Abstract
Cardiolipins (CL) represent unique phospholipids of bacteria and eukaryotic mitochondria with four acyl chains and two phosphate groups that have been implicated in numerous functions from energy metabolism to apoptosis. Many proteins are known to interact with CL, and several cocrystal structures of protein-CL complexes exist. In this work, we describe the collection of the first systematic and, to the best of our knowledge, the comprehensive gold standard data set of all known CL-binding proteins. There are 62 proteins in this data set, 21 of which have nonredundant crystal structures with bound CL molecules available. Using binding patch analysis of amino acid frequencies, secondary structures and loop supersecondary structures considering phosphate and acyl chain binding regions together and separately, we gained a detailed understanding of the general structural and dynamic features involved in CL binding to proteins. Exhaustive docking of CL to all known structures of proteins experimentally shown to interact with CL demonstrated the validity of the docking approach, and provides a rich source of information for experimentalists who may wish to validate predictions.
Introduction
Cardiolipin (CL) is an unusual anionic phospholipid with two negatively charged phosphate groups in its polar head and four hydrophobic acyl chains (1,2). CL is the signature phospholipid of the inner bacterial membrane and the inner mitochondrial membranes of eukaryotic cells where its presence is important for the proper structural arrangements and functioning of a number of proteins (3,4). In addition to its structural role for the proteins of the electron transport chain, it also plays roles in different stages of apoptosis (5), adenylate translocase ion transport (6), and translocation by NDPK-D (7). Lately, extramitochondrial functions of CL in regulating the process of mitophagy (8–11) and orchestrating the NLRP3 inflammasome and maturation of IL-1β (12,13) have been documented. Not surprisingly, CL has been implicated in a number of different diseases, such as Barth syndrome, diabetes, heart disease, radiation damage, Parkinson’s disease, and others. Some of these roles are membrane related, i.e., aberrant CL concentrations and distributions were shown to affect the structure, dynamics, and permeability of the mitochondrial membranes and CL is known to appear on the surface of the cell and also on the plasma membrane during apoptosis. On the other hand, interactions with proteins are crucial for understanding CL’s functions and dysfunctions. Numerous proteins have been reported to interact with CL (reviewed in (14,15)), and in cases where cocrystal structures are known, individual CL binding patches (BPs) have been identified. However, when there is no structure, the location and mode of binding can be difficult to establish. For example, for cytochrome c at least three distinct BPs have been proposed (16–21). Here, we aimed to retrieve a comprehensive list of all known CL-protein pairs to date. The properties of the BPs were investigated in detail using molecular modeling approaches for the available cocrystal structures.
Molecular modeling has been used extensively to predict interactions between small molecules and proteins, essential for example in the drug discovery process (22). Identifying and studying the BPs of specific ligands to a group of proteins helps understanding of the molecular mechanisms governing such interactions (23). In turn, this knowledge may help in the design of proteins with specific functions (24,25), and in development of inhibitors and drugs targeting the identified binding region (26). A wide variety of approaches have been developed and employed to address the BP identification problem in silico (recently reviewed in (27)). Most detailed methods, such as the 3D QSAR relationship, require abundant previous structural knowledge on the binding regions of the ligand to its target. These methods have been successfully used, e.g., for G-protein coupled receptors (28). However, when computational resources and structural information are limited, patch analysis of the biophysical properties of the binding sites become particularly informative (29). Here, we analyzed the trends of amino acid distribution and secondary structure usage in CL-BPs, to differentiate between the regions that bind the polar headgroup and the acyl chains of the CL molecules, respectively. Additionally, taking advantage of the recently discovered overrepresentation of certain supersecondary structural motifs in protein-protein interaction interfaces (30), we searched for such motifs in CL-BPs. Finally, where experimental structural data was not available for analysis, computational predictions by docking methods were performed to provide valuable insight into the binding modes of the ligand to its target (31,32). We therefore also conducted a comprehensive analysis of the potential validity of such predictions in the case of CL-binding proteins.
Materials and Methods
Preparing a database of known CL-binding proteins
A list of proteins experimentally demonstrated to interact with CL (the gold standard), was compiled using four different strategies: 1) extensive literature search, 2) retrieval from the UniProtKB database (33), 3) retrieval from the Protein Data Bank (PDB) (34), and 4) search for the term cardiolipin binding in the Gene Ontology browser—Quick GO (35), as follows. In addition, where available, the PDB entry was obtained from the Cross-reference section of the protein description in the UniProtKB database.
Extensive literature search
Starting with previous reviews of CL-binding proteins (14,15) we extended the search to publications in Pubmed using cardiolipin binding as the search term. Different attributes of the CL-binding proteins detailed in the identified publications (such as their name, gene name, organism name, EC number, PDB entries) were then used to search UniProtKB. The UniProt entry with the best match of the protein attributes was included in the gold standard.
Retrieval from the UniProtKB database
The keyword cardiolipin was used to search the UniProtKB database, resulting in further identification of proteins that interact with CL. Such interactions were later verified by literature review.
Retrieval from the PDB
Additional CL-protein interactions were identified by searching for structures containing CL in the PDB files (34). The chain IDs in the remarks were used to retrieve the UniProt entry of the protein.
Gene ontology cardiolipin binding term search
The GO:1901612 ID, which corresponds to the gene ontology term cardiolipin binding, was used to search the Quick GO browser. The search results revealed additional CL-binding proteins, which were recorded along with the UniProtKB references provided in the Quick GO search results. Literature search was used to verify the interactions of these proteins with CL.
Predicting 3D structures of proteins in the gold standard without known structures
For proteins without annotated PDB entries, 3D models were obtained using the online protein structure homology-modeling server—SWISS-MODEL (36,37). Models having QMEAN Z-scores >−2.0 were selected and included for obtaining structure-based features of the protein. For models having a QMEAN4 score <−2.0 and Torsion Z-score >−2.0, the Optimize plugin (38) for the PyMol software (10,39), was used to optimize the models. The optimized models were then uploaded to the QMEAN server (40,41) for reevaluation. The models with a QMEAN Z-score >−2.0 were included for obtaining structure-based features of the protein.
Homology assessment
To avoid overrepresentation of close homologs in the analysis of BPs thus ensuring no redundant information about CL-binders is included in the gold standard, we removed proteins sharing >90% sequence identity using the CD-HIT Suite (42).
Definition of BPs
CL-BPs were defined as the set of amino acids in the interacting protein chain containing at least one atom within a distance of 5 Å to any CL atom, for each protein chain/CL molecule pair. This definition was narrowed further to define three subsets of BPs to consider the specific nature of negative electric charges in the CL polar head and hydrophobic interactions involving the acyl chains. We thus defined phosphate binding patches (PBPs) as the set of amino acids in the interacting protein chain containing at least one atom within a range distance of 5 Å to any phosphorous atom in the CL molecule. Analogously, we defined hydroxyl binding patches (OBPs) as the set of amino acids in the interacting protein chain containing at least one atom within a range distance of 5 Å to the oxygen atom in the central glycerol moiety in CL molecule. Hence, a PBP or a OBP will always be a subset of the corresponding general BP given by the first definition in this subsection. Acyl binding patches (ABPs) were defined as the asymmetric difference set operation between the BP and the union of PBP and OBP for each CL-protein chain pair (i.e., an ABP is defined by the subset of residues in the BP neither belonging to the corresponding PBP nor to the corresponding OBP). All these definitions were applied to each crystallographic structure obtained from the PDB that contained 3D coordinates for both the CL molecule and its binding protein.
Sequence- and structure-based features
For proteins with known 3D structure, their amino acid composition (primary structure) was obtained directly from the corresponding PDB file. The assignment of secondary structure in those proteins, and the solvent accessibility was obtained by using DSSP 2.0.4 (43). We categorized the different annotations of DSSP as follows: H, G, and I as Helix; E and B as strand; any other annotation as Coiled coil. Whenever DSSP was unable to produce an assignment for a particular amino acid, this amino acid was excluded from the secondary structure analysis. Finally, the assignment of supersecondary structures or loops on such proteins was obtained from Markov Clustering classification terms listed in the ArchDB database (44). Relative accumulated frequencies and propensities of amino acids and secondary structures, and relative accumulated frequencies of loops were computed and represented using R (45) (see Metrics section below). Such computation was applied to five different data sets: 1) the complete protein chain, 2) BPs, 3) PBPs, 4) OBPs, and 5) ABPs. For this purpose, a loop was considered to be included in the relevant protein region for a given analysis scope if at least one amino acid of the loop belonged to such a region.
Metrics
The relative frequency of an element in a population is given by
| (1) |
where N(i) represents the total number of observations of such element and T the total number of elements in the population. In this work, we refer to the relative accumulated frequencies of a certain amino acid as the total number of observed instances of such amino acid in a given region (i.e., whole chain or any type of binding patch) over the total number of amino acids (any one) in the same region. For secondary structures, we consider the total number of residues within a certain structural type in a given region over the total number of residues in such structural type on the same region. For loops, the relative accumulated frequency would be the number of instances of a particular loop type in a certain region over the total number of loops in that region.
Propensity of a certain amino acid or secondary structure element is defined as in (29):
| (2) |
where N(i)surf is the total count of exposed instances of such amino acid or of residues in such secondary structure element, and Tsurf is the total number of exposed amino acids. We considered a residue exposed if its relative solvent accessibility (46) was higher than 25% (47). Within a defined set of protein crystals, every amino acid or secondary structural element will have a characteristic N(i)surf/Tsurf ratio, K(i). Compared to frequencies, the computation of propensities takes into account the natural inclination of a certain element (amino acid or secondary structural element) to be exposed in the protein surface. Hence, propensities will highlight, among the studied population, elements which are abundant in relation to the natural tendency of such element to be exposed. To study BPs, this measure can pinpoint key elements for the interaction (48).
Statistical significance of differences between the different measures analyzed was assessed using the Chi-squared test as implemented in R (45). The null model considered the compared elements have equal F(i) (see Eq. 1) and equal K(i) (see Eq. 2). P-values from these Chi-squared tests are given throughout this work. For a given comparison, a p-value <= 0.05 denotes statistically significant differences, whereas a p-value >0.05 denotes that the differences observed between the two analyzed elements could also be observed by chance. To stress this breakpoint, we illustrate p-values in a double colored scale, linear values from dark red (1) to light pink (0.05) to denote nonsignificant p-values and a logarithmic scale ranging from light blue (0.05) to dark blue (0) for significant p-values.
Docking
Proteins were classified into two groups for structural analysis, 1) CL cocrystal structures, i.e., proteins having a PDB entry with a CL structure present in the PDB file; 2) proteins having a PDB entry, but no CL structures included; 3) proteins only having a Swiss model entry. Docking was essential for proteins belonging to the second and third category, but it was also performed for the proteins in the first category to test the compatibility of the docking results with the CL-binding sites given in the PDB files. AutoDock Tools (49) was used to obtain PDBQT files of the receptors (proteins) and ligand (CL), and for determining the dimensions of the grid box for each protein chain, which acts as the search space for docking the ligand. The polar hydrogens were added to the protein chain molecule followed by computing Gasteiger charges for the molecule. The result was used to compute the corresponding PDBQT file. The CL species selected as ligand in all cases irrespective of the CL species bound in the cocrystal structures was 1,1′,2,2′-tetralinoleoyl-CL (TLCL), as it is an oxidizable species of CL and the most abundant CL species found in mammalian mitochondria (50). AutoDock Vina (51) was employed to run molecular docking simulations. Default parameters were used and the docking specific parameters are provided in File S5. Results were analyzed using PyMol (39) and all pymol sessions are provided in Files S3 and S5 for single chain and multiple chain dockings, respectively. The top nine different docking poses of CL were manually classified into binding sites, i.e., those modes of the docked CL that bound the receptor at the same location were grouped into a common binding site. For proteins having CL costructures in the PDB file, overlap of each predicted binding site with each crystallographic CL site was recorded. An overlap was identified whenever the phosphate heads of the docked CL were in proximity to those of the CL costructure and both the molecules shared similar orientations of the acyl chains. Proteins with no overlapping predicted site to the crystallographic one were redocked and this time the top 50 different docking poses were considered for manual classification into binding sites and posterior analysis.
Protein chains were docked individually, i.e., if other chains were present in the pdb file, they were removed for docking. Therefore, in a few isolated cases, we found some of the CL docking poses to be partially overlapping with these other chains. These instances were recorded as clashes. We also retrieved the amino acid residues of each receptor protein at 5 Å distance from each CL docking pose.
Results
Identification of CL-binding proteins
We identified a total of 62 CL-binding proteins by different procedures described in the Materials and Methods section, and these proteins are listed in Table 1. Detailed information about these gold standard proteins is provided in Table S1. 26 of them were identified from extensive literature search, 12 were retrieved from the UniprotKB database, 22 were obtained from the PDB, and 2 from GO term search. Crystallographic data containing CL molecules and their protein binding partners were available for 22 of these proteins, represented in 34 different PDB protein chains. For another 21, crystallographic data was available but not in the presence of the CL binding molecule (30 PDB protein chains). 3D structure was assigned to six proteins using homology modeling. We were not able to assign 3D structure to the 13 remaining proteins. To avoid overrepresentation of near identical structures, we pruned our analysis set of proteins sharing >90% sequence identity with others using CD-hit (42). See Table S1 for details on which protein chains were removed. This procedure resulted in a set containing 21 proteins with CL cocrystals observed in 15 different PDB records.
Table 1.
Proteins in gold standard
| Protein Name | Mb | CL | PDB_Chain | Ref. |
|---|---|---|---|---|
| Alpha-MGlcDAG synthase | Y | NA | 1Z2T_A | (52) |
| Phospholipase A2 | N | NA | 1POC_A | (53) |
| Cyt. c oxidase sub. 1 | Y | NA | 2OCC_A | (54) |
| Cyt. c oxidase sub. 7A1, mit. | Y | NA | 2OCC_J | (54) |
| Cyt. c oxidase sub. 7C, mit. | Y | NA | 2OCC_L | (54) |
| ADP/ATP translocase 1 | Y | CDL | 1OKC_A | (55) |
| Cyt. b-c1 complex sub. 1, mit. | Y | CDL | 1SQP_A | (56) |
| Cyt. c1, heme protein, mit. | Y | CDL | 1SQP_D, | (56), |
| 2A06_D, Q | (57) | |||
| Cytochrome b | Y | CDL | 2A06_C, P | (57) |
| Adrenodoxin, mit. | Y | NA | 2BT6_A, B | (58,59) |
| ATP synthase sub. beta, mit. | Y | NA | 2CK3_D, E, F | (60) |
| Cyt. b-c1 complex subunit 8 | Y | CDL | 2YBB_G, | (61), |
| 1SQP_G | (56) | |||
| Cholesterol desmolase | Y | NA | 3MZS_A,B,C,D | (58,62) |
| Protein S100-B | N | NA | 3RM1_A | (63) |
| Cyt. c oxidase sub. 6A2, mit. | Y | CDL | 3WG7_G, T | (64) |
| Cyt. c oxidase sub. 3 | Y | CDL | 3WG7_C, P | (64) |
| FDH-N sub. gamma | Y | CDL | 1KQF_C | (65) |
| FDH-N sub. beta | Y | CDL | 1KQG_B | (65) |
| Cyt. c | Y | NA | 3O1Y_A, B, C | (66) |
| Chr. replication initiator prot. DnaA | Y | NA | 1J1V_A | (67) |
| Cyt. b-556 | Y | CDN | 2ACZ_C | (68) |
| DNA topoisomerase 1 | N | NA | 1MW9_X | (69) |
| Cyt. b | Y | CDL | 3TGU_C, P | (70) |
| Mit. ubiquinol-cyt. c reductase ubiquinone-binding prot. qp-c | Y | CDL | 3TGU_G | (70) |
| Mit. cyt. c1, heme protein | Y | CDL | 3TGU_Q | (70) |
| Prohibitin | Y | NA | 1LU7_A | (71) |
| Beta-2-glycoprotein 1 | N | NA | 3OP8_A, B | (72,73) |
| ABC transporter 10 prot. | Y | CDL | 3ZDQ_A | (74) |
| Acyl-CoA thioesterase THEM5 | Y | NA | 4AE7_A | (75) |
| Caspase-8 | Y | NA | 4JJ7_A | (76) |
| Apoptosis regulator BAX | Y | NA | 4BD2_A | (77) |
| Uncharacterized protein | Y | CDL | 4A2N_B | (78) |
| BH3-interacting domain death agonist | Y | NA | 2VOI_B | (79,80) |
| Antigen-presenting glycoprot. CD1d1 | Y | CD4 | 3MA7_A, C | (81) |
| Neutral ceramidase | N | NA | 2zws_A | (82) |
| Carnitine/acylcarnitine translocase | Y | NA | 2BMN_A | (83) |
| Reaction center prot. M chain | Y | CDL | 4LWY_M | (84) |
| Reaction center prot. M chain | Y | CDL | 4N7L_M | (85) |
| ADP,ATP carrier prot. 2 | Y | CDL | 4C9G_A | (86) |
| ADP,ATP carrier prot. 3 | Y | CDL | 4C9J_A, B | (86) |
| Cyt. b | Y | CDL, | 1KB9_C, | (87), |
| CN3, | 3CX5_N, | (88), | ||
| CN5 | 3CX5_C | (88) | ||
| Cyt. c1, heme prot. mit. | Y | CDL, | 1P84_D, | (89), |
| CN3 | 3CX5_D | (88) | ||
| Sapecin | N | NA | 1L4V_A | (90) |
| Malate dehydrogenase, mit. | Y | NA | model | (59) |
| Thiosulfate sulfurtransferase | Y | NA | model | (91) |
| Tafazzin | Y | NA | model | (76) |
| Serine/threonine-prot. kinase N1 | Y | NA | model | (92) |
| Creatine kinase S-type, mit. | Y | NA | model | (93) |
| Mit. glycerol-3-phosphate dehydrogenase | Y | NA | model | (94) |
| NADH dehydrogenase sub. 1 | Y | NA | NA | (95) |
| Phosphate carrier prot mit. | Y | NA | NA | (96) |
| Prot. UPS1, mit. | Y | NA | NA | (97) |
| Prot. UPS2, mit. | Y | NA | NA | (97) |
| Lysocardiolipin acyltransferase 1 | Y | NA | NA | (98) |
| Phospholipid scramblase 3 | Y | NA | NA | (99,100) |
| Stomatin-like prot. 2, mit. | Y | NA | NA | (101) |
| MICOS complex subunit MIC27 | Y | NA | NA | (102) |
| Phospholipid-transporting ATPase IC | Y | NA | NA | (103) |
| Glycerol-3-phosphate acyltransf. 1, mit. | Y | NA | NA | (104) |
| Carbamoyl-phosphate sinthase I | Y | NA | NA | (105) |
| Neoverrucotoxin subunit alpha | N | NA | NA | (106) |
| AMP deaminase 1 isoform M | N | NA | NA | (107) |
The column Protein name shows the UniProtKB recommended name of the protein. Some records use alternate and short names as provided in UniProtKB for convenience. Recurring words are abbreviated: mit. for mitochondrial, cyt. for cytochrome, sub. for subunit, chr. for chromosomal, prot. for protein, and transf. for transferase. The column Mb indicates if protein is a membrane or nonmembrane protein (Y and N, respectively). CL shows the structure ID(s) of the cardiolipin costructure(s) given in the PDB file of the protein. PDB_chain indicates the chain of the protein’s PDB structure known to interact with cardiolipin. Records of proteins included in the nonredundant cocrystals subset are shown in bold face. Note that the reaction center protein M chain in PDB 4NL7 was excluded from this subset because it is 100% identical to reaction center protein M chain in PDB 4LWY, although both proteins are from different organisms.
Identification and analysis of CL-BPs
As some of the protein chains had more than one CL molecule bound, a total of 27 CL-BPs were identified in 21 protein cocrystal structures with CL. Within BPs, we differentiated between regions with preference for binding of the polar headgroup (PBPs and OBPs) of CL molecule and regions with preference for binding the acyl chains of CL (ABPs) (see Materials and Methods section). 15 of the 21 proteins were also mapped to supersecondary structures in the ArchDB database, referred hereinafter as loops (44). Loops were mapped to 14 BPs, 12 ABPs, 5 OBPs, and 9 PBPs.
We have used the set of CL-binding protein cocrystal structures to analyze the characteristics of CL-BPs in proteins in terms of their amino acid composition (Figs. 1 and S1), secondary structure (Figs. 2 and S2), and supersecondary structure/loops (Fig. 3). Finally, we compared the values of these three features in the CL-BPs, ABPs, OBPs, and PBPs to the values obtained from the complete CL-interacting proteins.
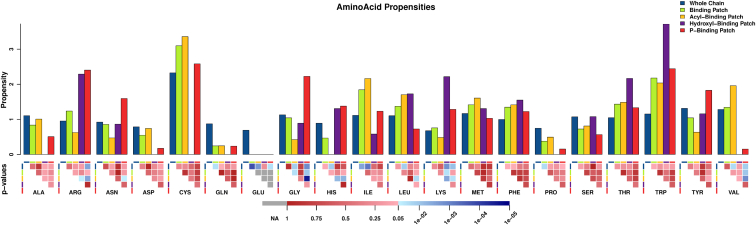
Figure 1.
Propensities of observed amino acids in all protein chains analyzed (blue), BPs (green), ABPs (dark yellow), OBPs (purple), and PBPs (red). The p-values for the statistical significance of differences between propensities of each amino acid in different BPs are shown in the lower panel. NA (gray) indicates that two empty sets were compared and hence no statistics can be computed. In red and using a linear scale, nonsignificant p-values ranging from 1 (darker red, indicates that greater differences could always be observed in a random set) to 0.05 (lighter red) are shown. In blue and using a logarithmic scale, significant p-values are shown, from 0.05 (light blue, differences observed could be reproduced by random only in 5 over 100 cases) to p-values near 0 (dark blue, the probability to observe such differences by chance is the lowest).
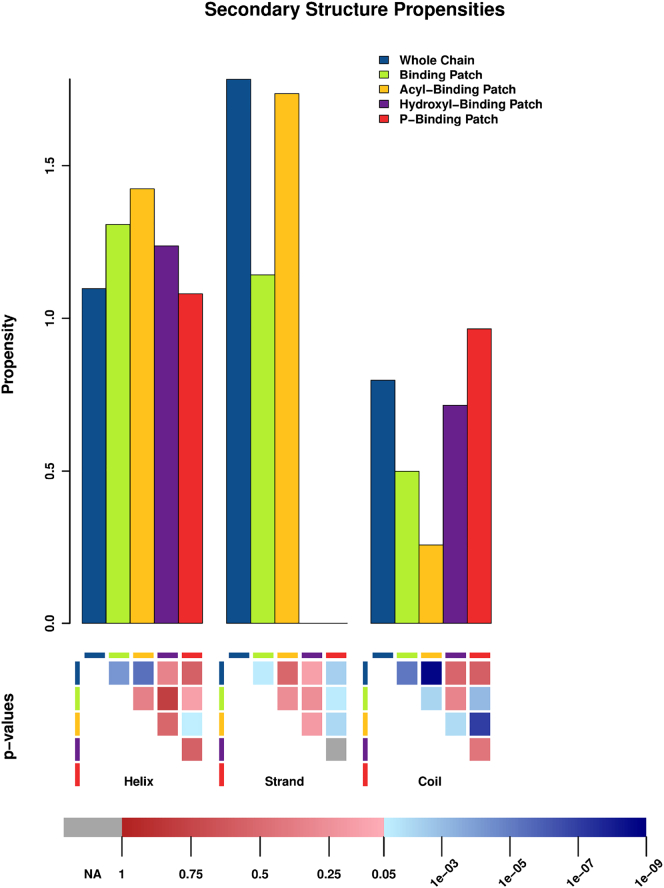
Figure 2.
Relative propensity of observed secondary structures in all protein chains (blue), BPs (green), ABPs (dark yellow), OBPs (purple), and PBPs (red). The p-values for the statistical significance of differences between propensities of each amino acid in different BPs are shown in the lower panel, using the same color legend described in Fig 1.
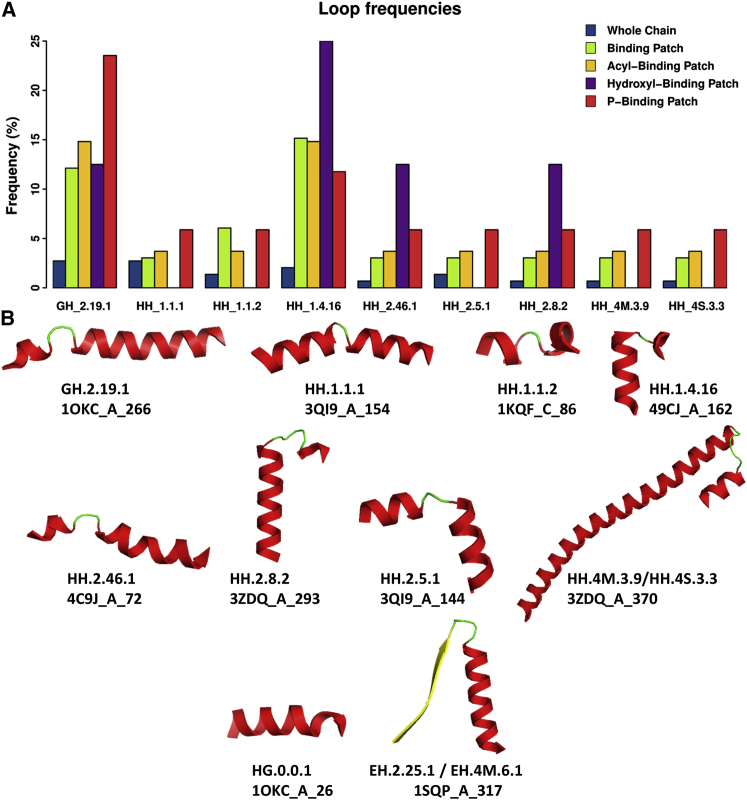
Figure 3.
(A) Relative frequencies of observed supersecondary motifs (loops) in all protein chains (blue), BPs (green), ABPs (dark yellow), OBPs (purple), and PBPs (red). (B) Cartoon images of the loops present in PBPs. Different secondary structure elements are shown in color (helices in red, strands in yellow, and coiled-coil regions in green). Each motif is identified in the first text line below by the code of its corresponding loop type(s). The PDB file from where each loop structure shown was obtained, protein chain and starting residue of the supersecondary structure are provided in the second line below each cartoon.
Electrostatic-driven amino acid bias trends in CL-binding regions
The amino acid composition shown in Fig. 1 indicates that there is an overrepresentation of positively charged residues in PBPs and OBPs, primarily Arg and Lys residues but also His, when compared to ABPs. This suggests an important role of electrostatic interactions in attracting CL molecules to their protein binders, as has been proposed for specific proteins previously (108–110). The fact that negatively charged residues (Asp and Glu) are underrepresented in BPs further supports this hypothesis. Strikingly, although Asp is only underrepresented in PBPs, demonstrating its incompatibility to interact with the negative polar head of CL, Glu is completely disallowed in the whole CL-binding region (Figs. 1 and S1), suggesting there exist further restrictions for the usage of this amino acid in CL-BPs.
Hydrophobic interactions with the acyl chains
As expected, hydrophobic amino acids dominate the ABPs (Figs. 1 and S1). In particular, Leu, Ile, and Val are most frequent. The accumulated frequencies of Leu, Ile, and Val alone represent 24% of the amino acid content of the studied proteins, and sum up to 15% in the PBPs. In comparison, these hydrophobic amino acids are overrepresented in BPs and ABPs, representing 31% and 40% of their amino acid content, respectively. Although these three amino acids are the most strikingly overrepresented ones, Phe, Thr, Met, and Trp are also more frequent in BPs when compared to all chains. Grouping amino acids by hydrophobicity alone is of course a simplification, and this is most clearly visible when looking at Tyr. Although a hydrophobic amino acid, it is rarely found in ABPs, either due to the hydrophilic OH group or its role in regulation, i.e., the potential to be phosphorylated, which would prevent CL binding. Significance values of frequencies and propensities comparisons between different amino acids are provided in File S1.
Flexibility trends in CL-binding regions
A remarkable bias was observed in the distribution of amino acids related to protein flexibility (Fig. 1): Gly, which imparts flexibility is highly overrepresented and Pro, which generally introduces rigidities in the 3D structure of proteins is highly underrepresented in PBSs. This observation suggests a requirement for higher flexibility in the region binding the polar headgroups of CL. The opposite is observed for the ABPs. Here, Gly is underrepresented, although Pro is also but less strongly underrepresented when compared to all chains. This indicates that structured regions favor the binding of the acyl chains.
Secondary structure analysis
The analysis of secondary structure compositions is shown in Figs. 2 and S2. Overall, helices are most frequent, but this is expected as the majority of known CL-binding proteins with structures are integral membrane proteins of the helical bundle type (Table 1). We therefore concentrated on the differences in the preference for secondary structures in BPs, PBPs, and ABPs as compared to the full chains. Although ABPs are mainly composed of helical secondary structures and only contain ∼10% unorganized regions, the percentage of residues with coiled-coil conformation remarkably rises to almost 40% in PBPs. Although this value is similar to that of the whole amino acid chain, the important fact is its increased percentage of coiled-coil conformation in BPs and especially PBPs when compared to ABPs (Fig. S2). In fact, propensities analysis reveal a reduced preference of ABPs for coiled-coil conformation (∼0.25) in comparison the PBPs, which display a normal trend for this secondary structure element (propensity ∼1) (Fig. 2). Although the intrinsic rigidity of Pro induces the disruption of regular secondary structures (111), the increased coiled-coil organization in PBPs cannot be attributed to this effect because the abundance of Pro is clearly diminished in such regions (Fig. 1). Hence, the increased preference for coiled-coil conformation in PBPs depicts the larger requirements for flexibility in the binding of the polar CL heads, further supporting the flexibility trends observed in the amino acid compositions (see above). In line with this conclusion is also the complete absence of strand secondary structure conformations in PBPs. In contrast to the PBPs, the ABPs prefer helices, and there is a clear trend even beyond the overall abundance of helices in our gold standard data set. Strand is also an acceptable structural element, although the percentage is no higher than observed in all chains. In line with this observation, coiled coil is strongly disfavored in ABPs. These observations also support the conclusions from amino acid preferences, that the acyl chains favor rigid structured elements to anchor on.
Supersecondary structure motifs in CL-binding regions
Recent studies show that loops, defined as supersecondary structural motifs formed by a coiled region flanked by two correlative regular secondary motifs (112), to be useful features in the prediction of protein-protein interactions (30). A database of such supersecondary structural motifs has been built (44). With the aim of identifying putative specific structural footprints of CL-binding sites, we extended our analysis of CL-binding regions to the loops in this database. 94 different types of loops were observed in regions of CL-binding proteins, 23 of which were located in different types of BPs (Tables S2 and S3). 20 of these 23 were represented in ABPs and only 12 of them were in PBPs. Moreover, three of the loop types are exclusive to PBPs, 11 are exclusive to ABPs, and 9 loop types are shared among PBPs and ABPs. The frequencies of these loops in the CL-binding proteins are shown in Fig. 3A, and cartoon images of these loop structures are depicted in Fig. 3B. Most frequent are loops GH_2.19.1 and HH_1.4.16. Most loops were observed more frequently in PBPs than in ABPs, with the exception of HH_1.4.16. PBP-exclusive loops are: 1) HG.0.1.1 for which two instances found in PBPs (1OKC_A residues 26–40 and 4N7L_M residues 82–91), and 2) EH.2.25.1 and EH.4M.6.1, which represent the same supersecondary structural motif and was observed once in PBPs (1SQP_A residues 317–348). All these loops represent hydrophobic residue-rich helices in their original structures and contain positively charged residues near the flexible region of the loop. Only the N-terminal β-strand of the EH motif in 1SQP_A is not in close proximity to the polar head of CL.
Of the nine loop types shared among PBPs and ABPs (Fig. 3A), GH.2.19.1 shows the most remarkable preference for PBPs. Four instances of this loop are present in three of the proteins included in our analysis, and all of them are also present in both PBPs and ABPs. This fact suggests a possible relationship between this supersecondary structure and the CL-binding function. Due to the lesser number of loops mapped in BPs than in proteins (23 and 94, respectively), and in PBPs than in ABPs (12 and 20, respectively), the relative frequency of this loop is progressively higher in BPs, ABPs, and PBPs. Remarkably, all structures clustered in this loop type contain a Gly residue in the second position of its flexible region (44), thus making it an extremely conserved residue. This Gly is preceded by a positively charged residue (Arg or Lys) in CL-binding proteins. A second positively charged residue is located near this position, allowing for the stabilization of the double negatively charged CL polar head. Fig. 4 illustrates these details in the example of the ATP/ADP carrier protein (PDB ID 1OKC), which has two different CL-binding sites. Strikingly, in both of the binding sites a GH.2.19.1 loop is present, showing that this particular protein adopted the same structural solution for binding two CL molecules. Thus, the presence of a GH.2.19.1 loop with a positively charged residue preceding a crucial flexibility conferring Gly in the phosphate binding region and a helix providing an anchor for acyl chain binding, may be considered a footprint for CL binding.
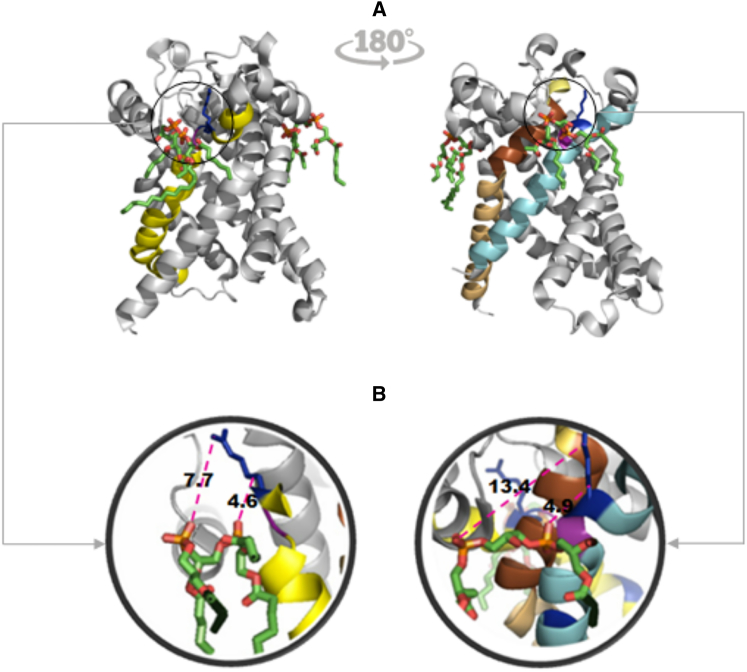
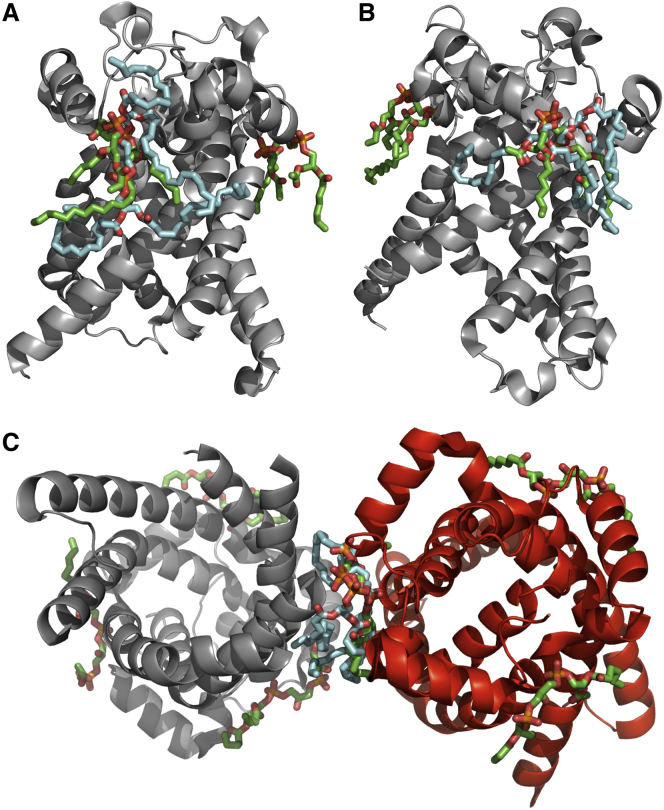
Figure 4.
Loops of ADP/ATP translocase 1 (PDB 1OKC chain A) interacting with two cardiolipin molecules. (A) One CL molecule (left) interacts with loop GH_2.19.1 (1OKC_A_66; residues 66–99; colored in yellow).The second CL molecule (right) interacts with a different instance of loop GH_2.19.1 (1OKC_A_266; residues 266–291; colored in cyan), showing that both binding sites adopt similar structural conformation. The second molecule also interacts with loop HH_1.1.2 (1OKC_A_4; residues 4–37; colored in light orange) and loop HG_0.1.1 (1OKC_A_26; residues 26–40; colored in pale yellow). Overlapping region between HH_1.1.2 and HG_0.1.1 (residues 26–37) is shown in brown). Positively charged amino acids Arg and Lys in loop GH_2.19.1 are shown in blue, and the conserved Gly in the loop motif is colored in purple. (B) Enlarged images of areas circled in (A) showing interaction between positively charged residues and the phosphate groups of CL in the two different binding sites, where CL binding is accomplished by different geometries. The geometry of binding is described by measured distances between two proximal oxygen atoms in the different CL phosphate groups to carbon beta and distal nitrogen in relevant Arg (4.6 Å and 7.7 Å, respectively, left) or Lys (4.9 Å and 13.4 Å, respectively, right).
Validity of predicted CL-binding sites
Approximately one-third of the proteins (22 of 62, 21 after removing sequence redundancy) in the gold standard data set have known CL bound cocrystal structures, and a further 21 have structures without CL (Table 1). This represents a potentially rich source of structural features for analysis and prediction of CL-binding sites. CL-protein docking predictions have been used successfully in the structural analysis of CL interactions previously (9,113–116) and have been useful in shedding light on the mechanisms governing such interactions. We therefore docked TLCL, the most abundant CL species in mitochondria, to all structures after removing all bound ligands in them, and analyzed the results (see Materials and Methods section). Of the top-ranked poses predicted by Autodock Vina, binding sites were manually clustered together and highlighted in File S2. In most cases, we predicted more than one binding site. However, checking for overlap with the known CL-binding sites in the 21 cocrystal structures (Table 2) indicated that the prediction of binding sites by docking is very effective. For 18 of the 21 analyzed proteins, the predicted CL docking poses overlapped with the crystallographic CL molecule annotated in the proteins’ structure. Fig. 5 illustrates this overlap for the ATP/ADP carrier protein in its monomeric and dimeric forms. Clashes (docking poses where CL molecule overlap with known protein-protein interaction interfaces, see Materials and Methods) were observed in 10 of the 18 binding sites, but seven of them had at least one overlapping docking pose that was free of clashes. The other 8 of 18 were completely free of clashes.
Table 2.
Overlap analysis for PDBs with CL costructure(s)
| UniProt | PDB | C. | CL ID | O. | Cl. | #DP | #OP | #CP | #NCP | #BS | #OS | #NCS | Homo-log Struct-ure | BE (kcal/mol) | RMSD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P0AEK7 | 1KQF | C | CDL | N | NA | 50 | NA | NA | NA | 5 | NA | NA | NA | NA | NA |
| P0AAJ3 | 1KQG | B | CDL | Y | Y | 9 | 9 | 8 | 1 | 1 | 1 | 1 | NA | −4.3 | 10.9 |
| P02722 | 1OKC | A | CDL | Y | N | 9 | 3 | 0 | 3 | 4 | 2 | 2 | NA | −4.0 | 11.6 |
| P31800 | 1SQP | A | CDL | N | NA | 50 | NA | NA | NA | 5 | NA | NA | NA | NA | NA |
| P13271 | 1SQP | G | CDL | Y | N | 9 | 1 | 0 | 1 | 3 | 1 | 1 | NA | −3.0 | 14.3 |
| P00157 | 2A06 | C | CDL | Y | Y | 50 | 5 | 2 | 3 | 5 | 1 | 1 | 2A06_P | −5.5 | 13.3 |
| P00125 | 2A06 | D | CDL | Y | Y | 9 | 4 | 4 | 0 | 3 | 1 | 0 | NA | −3.5 | 12.8 |
| P69054 | 2ACZ | C | CDN | Y | N | 50 | 1 | 0 | 1 | 8 | 1 | 1 | NA | −5.2 | 11.8 |
| P00163 | 3CX5 | C | CN5 | Y | Y | 50 | 4 | 1 | 3 | 8 | 1 | 1 | NA | −6.8 | 17.5 |
| P07143 | 3CX5 | D | CN3 | Y | Y | 9 | 9 | 9 | 0 | 1 | 1 | 0 | NA | −4.5 | 15.9 |
| P11609 | 3MA7 | A | CD4 | Y | N | 9 | 7 | 0 | 7 | 2 | 1 | 1 | NA | −7.0 | 12.6 |
| P18946 | 3TGU | C | CDL | Y | Y | 50 | 10 | 1 | 9 | 8 | 1 | 1 | 3TGU_P | −4.8 | 12.7 |
| D0VX32 | 3TGU | G | CDL | Y | Y | 50 | 5 | 4 | 1 | 5 | 1 | 1 | NA | −2.9 | 14.7 |
| D0VX26 | 3TGU | Q | CDL | Y | Y | 50 | 1 | 1 | 0 | 10 | 1 | 0 | NA | −5.0 | 12.0 |
| P00415 | 3WG7 | C | CDL | Y | Y | 50 | 4 | 1 | 3 | 6 | 1 | 1 | NA | −5.8 | 12.5 |
| P07471 | 3WG7 | G | CDL | N | NA | 50 | NA | NA | NA | 6 | NA | NA | NA | NA | NA |
| Q9NRK6 | 3ZDQ | A | CDL | Y | N | 9 | 3 | 0 | 3 | 2 | 1 | 1 | NA | −5.8 | 13.5 |
| Q8TMG0 | 4A2N | B | CDL | Y | N | 9 | 2 | 0 | 2 | 3 | 1 | 1 | NA | −4.6 | 13.4 |
| P18239 | 4C9G | A | CDL | Y | N | 50 | 3 | 0 | 3 | 8 | 1 | 1 | NA | −3.5 | 12.4 |
| P18238 | 4C9J | A | CDL | Y | N | 50 | 2 | 0 | 2 | 8 | 2 | 2 | NA | −3.4 | 14.5 |
| P0C0Y9 | 4LWY | M | CDL | Y | Y | 50 | 4 | 1 | 3 | 8 | 1 | 1 | NA | −5.8 | 13.6 |
The column UniProt shows the UniProt ID of the protein. PDB indicates the PDB structure ID. C. lists the chain of the PDB structure, which corresponds to the UniProt ID. CL ID indicates the structure ID of the CL molecule that binds with the protein. O. shows whether the crystallographic CL molecule overlaps with any of the docked CLs or not (Y and N, respectively); Cl. indicates whether any clash has occurred between the overlapping docking poses and other chains of the PDB structure or not (Y and N, respectively); #DP gives the number of docking poses of the docked CL; #OP indicates the number of docking poses of the docked CL overlapping with the CL crystallographic costructure; #CP shows the number of overlapping modes that clash with other chains of the PDB structure; #NCP Number of overlapping modes which do not clash with any chain of the PDB structure; #BS shows the number of different manually clustered binding sites of the docked CL; #OS indicates the number of binding sites including at least one pose overlapping with the CL crystallographic costructure; #NCS shows the number of binding sites including at least one pose which do not clash with any chain of the PDB structure; Homolog structure indicates an alternate structure file for the same protein (UniProt ID) in which an overlap between crystallographic CL and any docked one was observed in cases where such overlap did not occur for the PDB and chain shown in columns 2 and 3. Crystal structures referred to in columns 2 and 3 were the ones included in the nonredundant cocrystals subset described in main text. BE kcal/mol shows the lowest binding energy of the docked CL’s overlapping docking pose. RMSD shows the root mean-square deviation between the lowest energy overlapping docked pose and the crystallographic pose of CL. NA here refers to no overlap observed. Proteins in which no overlap between the crystallographic CL molecule and any docked one was observed show NA in Cl., #DP, #OP, #CP, #NCP, #BS, #OS, #NCS, and BE kcal/mol columns.
Figure 5.
Overlap of docked CL molecule with CL costructures of 1OKC and 4C9J. The docked CL is colored in cyan and the CL costructures are colored in green. (A) Overlap with CDL 800 molecule of 1OKC; (B) Overlap with CDL 801 molecule of 1OKC; (C) Overlap with CDL 802 molecule of 4C9J chain A (red).
Only TLCL was docked in this study, although there were five different crystallographic identities of CL species (CDL, CN3, CN5, CDN, and CD4). The fact that docking was still able to identify the known CL-binding site despite the different CL species used in docking provides important evidence that docking with AutoDock Vina (51) is a highly reliable method for identification of general CL-binding sites in proteins. All of the poses obtained in the docking are available in the Supporting Material as PyMol (39) sessions (File S3), not only for the CL-cocrystal structures but also structures lacking CL. This provides a rich source of predictions that experimentalists can use to verify CL-binding sites.
Comparison of studied trends in crystallographic CL-protein complexes to docking and negative data sets
Taking advantage of all docked poses, we defined four new sets of CL-binding protein costructures. The first one contains all docking results in Table 2. The second and third ones respectively contain the particular docking poses overlapping (positive docking control) and nonoverlapping (negative control) with the crystallographic structure as defined in Table 2 and Supporting Material File S2. The fourth data set contains the results of docking TLCL to 30 nonredundant proteins for which no cocrystal was available. Results of this comparison are provided in Figs. S3–S10 (amino acid and secondary structure elements propensity values), and in File S4 (statistical significance for differences between trends in crystallographic complexes and trends in the different docking data sets defined previously) in the Supporting Material. Remarkably, docking results by Autodock Vina of overlapping binding sites (docking positives) reproduce the electrostatic and hydrophobic features described for crystallographic complexes, but not the flexibility trends. For instance, docking positives as well as the overall docking results show an enrichment of positively charged residues in PBPs and OBPs, whereas in ABPs hydrophobic residues are predominant. If secondary structural elements are considered, the ABPs preference for strand conformation (compared to PBPs and OBPs) is then confirmed. However, not even the controlled positives from docking experiments are able to reproduce the unique trend for Gly in PBPs or the preference for coiled-coil conformation in PBPs and OBPs seen in the experimentally observed crystal structure binding sites.
Discussion
In this work we made three contributions to the field of CL binding to proteins. 1) We collected a gold standard data set of all known CL binding proteins. This is the only systematic and to the best of our knowledge comprehensive list available to date. 2) Our BP analysis of the compiled set of cocrystal structures not only confirmed for some CL binding proteins the experimentally validated expectation that hydrophobicity and charge play a role in binding, but we also revealed a new, to our knowledge, and unexpected feature important for binding: flexibility. Thus, a typical binding patch is composed of positively charged or nonpolar residues in PBPs and ABPs, respectively, with a remarkable overrepresentation of Arg and Lys in PBPs and of Leu, Ile, and Val in ABPs. In contrast, negatively charged residues (Asp and Glu) are either underrepresented or disallowed in PBPs. Furthermore, the region binding the polar headgroups of the CL molecule shows a differential trend toward increased flexibility, with a higher content of Gly and coiled-coil secondary structure, and a lower content of Pro. The only and remarkable case in which crystallographic data did not comply with these trends was that of the hydrophobic residue Tyr, overrepresented in PBPs. Tyr is a widely used residue in signaling due to its ability of being phosphorylated. Such phosphorylation may prevent CL binding and could therefore be disfavored here. In addition, Tyr usually participates in hydrogen bonding and it may have other roles besides binding. For example, it may participate in structural arrangements essential for catalytic functions, similar to Tyr-67 in cytochrome c (117). Finally, in the investigation of loop supersecondary structural motifs (44) in BPs, loop GH.2.19.1., characteristic for the mitochondrial ATP/ADP carrier, appeared particularly prominently. It is tempting to speculate that this loop containing a positively charged residue preceding the crucial flexibility conferring Gly in the PBP region, and a helix providing an anchor for acyl chain binding, may be considered a footprint for CL binding. 3) We carried out an exhaustive docking of TLCL to all known structures of proteins experimentally shown to interact with CL. This not only demonstrates the validity of the docking approach, but also provided new, to our knowledge, insights for designing further experiments.
CL binding has initially been thought of as being dominated by electrostatic forces (17,19,118), and our analysis supports this notion. However, numerous studies have proposed additional hydrophobic forces to contribute to binding (19,21,66), a hypothesis also supported by our findings. A popular model combining these two features is a two-step process where electrostatic attraction initiates the interaction of CL with protein, and hydrophobic contacts further consolidate the binding to be of high affinity (20,66). Recently, the role of electrostatic attraction has been challenged (86) because the crystal structures of the yeast mitochondrial ATP/ADP carrier (PDBs 4C9G, 4C9J) show that the lateral chains of the positively charged residues preceding the central conserved Gly are oriented in a plane perpendicular to that defined by the phosphate groups in the CL molecule, and pointing in the direction opposite to the phosphates. This led to the speculation that instead of direct electrostatic interaction, CL interacts with the positively charged end of the helix dipoles, which correspond to the C-terminal secondary structure in loops GH.2.19.1 shown in Fig. 4. Further insight can be gained from analysis of the cattle mitochondrial ATP/ADP carrier (PDB 1OKC) (55), which contains two different CL-BPs. In one of them (Fig. 4, right), the lateral chain of the preceding Lys is oriented analogous to the yeast mitochondrial ATP/ADP carrier, whereas the other binding site shows the lateral chain of the Arg preceding Gly to be parallel to the plane defined by the phosphate groups in the CL molecule (Fig. 4, left). Our finding that flexibility appears to be an important feature for binding of the phosphate groups, would suggest that a wide spectrum of geometries of amino acid side chains can be involved in CL binding to the same supersecondary motif. The high flexibility in these areas would make it likely that the Lys side chains are not fixed in space (either parallel or perpendicular) but are likely to be dynamic and able to adopt multiple conformations. This view is compatible with both models, direct electrostatic interactions of the phosphate groups with positively charged amino acids or with the electric dipole created by the helix. It is remarkable that loop GH.2.19.1, which we propose here as a footprint of CL binding, is compatible with all features, the previously proposed positive charge and/or the electric helix dipole and the hydrophobic anchor. In addition, we propose a direct role of flexibility in facilitating binding.
An important issue to address in the future is the role of multiple chains in binding CL. The majority of CL cocrystal structures contain multiple chains (14 of 18 pdb files) and even though the HETATM field in the pdb code identifies each CL molecule with a specific chain, there are contributions of neighboring chains to the binding pockets. This can yield further insight into binding but can also help improve the docking. Docking of the large protein complexes becomes highly challenging and multiple grid boxes are required to cover the docking space. For illustration purposes, we selected three pdb files containing multiple chains (File S6) and compared docking of the single chains with docking of all chains (File S7). The comparison indicates that highly complementary information can be obtained by the two approaches, requiring further in-depth investigation for the contribution of interfaces to CL-binding pockets in the future.
Although we have observed clear trends in BP properties in CL-binding proteins, we have not yet explored specificity. For example, other anionic phospholipids or even other doubly negatively charged small molecules such as GDP or ADP may show similar general structural and dynamics features in their respective binding pockets. Hence, it is premature to regard the trends described here as specific to CL binding. Conversely, we cannot ascertain if all CL binding modes will be the same. In particular, there may be different modes depending on the role of the CL interaction with proteins. For example, in respirasomes, CL acts as a staple between the subunits. In contrast, in cytochrome c, CL appears to remain membrane bound while interacting with the protein. In some proteins, CL binding has no demonstrated effect on function. Thus, different functional roles of CL binding may be accomplished by yet unidentified modes of binding and these may be specific to a given role.
At least for some types of CL-binding proteins—those represented by the current data set—our studies open the door for developing a generic algorithm for qualitatively predicting protein-CL interactions and for stratifying computed binding sites from docking. By comparing a predicted binding site for the presence of specific amino acid types, secondary structures and supersecondary structures, it might be possible to accurately select the actual binding pose of CLs from several high-ranking docking poses. Furthermore, the availability of a gold standard data set can be used in combination with suitable features to develop a general classification model that can be applied at the system-wide level to predict if a given protein may interact with CL or not. Currently, work is underway to develop such a classifier.
Author Contributions
J.K.S. initiated and oversaw the study. J.P.I. and J.K.S. designed the research and interpreted the results. J.P.I., H.D., and D.M. generated and analyzed the data presented. N.K. and V.E.K. contributed critical discussion. J.P.I., H.D., and J.K.S. wrote the article and all authors participated in editing. All authors read and accepted this manuscript.
Acknowledgments
The authors thank the Human Frontier Science Program and National Institutes of Health (grant NIH U19AIO68021) for funding this study.
Editor: Ozlem Keskin.
Footnotes
Joan Planas-Iglesias and Himal Dwarakanath contributed equally to this work.
Ten figures, three tables, and seven supporting files are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00769-9.
Supporting Material
This file shows the statistical significance of the comparison of frequencies and propensities of the defined binding patches between different amino acids or secondary structure elements.
This file shows details on which docking poses were clustered to define binding sites and their overlap with crystallographic CL and clashes with the rest of the structure.
This file contains PyMol (58) sessions illustrating structural details of the results of each individual docking.
This file shows the statistical significance of the comparison of frequencies and propensities of the defined binding patches between crystallographic complexes and docking results.
This file presents the center of each protein chain coordination and the dimensions of the grid-box, which are used in Vina docking software.
This file contains the PyMol (58) sessions illustrating structural details of the results of three exemplary multi-chain dockings.
This file provides an analysis of the results from multiple chain docking (File S6) with the corresponding single chain dockings (within File S3).
References
- 1.Daum G. Lipids of mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 2.Schlame M., Brody S., Hostetler K.Y. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur. J. Biochem. 1993;212:727–735. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- 3.Belikova N.A., Tyurina Y.Y., Kagan V.E. Heterolytic reduction of fatty acid hydroperoxides by cytochrome c/cardiolipin complexes: antioxidant function in mitochondria. J. Am. Chem. Soc. 2009;131:11288–11289. doi: 10.1021/ja904343c. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y., Kelley R.I., Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J. Biol. Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper R.H., Vaz F.M. Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 2008;65:2493–2506. doi: 10.1007/s00018-008-8030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rück A., Dolder M., Brdiczka D. Reconstituted adenine nucleotide translocase forms a channel for small molecules comparable to the mitochondrial permeability transition pore. FEBS Lett. 1998;426:97–101. doi: 10.1016/s0014-5793(98)00317-2. [DOI] [PubMed] [Google Scholar]
- 7.Francois-Moutal L., Marcillat O., Granjon T. Structural comparison of highly similar nucleoside-diphosphate kinases: molecular explanation of distinct membrane-binding behavior. Biochimie. 2014;105:110–118. doi: 10.1016/j.biochi.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Chu C.T., Bayır H., Kagan V.E. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: implications for Parkinson disease. Autophagy. 2014;10:376–378. doi: 10.4161/auto.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu C.T., Ji J., Kagan V.E. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan V.E., Chu C.T., Bayir H. Cardiolipin asymmetry, oxidation and signaling. Chem. Phys. Lipids. 2014;179:64–69. doi: 10.1016/j.chemphyslip.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X.X., Tsoi B., Li Y.F., Kurihara H., He R.R. Cardiolipin and its different properties in mitophagy and apoptosis. J. Histochem. Cytochem. 2015;63:301–311. doi: 10.1369/0022155415574818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iyer S.S., He Q., Sutterwala F.S. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill L.A. Cardiolipin and the Nlrp3 inflammasome. Cell Metab. 2013;18:610–612. doi: 10.1016/j.cmet.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Schlame M., Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta. 2009;1788:2080–2083. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlame M., Rua D., Greenberg M.L. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 16.Kawai C., Prado F.M., Nantes I.L. pH-Dependent interaction of cytochrome c with mitochondrial mimetic membranes: the role of an array of positively charged amino acids. J. Biol. Chem. 2005;280:34709–34717. doi: 10.1074/jbc.M412532200. [DOI] [PubMed] [Google Scholar]
- 17.Kostrzewa A., Páli T., Marsh D. Membrane location of spin-labeled cytochrome c determined by paramagnetic relaxation agents. Biochemistry. 2000;39:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- 18.Rytömaa M., Kinnunen P.K. Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J. Biol. Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 19.Rytömaa M., Kinnunen P.K. Reversibility of the binding of cytochrome c to liposomes. Implications for lipid-protein interactions. J. Biol. Chem. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 20.Sinibaldi F., Howes B.D., Santucci R. Extended cardiolipin anchorage to cytochrome c: a model for protein-mitochondrial membrane binding. J. Biol. Inor. Chem. 2010;15:689–700. doi: 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 21.Tuominen E.K., Wallace C.J., Kinnunen P.K. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. J. Biol. Chem. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 22.Jacob L., Vert J.P. Protein-ligand interaction prediction: an improved chemogenomics approach. Bioinformatics. 2008;24:2149–2156. doi: 10.1093/bioinformatics/btn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sael L., Kihara D. Detecting local ligand-binding site similarity in nonhomologous proteins by surface patch comparison. Proteins. 2012;80:1177–1195. doi: 10.1002/prot.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Looger L.L., Dwyer M.A., Hellinga H.W. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 25.Bonet J., Segura J., Fernandez-Fuentes N. Frag‘r’Us: knowledge-based sampling of protein backbone conformations for de novo structure-based protein design. Bioinformatics. 2014;30:1935–1936. doi: 10.1093/bioinformatics/btu129. [DOI] [PubMed] [Google Scholar]
- 26.Joseph-McCarthy D., Campbell A.J., Moustakas D. Fragment-based lead discovery and design. J. Chem. Inf. Model. 2014;54:693–704. doi: 10.1021/ci400731w. [DOI] [PubMed] [Google Scholar]
- 27.Konc J., Janežič D. Binding site comparison for function prediction and pharmaceutical discovery. Curr. Opin. Struct. Biol. 2014;25:34–39. doi: 10.1016/j.sbi.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez D., Gutiérrez-de-Terán H. Computational approaches for ligand discovery and design in class-A G protein-coupled receptors. Curr. Pharm. Des. 2013;19:2216–2236. doi: 10.2174/1381612811319120009. [DOI] [PubMed] [Google Scholar]
- 29.Jones S., Thornton J.M. Prediction of protein-protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 30.Planas-Iglesias J., Bonet J., Oliva B. Understanding protein-protein interactions using local structural features. J. Mol. Biol. 2013;425:1210–1224. doi: 10.1016/j.jmb.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Sousa S.F., Ribeiro A.J., Ramos M.J. Protein-ligand docking in the new millennium–a retrospective of 10 years in the field. Curr. Med. Chem. 2013;20:2296–2314. doi: 10.2174/0929867311320180002. [DOI] [PubMed] [Google Scholar]
- 32.Brewerton S.C. The use of protein-ligand interaction fingerprints in docking. Curr. Opin. Drug Discov. Devel. 2008;11:356–364. [PubMed] [Google Scholar]
- 33.Consortium T.U. Activities at the Universal Protein Resource (UniProt) Nucl. Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman H.M., Westbrook J., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Binns D., Dimmer E., Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasini M., Bienert S., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordoli L., Kiefer F., Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 38.O’Boyle N.M., Banck M., Hutchison G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC, New York, NY.
- 40.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benkert P., Künzli M., Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Y., Niu B., Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joosten R.P., te Beek T.A., Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonet J., Planas-Iglesias J., Oliva B. ArchDB 2014: structural classification of loops in proteins. Nucleic Acids Res. 2014;42:D315–D319. doi: 10.1093/nar/gkt1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A language and environment for statistical computing. [Google Scholar]
- 46.Adamczak R., Porollo A., Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004;56:753–767. doi: 10.1002/prot.20176. [DOI] [PubMed] [Google Scholar]
- 47.Adamczak R., Porollo A., Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 48.Keskin O., Tsai C.J., Wolfson H., Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyurina Y.Y., Kini V., Kagan V.E. Mechanisms of cardiolipin oxidation by cytochrome c: relevance to pro- and antiapoptotic functions of etoposide. Mol. Pharmacol. 2006;70:706–717. doi: 10.1124/mol.106.022731. [DOI] [PubMed] [Google Scholar]
- 51.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berg S., Edman M., Wieslander A. Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J. Biol. Chem. 2001;276:22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 53.Hovius R., Thijssen J., de Kruijff B. Phospholipid asymmetry of the outer membrane of rat liver mitochondria. Evidence for the presence of cardiolipin on the outside of the outer membrane. FEBS Lett. 1993;330:71–76. doi: 10.1016/0014-5793(93)80922-h. [DOI] [PubMed] [Google Scholar]
- 54.Arnarez C., Marrink S.J., Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci. Rep. 2013;3:1263–1270. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pebay-Peyroula E., Dahout-Gonzalez C., Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 56.Esser L., Quinn B., Xia D. Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex. J. Mol. Biol. 2004;341:281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 57.Huang L.S., Cobessi D., Berry E.A. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J. Mol. Biol. 2005;351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pember S.O., Powell G.L., Lambeth J.D. Cytochrome P-450scc-phospholipid interactions. Evidence for a cardiolipin binding site and thermodynamics of enzyme interactions with cardiolipin, cholesterol, and adrenodoxin. J. Biol. Chem. 1983;258:3198–3206. [PubMed] [Google Scholar]
- 59.Ou W.J., Ito A., Omura T. Specific binding of mitochondrial protein precursors to liposomes containing cardiolipin. J. Biochem. 1988;103:589–595. doi: 10.1093/oxfordjournals.jbchem.a122312. [DOI] [PubMed] [Google Scholar]
- 60.Eble K.S., Coleman W.B., Cunningham C.C. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 1990;265:19434–19440. [PubMed] [Google Scholar]
- 61.Althoff T., Mills D.J., Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lambeth J.D. Cytochrome P-450scc. Cardiolipin as an effector of activity of a mitochondrial cytochrome P-450. J. Biol. Chem. 1981;256:4757–4762. [PubMed] [Google Scholar]
- 63.Zolese G., Tangorra A., Donato R. Interaction of S-100b protein with cardiolipin vesicles as monitored by electron spin resonance, pyrene fluorescence and circular dichroism. Cell Calcium. 1988;9:149–157. doi: 10.1016/0143-4160(88)90018-8. [DOI] [PubMed] [Google Scholar]
- 64.Hirata K., Shinzawa-Itoh K., Ago H. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nat. Methods. 2014;11:734–736. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- 65.Jormakka M., Törnroth S., Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 66.Kalanxhi E., Wallace C.J. Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem. J. 2007;407:179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yung B.Y., Kornberg A. Membrane attachment activates dnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horsefield R., Yankovskaya V., Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 2006;281:7309–7316. doi: 10.1074/jbc.M508173200. [DOI] [PubMed] [Google Scholar]
- 69.Mizushima T., Natori S., Sekimizu K. Inhibition of Escherichia coli DNA topoisomerase I activity by phospholipids. Biochem. J. 1992;285:503–506. doi: 10.1042/bj2850503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao G.F., Wang F., Yang G.F. Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex. J. Am. Chem. Soc. 2012;134:11168–11176. doi: 10.1021/ja3001908. [DOI] [PubMed] [Google Scholar]
- 71.Thuaud F., Ribeiro N., Désaubry L. Prohibitin ligands in cell death and survival: mode of action and therapeutic potential. Chem. Biol. 2013;20:316–331. doi: 10.1016/j.chembiol.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kertesz Z., Yu B.B., Sim R.B. Characterization of binding of human beta 2-glycoprotein I to cardiolipin. Biochem. J. 1995;310:315–321. doi: 10.1042/bj3100315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuura E., Igarashi Y., Koike T. Heterogeneity of anticardiolipin antibodies defined by the anticardiolipin cofactor. J. Immunol. 1992;148:3885–3891. [PubMed] [Google Scholar]
- 74.Shintre C.A., Pike A.C., Carpenter E.P. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. USA. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuravleva E., Gut H., Hemmings B.A. Acyl coenzyme A thioesterase Them5/Acot15 is involved in cardiolipin remodeling and fatty liver development. Mol. Cell. Biol. 2012;32:2685–2697. doi: 10.1128/MCB.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalvez F., Schug Z.T., Gottlieb E. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 2008;183:681–696. doi: 10.1083/jcb.200803129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sani M.A., Dufourc E.J., Gröbner G. How does the Bax-alpha1 targeting sequence interact with mitochondrial membranes? The role of cardiolipin. Biochim. Biophys. Acta. 2009;1788:623–631. doi: 10.1016/j.bbamem.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 78.Yang J., Kulkarni K., Barford D. Mechanism of isoprenylcysteine carboxyl methylation from the crystal structure of the integral membrane methyltransferase ICMT. Mol. Cell. 2011;44:997–1004. doi: 10.1016/j.molcel.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y., Tjandra N. Structural insights of tBid, the caspase-8-activated Bid, and its BH3 domain. J. Biol. Chem. 2013;288:35840–35851. doi: 10.1074/jbc.M113.503680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gonzalvez F., Pariselli F., Petit P.X. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 2005;12:614–626. doi: 10.1038/sj.cdd.4401571. [DOI] [PubMed] [Google Scholar]
- 81.Dieudé M., Striegl H., Rauch J. Cardiolipin binds to CD1d and stimulates CD1d-restricted γδ T cells in the normal murine repertoire. J. Immunol. 2011;186:4771–4781. doi: 10.4049/jimmunol.1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kita K., Sueyoshi N., Ito M. Activation of bacterial ceramidase by anionic glycerophospholipids: possible involvement in ceramide hydrolysis on atopic skin by Pseudomonas ceramidase. Biochem. J. 2002;362:619–626. doi: 10.1042/0264-6021:3620619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Indiveri C., Tonazzi A., Palmieri F. Kinetic characterization of the reconstituted carnitine carrier from rat liver mitochondria. Biochim. Biophys. Acta. 1991;1065:231–238. doi: 10.1016/0005-2736(91)90235-z. [DOI] [PubMed] [Google Scholar]
- 84.Kljashtorny V.G., Fufina T.Y., Gabdulkhakov A.G. Molecular dynamic studies of reaction centers mutants from Rhodobacter sphaeroides and his mutant form L(M196)H+H(M202)L. Crystallogr. Rep. 2014;59:536–541. [Google Scholar]
- 85.Saer R.G., Pan J., Beatty J.T. Structural and kinetic properties of Rhodobacter sphaeroides photosynthetic reaction centers containing exclusively Zn-coordinated bacteriochlorophyll as bacteriochlorin cofactors. Biochim. Biophys. Acta. 2014;1837:366–374. doi: 10.1016/j.bbabio.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 86.Ruprecht J.J., Hellawell A.M., Kunji E.R. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. USA. 2014;111:E426–E434. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lange C., Nett J.H., Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Solmaz S.R., Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J. Biol. Chem. 2008;283:17542–17549. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 89.Palsdottir H., Lojero C.G., Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J. Biol. Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 90.Komano H., Homma K., Natori S. Involvement of sapecin in embryonic cell proliferation of Sarcophaga peregrina (flesh fly) FEBS Lett. 1991;289:167–170. doi: 10.1016/0014-5793(91)81061-c. [DOI] [PubMed] [Google Scholar]
- 91.Zardeneta G., Horowitz P.M. Physical characterization of a reactivatable liposome-bound rhodanese folding intermediate. Biochemistry. 1993;32:13941–13948. doi: 10.1021/bi00213a025. [DOI] [PubMed] [Google Scholar]
- 92.Morrice N.A., Gabrielli B., Wettenhall R.E. A cardiolipin-activated protein kinase from rat liver structurally distinct from the protein kinases C. J. Biol. Chem. 1994;269:20040–20046. [PubMed] [Google Scholar]
- 93.Müller M., Moser R., Carafoli E. Cardiolipin is the membrane receptor for mitochondrial creatine phosphokinase. J. Biol. Chem. 1985;260:3839–3843. [PubMed] [Google Scholar]
- 94.Beleznai Z., Jancsik V. Role of cardiolipin in the functioning of mitochondrial L-glycerol-3-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 1989;159:132–139. doi: 10.1016/0006-291x(89)92414-5. [DOI] [PubMed] [Google Scholar]
- 95.Fry M., Green D.E. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J. Biol. Chem. 1981;256:1874–1880. [PubMed] [Google Scholar]
- 96.Cheneval D., Müller M., Carafoli E. The mitochondrial phosphate carrier reconstituted in liposomes is inhibited by doxorubicin. FEBS Lett. 1983;159:123–126. doi: 10.1016/0014-5793(83)80429-3. [DOI] [PubMed] [Google Scholar]
- 97.Osman C., Haag M., Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Y., Chen Y.Q., Cao G. The microsomal cardiolipin remodeling enzyme acyl-CoA lysocardiolipin acyltransferase is an acyltransferase of multiple anionic lysophospholipids. J. Lipid Res. 2009;50:945–956. doi: 10.1194/jlr.M800567-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Inuzuka T., Inokawa A., Maki M. ALG-2-interacting Tubby-like protein superfamily member PLSCR3 is secreted by an exosomal pathway and taken up by recipient cultured cells. Biosci. Rep. 2013;33:e00026. doi: 10.1042/BSR20120123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Van Q., Liu J., Hatch G.M. Phospholipid scramblase-3 regulates cardiolipin de novo biosynthesis and its resynthesis in growing HeLa cells. Biochem. J. 2007;401:103–109. doi: 10.1042/BJ20060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Christie D.A., Lemke C.D., Madrenas J. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol. Cell. Biol. 2011;31:3845–3856. doi: 10.1128/MCB.05393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weber T.A., Koob S., Reichert A.S. APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS One. 2013;8:e63683. doi: 10.1371/journal.pone.0063683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ray N.B., Durairaj L., Mallampalli R.K. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat. Med. 2010;16:1120–1127. doi: 10.1038/nm.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vancura A., Haldar D. Purification and characterization of glycerophosphate acyltransferase from rat liver mitochondria. J. Biol. Chem. 1994;269:27209–27215. [PubMed] [Google Scholar]
- 105.Brandt M.A., Powers-Lee S.G. The interaction of cardiolipin with rat liver carbamoyl phosphate synthetase I. Arch. Biochem. Biophys. 1991;290:14–20. doi: 10.1016/0003-9861(91)90585-7. [DOI] [PubMed] [Google Scholar]
- 106.Ueda A., Suzuki M., Shiomi K. Purification, properties and cDNA cloning of neoverrucotoxin (neoVTX), a hemolytic lethal factor from the stonefish Synanceia verrucosa venom. Biochim. Biophys. Acta. 2006;1760:1713–1722. doi: 10.1016/j.bbagen.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 107.Purzycka-Preis J., Zydowo M.M. Regulatory effect of pig heart phospholipids on heart muscle AMP-deaminase. Int. J. Biochem. 1987;19:565–568. doi: 10.1016/0020-711x(87)90142-x. [DOI] [PubMed] [Google Scholar]
- 108.Milon L., Meyer P., Lacombe M.L. The human nm23-H4 gene product is a mitochondrial nucleoside diphosphate kinase. J. Biol. Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 109.Schlattner U., Tokarska-Schlattner M., Epand R.M. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochim. Biophys. Acta. 2009;1788:2032–2047. doi: 10.1016/j.bbamem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 110.Vacheron M.J., Clottes E., Vial C. Mitochondrial creatine kinase interaction with phospholipid vesicles. Arch. Biochem. Biophys. 1997;344:316–324. doi: 10.1006/abbi.1997.0181. [DOI] [PubMed] [Google Scholar]
- 111.MacArthur M.W., Thornton J.M. Influence of proline residues on protein conformation. J. Mol. Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 112.Oliva B., Bates P.A., Sternberg M.J. An automated classification of the structure of protein loops. J. Mol. Biol. 1997;266:814–830. doi: 10.1006/jmbi.1996.0819. [DOI] [PubMed] [Google Scholar]
- 113.Kagan V.E., Tyurina Y.Y., Bayir H. Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxid. Redox Signal. 2015;22:1667–1680. doi: 10.1089/ars.2014.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tyurina Y.Y., Poloyac S.M., Kagan V.E. A mitochondrial pathway for biosynthesis of lipid mediators. Nat. Chem. 2014;6:542–552. doi: 10.1038/nchem.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kapralov A.A., Yanamala N., Kagan V.E. Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochim. Biophys. Acta. 2011;1808:2147–2155. doi: 10.1016/j.bbamem.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atkinson J., Kapralov A.A., Kagan V.E. A mitochondria-targeted inhibitor of cytochrome c peroxidase mitigates radiation-induced death. Nat. Commun. 2011;2:497. doi: 10.1038/ncomms1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kagan V.E., Bayir H.A., Borisenko G. Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic. Biol. Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beyer K., Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file shows the statistical significance of the comparison of frequencies and propensities of the defined binding patches between different amino acids or secondary structure elements.
This file shows details on which docking poses were clustered to define binding sites and their overlap with crystallographic CL and clashes with the rest of the structure.
This file contains PyMol (58) sessions illustrating structural details of the results of each individual docking.
This file shows the statistical significance of the comparison of frequencies and propensities of the defined binding patches between crystallographic complexes and docking results.
This file presents the center of each protein chain coordination and the dimensions of the grid-box, which are used in Vina docking software.
This file contains the PyMol (58) sessions illustrating structural details of the results of three exemplary multi-chain dockings.
This file provides an analysis of the results from multiple chain docking (File S6) with the corresponding single chain dockings (within File S3).