHB-SD. The HB fragment was cloned into the same vector to obtain a recombinant plasmid of pHB as control. HB-SDY fragment was inserted into the vector of pPIC9 between restriction endonucleases Eco RI and Not I. The digested fragment shown in Figure 2 was the same as designed in base pair numbers. The recombinant vector was denominated as pHB-SDY after its sequence was confirmed by DNA sequencing. HBY fragment was cloned into the pPIC9 to obtain recombinant vector of pHBY as control.
Figure 2.
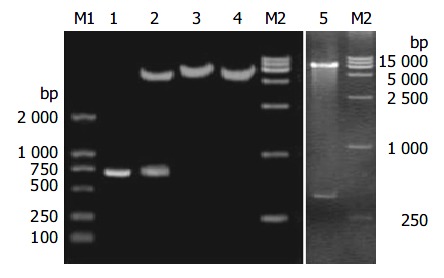
RFLP analysis of recombinant vectors of pHB-SD and pHB-SDY. Lane M1: DNA marker; Lane 1: HB-SD fragment; Lane 2: pHB-SD candidate digested by restriction endonu-clease Hind III and Eco RI; Lane 3: pHB-SD candidate digested by restriction endonuclease Bam HI; Lane 4: pcDNA3 digested by restriction endonuclease Bam HI; Lane 5: pHB-SDY candi-date digested by restriction endonuclease Not I and Eco RI; Lane M2: DNA marker.
Expression of recombinant plasmid pHB-SDY in yeast
MutS transformants for pHB-SDY and its control pHBY were successfully selected though growing on plates of minimal dextrose and minimal methanol. The integrant screening results of transformants are shown in Figure 3. The PCR product of Mut+ transformants without gene of interest was 2.2 kb (Figure 3A). The PCR product of pPIC9 alone was 492 bp (Figure 3A). The PCR products of integrants of pHBY and pHB-SDY were 1200 bp and 1179 bp respectively. These transformants shown in Figure 3A were recombinants with the genes of interest, and belonged to Muts transformants since they had no bands of wild-type AOX1 gene. The results of Coomassie-stained SDS-PAGE analysis of the protein expression of Muts transformants with pHBY and pHB-SDY are shown in Figure 3B. No recombinant protein was visible. The results of HBsAg antigenicity detection using ELISA and immune dot blotting are shown in Figure 4. The expression of transformants with pHBY was intracellular. The recombinant protein of pHB-SDY could not be detected by ELISA, and only weakly demonstrated by immune dot blotting assay. The IMx results are shown in Table 2. The ratio of sample value/cut off value of pHB-SDY was much lower than that of pHBY (P < 0.01, t = 5.02).
Figure 3.
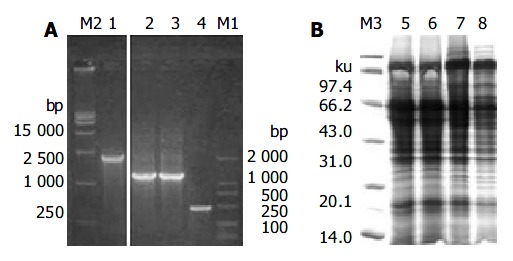
Screening integrants and SDS-PAGE analysis. A: Screening integrants using PCR with AOX1 primers. Lane M2: DNA marker; Lane 1: PCR products of Mut+ transformant without gene of interest; Lane 2: PCR products of Muts transformants with pHBY; Lane 3: PCR products of Muts transformants with pHB-SDY; Lane 4: PCR products of pPIC9 alone. Lane M1 were DNA markers. B: Coomassie-stained SDS-PAGE analysis of the recombinant proteins of pHB-SDY and pHBY. Lane M3: protein marker; Lane 5: cell lysate super-natants of yeast alone; Lane 6: yeast transformed with pPIC9; Lane 7: yeast transformed with pHBY; Lane 8: yeast trans-formed with pHB-SDY.
Figure 4.
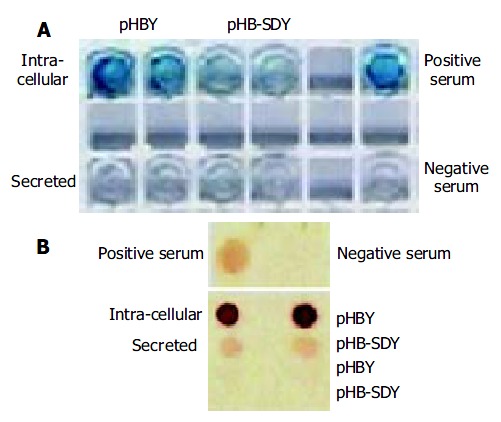
Antigenicity analysis of recombinant proteins using ELISA and immune dot blotting assay. (A) ELISA, (B) immune dot blotting assay.
Table 2.
Antigenicity of HBsAg with second-loop deletion of α determinant
| Groups | HBsAg titer by IMx S/CO value |
Anti-HBs responses in DNA immunization |
|
| Positive rate | mIU/L (GMT ± SE) | ||
| Empty control | - | 0/5 | - |
| pcDNA3 | - | 0/5 | - |
| pHB | 1 589 ± 234.5b | 6/10 | 27.60 ± 17.3c |
| pHB-SD | 106 ± 26.8 | 2/10 | 4.56 ± 3.52 |
S/CO: the ratio of sample against cut off value. GMT: geomet-ric mean titer.
P < 0.01, t = 5.02 vs groups of pHB-SD;
P = 0.02, t = 2.7 vs groups of pHB-SD.
DNA immunization
All mice were alive after the inoculation schedule was finished. The anti-HBs levels are shown in Table 2. The anti-HBs was negative in groups of normal saline and plasmid pcDNA3. The positive rate of anti-HBs was 2/10 in pHB-SD group and 6/10 in pHB group. The amount of anti-HBs induced in groups of HBV S gene with deleted second-loop of the α determinant was less than that in the complete HBV S gene groups (P = 0.02, t = 2.7).
DISCUSSION
The hosts usually do not respond to the subdominant epitopes in both vaccinated individuals and patients because of the dominant negative mechanism. Since they usually have no escape mutations and are not tolerant in patients, the subdominant epitopes have been widely used to overcome immunological tolerance in the fields of tumor and chronic infections[24-27]. Many subdominant epitopes were successfully responded by the hosts with chronic infections or tumor carrying patients[24-29]. However, the most data were limited in CD8 + cells or cytotoxic T lymphocytes. There are few such literatures about the epitopes of B lymphocytes. As a protective antigen, HBsAg is of great significance for prevention of HBV infection. It has been confirmed that there are more than three B-lymphocyte epitopes in HBsAg. The second-loop of α determinant is the strongest one among them[30]. It is the dominant epitope, and the rest ones are the subdominant epitopes.
To completely destroy the dominant epitope may be able to eliminate the dominant negative mechanism, and let the subdominant epitopes be responded by the hosts. In our study even after deletion of the second-loop of HBsAg α determinant, the recombinant protein expressed in yeasts still had a weak antigenicity. Though it escaped the monoclonal anti-HBs derived from the α determinant, the recombinant protein might react with polyclonal anti-HBs or monoclonal anti-HBs derived from other parts of HBsAg because it could be detected by immune dot blotting and Abbott IMx kits, in which the demonstrating antibodies consisted of monoclonal anti-HBs derived from α determinant and the rest part of HBsAg[31]. These results are similar to that of the α determinant variants that could be detected by monoclonal antibody from the rest part of HBsAg too[22]. Mammalian expression vector with deleted HBV S gene could raise a weak anti-HBs response to DNA immunization. It was not sure that this anti-HBs was protective. However, it might be able to react with HBV particles since the anti-HBs derived from the first-loop of the α determinant of HBsAg could be accessible on native HBsAg[21]. These suggested that it was possible to develop a subdominant vaccine for the prevention of immune escape mutant infections of HBV.
The response rate of recombinant plasmids was very low in our study. It might be the nature of DNA immunization because this phenomenon also occurred in researches of other scientists[32]. The deleted HBV S gene was successfully expressed in yeasts. However, the expression conditions need to be improved. The exact value of deleted HBsAg should be evaluated after purified recombinant proteins are obtained in the future. The effect of anti-HBs raised by subdominant epitopes of HBsAg on the infectivity of HBV should also be evaluated in the future.
HBsAg with deleted second-loop of the α determinant still has antigenicity, and can also raise weak anti-HBs response in mice to DNA immunization, suggesting that it is possible to develop a subdominant vaccine for preventing infections of immune escape mutants of HBV.
Footnotes
Supported by the National Natural Science Foundation of China, No. 39970677 and the Science Foundation of Guangdong Province, No. 99M04801G
Edited by Kumar M and Wang XL Proofread by Xu FM
References
- 1.Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395–403. doi: 10.1016/s1473-3099(02)00315-8. [DOI] [PubMed] [Google Scholar]
- 2.Huang P, Ye G, Zhong J, Sha Q. Assessment of current epidemiological status of viral hepatitis in Guangdong Province, China. Southeast Asian J Trop Med Public Health. 2002;33:832–836. [PubMed] [Google Scholar]
- 3.Kane MA. Global control of primary hepatocellular carcinoma with hepatitis B vaccine: the contributions of research in Taiwan. Cancer Epidemiol Biomarkers Prev. 2003;12:2–3. [PubMed] [Google Scholar]
- 4.Tao Q, Feng B. Prevention and therapy of hepatitis B. Chin Med J (Engl) 1999;112:942–946. [PubMed] [Google Scholar]
- 5.André FE, Zuckerman AJ. Review: protective efficacy of hepatitis B vaccines in neonates. J Med Virol. 1994;44:144–151. doi: 10.1002/jmv.1890440206. [DOI] [PubMed] [Google Scholar]
- 6.He C, Nomura F, Itoga S, Isobe K, Nakai T. Prevalence of vaccine-induced escape mutants of hepatitis B virus in the adult population in China: a prospective study in 176 restaurant employees. J Gastroenterol Hepatol. 2001;16:1373–1377. doi: 10.1046/j.1440-1746.2001.02654.x. [DOI] [PubMed] [Google Scholar]
- 7.Jeantet D, Chemin I, Mandrand B, Zoulim F, Trepo C, Kay A. Characterization of two hepatitis B virus populations isolated from a hepatitis B surface antigen-negative patient. Hepatology. 2002;35:1215–1224. doi: 10.1053/jhep.2002.32710. [DOI] [PubMed] [Google Scholar]
- 8.Kim KH, Lee KH, Chang HY, Ahn SH, Tong S, Yoon YJ, Seong BL, Kim SI, Han KH. Evolution of hepatitis B virus sequence from a liver transplant recipient with rapid breakthrough despite hepatitis B immune globulin prophylaxis and lamivudine therapy. J Med Virol. 2003;71:367–375. doi: 10.1002/jmv.10503. [DOI] [PubMed] [Google Scholar]
- 9.Poovorawan Y, Theamboonlers A, Chongsrisawat V, Sanpavat S. Molecular analysis of the a determinant of HBsAg in children of HBeAg-positive mothers upon failure of postexposure prophylaxis. Int J Infect Dis. 1998;2:216–220. doi: 10.1016/s1201-9712(98)90056-x. [DOI] [PubMed] [Google Scholar]
- 10.Levicnik-Stezinar S. Hepatitis B surface antigen escape mutant in a first time blood donor potentially missed by a routine screening assay. Clin Lab. 2004;50:49–51. [PubMed] [Google Scholar]
- 11.Chakravarty R, Neogi M, Roychowdhury S, Panda CK. Presence of hepatitis B surface antigen mutant G145R DNA in the peripheral blood leukocytes of the family members of an asymptomatic carrier and evidence of its horizontal transmission. Virus Res. 2002;90:133–141. doi: 10.1016/s0168-1702(02)00147-8. [DOI] [PubMed] [Google Scholar]
- 12.Thakur V, Kazim SN, Guptan RC, Malhotra V, Sarin SK. Molecular epidemiology and transmission of hepatitis B virus in close family contacts of HBV-related chronic liver disease patients. J Med Virol. 2003;70:520–528. doi: 10.1002/jmv.10426. [DOI] [PubMed] [Google Scholar]
- 13.Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237–247. doi: 10.1007/BF02256597. [DOI] [PubMed] [Google Scholar]
- 14.Koyanagi T, Nakamuta M, Sakai H, Sugimoto R, Enjoji M, Koto K, Iwamoto H, Kumazawa T, Mukaide M, Nawata H. Analysis of HBs antigen negative variant of hepatitis B virus: unique substitutions, Glu129 to Asp and Gly145 to Ala in the surface antigen gene. Med Sci Monit. 2000;6:1165–1169. [PubMed] [Google Scholar]
- 15.Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, Zhou F, Waters J, Karayiannis P, Luo K. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027–1034. doi: 10.1053/jhep.2001.28708. [DOI] [PubMed] [Google Scholar]
- 16.Karthigesu VD, Allison LM, Fortuin M, Mendy M, Whittle HC, Howard CR. A novel hepatitis B virus variant in the sera of immunized children. J Gen Virol. 1994;75(Pt 2):443–448. doi: 10.1099/0022-1317-75-2-443. [DOI] [PubMed] [Google Scholar]
- 17.Chen HB, Fang DX, Li FQ, Jing HY, Tan WG, Li SQ. A novel hepatitis B virus mutant with A-to-G at nt551 in the surface antigen gene. World J Gastroenterol. 2003;9:304–308. doi: 10.3748/wjg.v9.i2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu H, Fujisawa T, Sogo T, Isozaki A, Inui A, Sekine I, Kobata M, Ogawa Y. Acute self-limiting hepatitis B after immunoprophylaxis failure in an infant. J Med Virol. 2002;66:28–33. doi: 10.1002/jmv.2107. [DOI] [PubMed] [Google Scholar]
- 19.Shizuma T, Hasegawa K, Ishikawa K, Naritomi T, Iizuka A, Kanai N, Ogawa M, Torii N, Joh R, Hayashi N. Molecular analysis of antigenicity and immunogenicity of a vaccine-induced escape mutant of hepatitis B virus. J Gastroenterol. 2003;38:244–253. doi: 10.1007/s005350300043. [DOI] [PubMed] [Google Scholar]
- 20.Oon CJ, Chen WN, Goh KT, Mesenas S, Ng HS, Chiang G, Tan C, Koh S, Teng SW, Toh I, et al. Molecular characterization of hepatitis B virus surface antigen mutants in Singapore patients with hepatocellular carcinoma and hepatitis B virus carriers negative for HBsAg but positive for anti-HBs and anti-HBc. J Gastroenterol Hepatol. 2002;17 Suppl:S491–S496. doi: 10.1046/j.1440-1746.17.s4.16.x. [DOI] [PubMed] [Google Scholar]
- 21.Ijaz S, Ferns RB, Tedder RS. A 'first loop' linear epitope accessible on native hepatitis B surface antigen that persists in the face of 'second loop' immune escape. J Gen Virol. 2003;84:269–275. doi: 10.1099/vir.0.18667-0. [DOI] [PubMed] [Google Scholar]
- 22.Jolivet-Reynaud C, Lésenéchal M, O'Donnell B, Becquart L, Foussadier A, Forge F, Battail-Poirot N, Lacoux X, Carman W, Jolivet M. Localization of hepatitis B surface antigen epitopes present on variants and specifically recognised by anti-hepatitis B surface antigen monoclonal antibodies. J Med Virol. 2001;65:241–249. doi: 10.1002/jmv.2026. [DOI] [PubMed] [Google Scholar]
- 23.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 24.Marastoni M, Bazzaro M, Micheletti F, Gavioli R, Tomatis R. Peptide analogues of a subdominant epitope expressed in ebv-associated tumors: synthesis and immunological activity. J Med Chem. 2001;44:2370–2373. doi: 10.1021/jm001136a. [DOI] [PubMed] [Google Scholar]
- 25.Hudrisier D, Riond J, Gairin JE. Molecular and functional dissection of the H-2Db-restricted subdominant cytotoxic T-cell response to lymphocytic choriomeningitis virus. J Virol. 2001;75:2468–2471. doi: 10.1128/JVI.75.5.2468-2471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barouch DH, Craiu A, Santra S, Egan MA, Schmitz JE, Kuroda MJ, Fu TM, Nam JH, Wyatt LS, Lifton MA, et al. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J Virol. 2001;75:2462–2467. doi: 10.1128/JVI.75.5.2462-2467.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson D, Bundell C, Robinson B. In vivo cross-presentation of a soluble protein antigen: kinetics, distribution, and generation of effector CTL recognizing dominant and subdominant epitopes. J Immunol. 2000;165:6123–6132. doi: 10.4049/jimmunol.165.11.6123. [DOI] [PubMed] [Google Scholar]
- 28.Tourdot S, Oukka M, Manuguerra JC, Magafa V, Vergnon I, Riché N, Bruley-Rosset M, Cordopatis P, Kosmatopoulos K. Chimeric peptides: a new approach to enhancing the immunogenicity of peptides with low MHC class I affinity: application in antiviral vaccination. J Immunol. 1997;159:2391–2398. [PubMed] [Google Scholar]
- 29.Chengalvala MV, Bhat RA, Bhat BM, Vernon SK, Lubeck MD. Enhanced immunogenicity of hepatitis B surface antigen by insertion of a helper T cell epitope from tetanus toxoid. Vaccine. 1999;17:1035–1041. doi: 10.1016/s0264-410x(98)00318-1. [DOI] [PubMed] [Google Scholar]
- 30.Maillard P, Pillot J. At least three epitopes are recognized by the human repertoire in the hepatitis B virus group a antigen inducing protection; possible consequences for seroprevention and serodiagnosis. Res Virol. 1998;149:153–161. doi: 10.1016/s0923-2516(98)80033-2. [DOI] [PubMed] [Google Scholar]
- 31.Shah DO, Coleman P, Chen J, Peterson B, Dimarco A, Stewart J. The detection of recombinant hepatitis B surface antigen from "vaccine escape mutants" in two HBsAg immunoassays. Clin Lab. 2000;46:161–163. [PubMed] [Google Scholar]
- 32.Geissler M, Tokushige K, Chante CC, Zurawski VR, Wands JR. Cellular and humoral immune response to hepatitis B virus structural proteins in mice after DNA-based immunization. Gastroenterology. 1997;112:1307–1320. doi: 10.1016/s0016-5085(97)70145-8. [DOI] [PubMed] [Google Scholar]


