Abstract
Introduction
Previous studies have shown that glutathione S-transferase P1 (GSTP1) was associated with chronic obstructive pulmonary disease (COPD). However, the association between GSTP1 Ile (105) Val gene polymorphism and COPD remains controversial. To drive a more precise estimation, we performed a meta-analysis based on published case–control studies.
Methods
An electronic search of PubMed, EMBASE, Cochrane library, Web of Science and China Knowledge Resource Integrated (CNKI) Database for papers on GSTP1 Ile (105) Val gene polymorphism and COPD risk was performed. The pooled odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of association in the homozygote model, heterozygote model, dominant model, recessive model and an additive mode. Statistical heterogeneity, test of publication bias and sensitivity analysis was performed. The software STATA (Version 13.0) was used data analysis.
Results
Overall, seventeen studies with 1892 cases and 2012 controls were included in this meta-analysis. The GSTP1 Ile (105) Val polymorphism showed pooled odds ratios for the homozygote comparison (OR = 1.501, 95%CI [0.862, 2.614]), heterozygote comparison (OR = 0.924, 95%CI [0.733, 1.165]), dominant model (OR = 1.003, 95%CI [0.756, 1.331]), recessive model (OR = 1.510, 95%CI [0.934, 2.439]), and an additive model (OR = 1.072, 95%CI [0.822, 1.398]).
Conclusions
In conclusion, the current meta-analysis, based on the most updated information, showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in any genetic models. The results of subgroup analysis also showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in Asian population and Caucasian population. Further studies involving large populations and careful control with age, sex, ethnicity, and cigarette smoking are greatly needed.
Keywords: Glutathione S-transferase P1, Ile (105) Val, GSTP1, Chronic obstructive pulmonary, COPD, Meta-analysis
1. Introduction
Chronic obstructive pulmonary disease (COPD), as a significant major cause of chronic morbidity and mortality worldwide, is characterized by incompletely reversible airflow limitation and persistent airway inflammation (Hanania and Marciniuk, 2011). Previous studies have demonstrated that chronic inflammation and varying degrees of emphysematous alveolar destruction are the key pathological features of the disease (Stockley et al., 2009). Some studies have revealed that an imbalance of endogenous proteinases and antiproteinases, inflammatory cells, proinflammatory mediators, and oxidative stress were responsible for the pathogenesis of COPD (Vestbo et al., 2013). Genetic factors and environmental exposures like tobacco smoke are also involved in the pathogenesis of COPD (Restrepo, 2015). Tobacco smoke is regarded the most important risk factor for COPD, and smokers account for 80–90% of all COPD patients (Wang et al., 2014a).
However, only 10–15% of smokers develop clinically significant COPD (Mannino et al., 2002; Salvi, 2014). Many COPD patients have a family history and several studies have showed that the individual's risk differences to tobacco smoke injury may be related to genetic factors and the genetic factors may also play an important role in the pathogenesis of COPD (Hoidal, 2001; Molfino, 2004). Therefore, it is widely believed that COPD results from an interaction between genetic factors and environmental exposures. A lot of candidate gene studies have been carried out to identify genetic susceptibility factors for COPD over the past few years (Wang et al., 2013a, 2014b; Castellucci et al., 2015; Cheng et al., 2015; Murphy et al., 2015).
Previous studies have demonstrated that the development of COPD may be associated with the genetic variation in the enzymes that detoxify cigarette smoke products, such as glutathione S-transferases (GSTs). GSTs, a functionally diverse family of enzymes, were involved in the conjugation of a wide range of electrophilic substances with glutathione, facilitating detoxification, metabolism and excretion of the smoke products. Of the six classes of GSTs, i.e. alpha (GSTA), mu (GSTM1), pi (GSTP1), theta (GSTT1), sigma and kappa, GSTP1 was expressed more abundantly in respiratory tissue (Cantlay et al., 1994). A number of studies have focused on the relationship between GSTP1 105Val/Val genotype and COPD risk in different ethnic populations with conflicting results, probably due to small sample sizes in those studies. Meta-analysis is a good statistical method to combine the results from multiple studies in an effort to increase power, improve estimates of the size of the effect and/or to resolve uncertainty when reports disagree. Yan et al. performed a meta-analysis included ten studies with a total of 1140 cases and 1263 controls and suggested a significant association between GSTP1 gene polymorphism and COPD risk (Yan et al., 2010). Based on the most updated information and the current available evidence, we performed this updated meta-analysis to drive a more precise estimation of GSTP1 Ile (105) Val gene polymorphism and COPD risk.
2. Materials and methods
2.1. Search strategy
This meta-analysis was performed according to the standard MOOSE guideline (Stroup et al., 2000).PubMed, EMBASE, Cochrane library, Web of science and China Knowledge Resource Integrated Database (until May 1, 2015) were searched using search terms as “(polymorphism OR variants OR mutation) AND COPD AND (GST OR Glutathione-S-transferase OR GSTP1)”. Studies published in English or in Chinese language were selected. Case–control studies containing available genotype frequencies of GSTP1 Ile (105) Val were chosen. Related reference articles were also searched to identify other relevant publications. Unpublished data were not included.
2.2. Inclusion and exclusion criteria
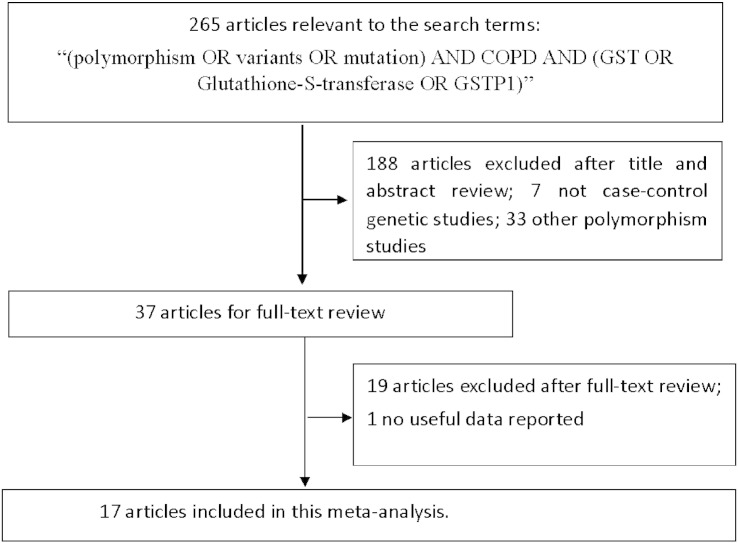
Eligible studies were selected following inclusion criteria: 1) GSTP1 Ile (105) Val polymorphism and COPD risk; 2) human case–control design; 3) application of standardized clinical or pathologic criteria for the diagnosis of COPD; 4) studies that reported the frequency of the GSTP1 Ile (105) Val gene polymorphism as number of cases and controls according to the three variant genotypes of either polymorphisms; and 5) published in English or Chinese. The criteria for the exclusion of studies are as follows: 1) not related to the GSTP1 Ile (105) Val gene polymorphism and COPD risk; 2) not a primary case–control study; 3) no usable or sufficient genotype data reported; 4) studies whose allele frequency in the control population deviated from the Hardy–Weinberg Equilibrium (HWE) at a p value equal or less than 0.01; 5) case reports, letter to Editor, book chapters or reviews. The study inclusion and exclusion procedures are summarized in Fig. 1.
Fig. 1.
Flow diagram of search strategy and study selection.
2.3. Data extraction
The data from all qualified studies were extracted by two investigators independently according to the selection standard listed above. Discrepancies were solved through discussion until agreement was reached. The following information was extracted: the first author's name, year of publication, Ethnicity, the source of control group evidence of Hardy-Weinberg equilibrium (HWE) in controls, the sample size, number of cases and controls with the three genotypes.
2.4. Statistical analysis
STATA software (Version 13.0) was used for all statistical analyses. Two-sided P values less than 0.05 were considered statistically significant. The strength of the association between the GSTP1 Ile (105) Val gene polymorphism and COPD risk was assessed by the odds ratios (ORs) with 95% CIs. The pooled ORs were calculated for the homozygote model (105Val/Val vs.105Ile/Ile), heterozygote model (105Ile/Val vs.105Ile/Ile), dominant model (105Val/Val + 105Ile/Val vs. 105Ile/Ile), recessive model (105Val/Val vs. 105Ile/Val + 105Ile/Ile), and an additive model (Val vs. Ile) (Yang et al., 2015; Yang and Liu, 2015). For the control groups for each study, the observed genotype frequencies of the GSTP1 Ile (105) Val polymorphism were evaluated for Hardy–Weinberg equilibrium (Wang et al., 2013b; Ma et al., 2014; Tian et al., 2015). Cochran's Q-statistic and the I2 metric were conducted to assess heterogeneity between studies, P < 0.10 and I2 > 50% were considered to indicate the existence of significant heterogeneity (Higgins and Thompson, 2002; Jackson et al., 2012). If the heterogeneity test result returned P > 0.1, the pooled ORs were analyzed using the random-effects model (DerSimonian and Laird, 1986), or else, the fixed effects model was used (Mantel and Haenszel, 1959). Sensitivity analyses were also performed after sequential removal of each study (Fang et al., 2014). We also tried to assess the source of heterogeneity by region, publication year, control source, and sample size (He et al., 2014; Xue et al., 2014). Lastly, Begg's funnel plot and Egger's test were used to examine statistically any publication bias (Peters et al., 2006).
3. Results
3.1. Characteristics of the included studies
In accordance with the inclusion criteria, seventeen case–control studies with 1892 cases and 2012 controls were included based on the search criteria for risk of COPD related to the GSTP1 polymorphism (Harries et al., 1997; Yim et al., 2002; Lu and He, 2002; Ishii et al., 2003; Xiao et al., 2003; Zhang et al., 2003; Cheng et al., 2004; Gaspar et al., 2004; Rodriguez et al., 2005; Hu et al., 2005; Calikoglu et al., 2006; Fang et al., 2006; Chan-Yeung et al., 2007; Vibhuti et al., 2007; Lakhdar et al., 2010; Zuntar et al., 2014; Wu et al., 2014). All of the 17 studies were published between 1997 and 2014. No overlap occurred between the studies based on case or control participation. The characteristics of all included studies are summarized in Table 1.
Table 1.
Characteristics of studies included in this meta-analysis.
| Author |
Year |
Ethnicity |
Source of controls |
Adjustment for smoking |
Case |
Control |
HWE(P) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 105Ile/Ile | 105Ile/Val | 105Val/Val | Total | 105Ile/Ile | 105Ile/Val | 105Val/Val | ||||||
| Harries et al. | 1997 | Caucasian | Healthy controls | No | 79 | 34 | 35 | 10 | 155 | 79 | 66 | 10 | 0.4396 |
| Ishii et al. | 1999 | Asian | Healthy controls | Yes | 53 | 42 | 11 | 0 | 50 | 26 | 22 | 2 | 0.3104 |
| Lu et al. | 2002 | Asian | Healthy controls | Yes | 97 | 70 | 22 | 5 | 67 | 41 | 24 | 2 | 0.4940 |
| Yim et al. | 2002 | Asian | Checkup | No | 89 | 63 | 24 | 2 | 94 | 57 | 35 | 2 | 0.1995 |
| Zhang et al. | 2003 | Asian | Healthy controls | Yes | 57 | 47 | 5 | 5 | 48 | 44 | 3 | 1 | 0.0110 |
| Cheng et al. | 2004 | Asian | Checkup | Yes | 184 | 97 | 78 | 9 | 212 | 99 | 98 | 15 | 0.1591 |
| Gaspa et al. | 2004 | Caucasian | Checkup | No | 75 | 35 | 35 | 5 | 90 | 47 | 36 | 7 | 0.9767 |
| Xiao et al. | 2004 | Asian | Healthy controls | Yes | 100 | 70 | 29 | 1 | 100 | 57 | 40 | 3 | 0.1959 |
| Hu et al. | 2005 | Asian | Healthy controls | Yes | 50 | 45 | 3 | 2 | 68 | 59 | 5 | 4 | 0.0000 |
| Rodriguez et al. | 2005 | Caucasian | Checkup | No | 98 | 52 | 36 | 10 | 198 | 97 | 88 | 13 | 0.2372 |
| Calikoglu et al. | 2006 | Caucasian | Healthy controls | Yes | 144 | 88 | 42 | 14 | 150 | 57 | 57 | 36 | 0.0059 |
| Fang et al. | 2006 | Asian | Healthy controls | No | 87 | 65 | 18 | 4 | 91 | 74 | 16 | 1 | 0.8972 |
| Vibhuti et al. | 2007 | Asian | Healthy controls | Yes | 202 | 105 | 75 | 22 | 136 | 90 | 42 | 4 | 0.7336 |
| Yeung et al. | 2007 | Asian | Healthy controls | Yes | 163 | 112 | 43 | 8 | 161 | 112 | 47 | 2 | 0.2280 |
| Lakhdar et al. | 2010 | Caucasian | Healthy controls | Yes | 234 | 81 | 104 | 49 | 182 | 84 | 79 | 19 | 0.9468 |
| Wu et al. | 2014 | Asian | Healthy controls | Yes | 150 | 113 | 18 | 19 | 150 | 132 | 11 | 7 | 0.0000 |
| Zuntar et al. | 2014 | Caucasian | Healthy controls | No | 30 | 10 | 16 | 4 | 60 | 34 | 25 | 1 | 0.1314 |
3.2. Results of the overall meta-analysis
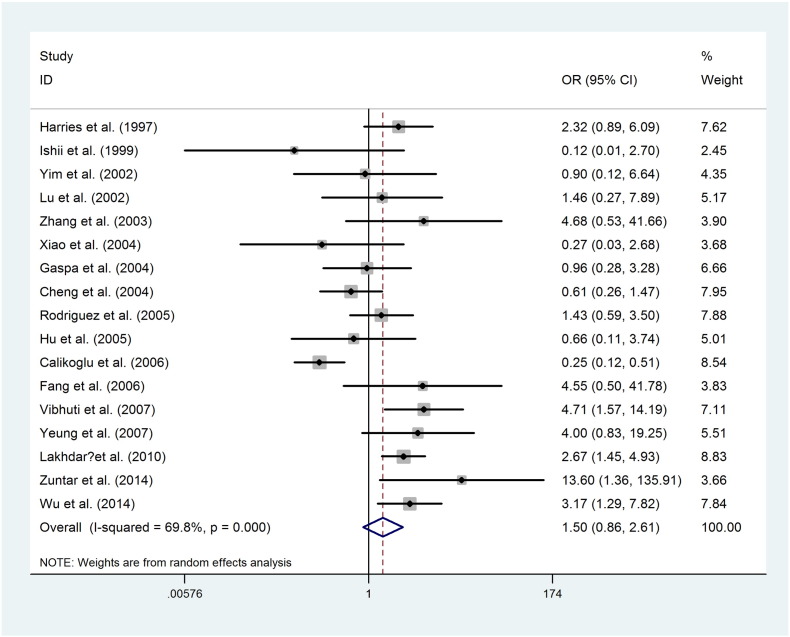
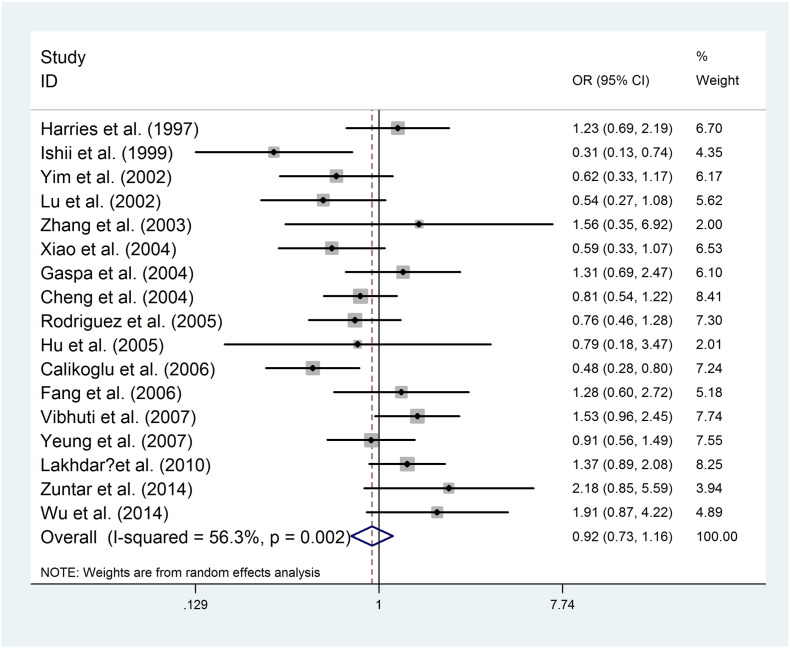
The main results of meta-analysis on the association between the GSTP1 Ile (105) Val polymorphism and COPD risk are listed in Table 2. The GSTP1 Ile (105) Val polymorphism showed pooled odds ratios for the homozygote comparison (Fig. 2, OR = 1.501, 95%CI [0.862, 2.614]), heterozygote comparison (Fig. 3, OR = 0.924, 95%CI [0.733, 1.165]), dominant model (OR = 1.003, 95%CI [0.756, 1.331]), recessive model (OR = 1.510, 95%CI [0.934, 2.439]), and an additive model (OR = 1.072, 95%CI [0.822, 1.398]).
Table 2.
Results of the overall meta-analysis.
| Contrast | OR,95% CI | Heterogeneity | Z and P |
|---|---|---|---|
| Homozygote | 1.501, [0.862, 2.614] | Chi-squared = 52.95 (d.f. = 16) p = 0.000, I-squared = 69.8% | z = 1.44, p = 0.151 |
| Heterozygote | 0.924, [0.733, 1.165] | Chi-squared = 36.65 (d.f. = 16) p = 0.002, I-squared = 56.3% | z = 0.67, p = 0.503 |
| Dominant | 1.003, [0.756, 1.331] | Chi-squared = 62.67 (d.f. = 16) p = 0.000, I-squared = 74.5% | z = 0.02, p = 0.984 |
| Recessive | 1.510, [0.934, 2.439] | Chi-squared = 41.41 (d.f. = 16) p = 0.000, I-squared = 61.4% | z = 1.68, p = 0.093 |
| Additive | 1.072, [0.822, 1.398] | Chi-squared = 85.96 (d.f. = 16) p = 0.000, I-squared = 81.4% | z = 0.51, p = 0.610 |
Fig. 2.
Random effect forest plot of homozygote model of GSTP1 gene polymorphism.
Fig. 3.
Random effect forest plot of heterozygote model of GSTP1 gene polymorphism.
3.3. Sub-group analysis
We performed a sub-group analysis stratified by ethnicity. There were 11 studies based on Asian population and 6 studies based on Caucasian population. The pooled OR was 1.586, 95%CI [0.814, 3.088] for Asian population, 1.476, 95%CI [0.558, 3.906] for Caucasian population in homozygote comparison. The subgroup analysis results for the all genetic models are listed in detail in Table 3.
Table 3.
Results of sub-group analysis.
| Ethnicity | Comparisons | Homozygote | Heterozygote | Dominant | Recessive | Additive |
|---|---|---|---|---|---|---|
| Caucasian | 6 | 1.476, [0.558, 3.906] | 1.040, [0.697, 1.552] | 1.110, [0.664, 1.854] | 1.393, [0.622, 3.118] | 1.131, [0.708, 1.806] |
| Asian | 11 | 1.586, [0.814, 3.088] | 0.858, [0.638, 1.154] | 0.948, [0.664, 1.353] | 1.663, [0.914, 3.025] | 1.038, [0.738, 1.460] |
| Overall | 17 | 1.501, [0.862,2.614] | 0.924, [0.733, 1.165] | 1.003, [0.756, 1.331] | 1.510, [0.934, 2.439] | 1.072, [0.822, 1.398] |
3.4. Heterogeneity test
There was a significant heterogeneity, in homozygote comparison: chi-squared = 52.95 (d.f. = 16) p = 0.000, I-squared = 69.8%, and in Heterozygote comparison: chi-squared = 36.65 (d.f. = 16) p = 0.002, I-squared = 56.3%. We assessed the source of heterogeneity by region, publication year, control source, and sample size. However, we did not observe any sources that contributed to the substantial heterogeneity. The meta-regression analysis did not yield any significant difference between subgroup analysis.
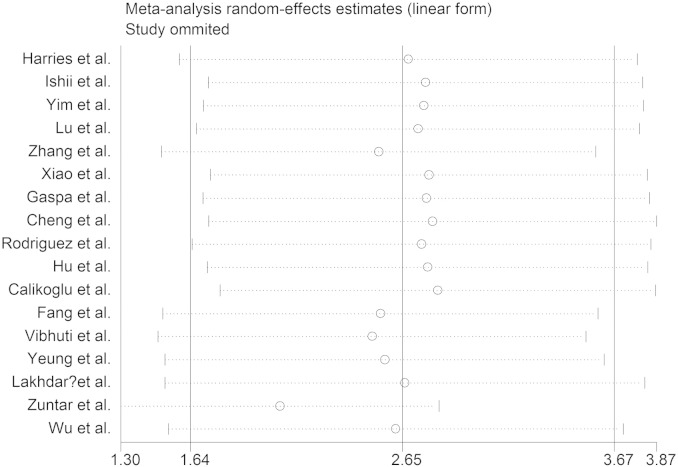
3.5. Sensitivity analysis
We conducted sensitivity analyses to ascertain the primary origin of the heterogeneity. Through sensitivity analysis, the current meta-analysis showed that no individual study had marked effect on the pooled ORs (Fig. 4).
Fig. 4.
Sensitivity analysis.
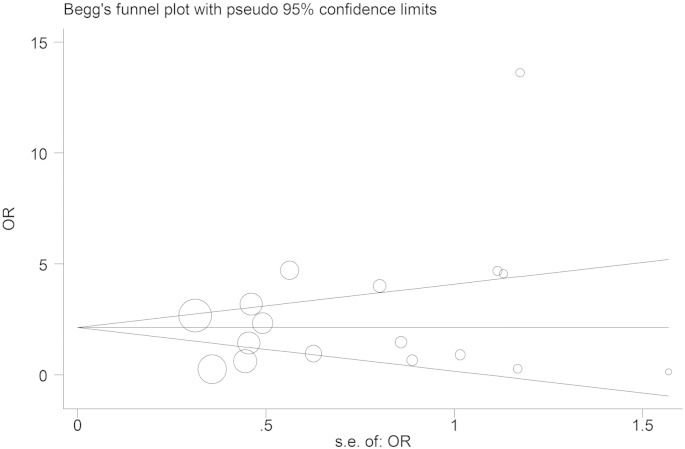
3.6. Publication bias
Funnel plot was generated to assess publication bias. Begg's test and Egger's test were performed to evaluate funnel plot symmetry statistically. The results showed no publication bias: Begg's test Pr > | z | = 0.343 and Egger's test P > | t | = 0.263 (Fig. 5).
Fig. 5.
Test for publication bias.
4. Discussion
COPD, one of the major health challenges, is the fourth leading cause of death globally presently, and it is predicted to become the third leading cause by 2030 (Hu et al., 2015). It is likely to be a complex interplay between genetic and environmental factors. A series of different genes were considered to play important roles in the metabolism of toxic substances in cigarette smoke, airway hyperresponsiveness, and the inflammatory response to cigarette smoke (Sandford et al., 2002).GSTs are derived from a superfamily of genes; they catalyze the conjugation of reactive chemical intermediates to soluble glutathione and may play a role in cellular defense by detoxifying various toxic substrates in cigarette smoke (Ketterer, 1988).Many previous studies have explored the association between GSTP1 105Val/Val genotype and COPD risk in different ethnic populations with conflicting results. As so far, only one meta-analyses, which was nested in case–control studies, have investigated the association of GSTP1 Ile (105) Val polymorphism and COPD susceptibility. Yan et al. performed a meta-analysis included ten studies with a total of 1140 cases and 1263 controls and suggested a significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk (Yan et al., 2010).Considering a series of new articles have been published we performed this updated meta-analysis to drive a more precise estimation of GSTP1 Ile (105) Val gene polymorphism and COPD risk.
According to the inclusion criteria 17 studies with 1892 cases and 2012 controls were included the current meta-analysis. To the best of our knowledge, the current meta-analysis is the largest one to investigate the association between GSTP1 Ile (105) Val gene polymorphism and COPD risk. The results showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in any genetic models. The results of subgroup analysis also showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in Asian population and Caucasian population. There was a significant heterogeneity, and we conducted sensitivity analyses to ascertain the primary origin of the heterogeneity. Through sensitivity analysis, the current meta-analysis showed that no individual study had significant effect on the pooled ORs. Funnel plot was generated to assess publication bias. Begg's test and Egger's test were performed to evaluate funnel plot symmetry statistically. No publication bias was detected in our meta-analysis.
Of course, we should be aware of that the hypothesis considering no association between GSTP1 Ile (105) Val gene polymorphism and COPD risk merely on the basis of the negative results in this study. If a putative genetic association is of small magnitude with point estimates less than 1.5, the small and underpowered studies may be unable to identify true genetic associations (Ioannidis, 2003; Ioannidis et al., 2006; Hindorff et al., 2009). Thus, more evidence is needed to support or deny such an association. By means of meta-analysis, a statistical technique for combining the results from independent studies, we drew a more reliable conclusion on the influence of GSTP1 Ile (105) Val gene polymorphism on COPD risk. However, COPD might be a result of multi-factors, future research should investigate not only individual genes, but also gene–gene interactions, other SNPs such as GSTM1, GSTT1.
Several potential limitations of this meta-analysis should be discussed: 1) although the funnel plot and Begg's Test showed no publication bias, selection bias may have occurred because only studies in English or Chinese were selected; 2) there was a significant heterogeneity. We assessed the source of heterogeneity by region, publication year, control source, and sample size. However, we did not observe any sources that contributed to the substantial heterogeneity. The meta-regression analysis did not yield any significant difference between subgroup analysis. Through sensitivity analysis, the current meta-analysis showed that no individual study had marked effect on the pooled ORs. However, this study also has some clear advantages: 1) this is the meta-analysis on the most updated information; 2) we performed a sub-group analysis stratified by ethnicity; 3) sensitivity analysis showed no individual study had marked effect on the overall results; 4) the scientific search and selection method significantly increased the reality of this meta-analysis; 5) no publication bias was detected.
In conclusion, the current meta-analysis, based on the most updated information, showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in any genetic models. The results of subgroup analysis also showed no significant association between GSTP1 Ile (105) Val gene polymorphism and COPD risk in Asian population and Caucasian population. Further studies involving large populations and careful control with age, sex, ethnicity, and cigarette smoking are greatly needed.
Conflict of interest statement
The authors declare that they have no conflict of interest.
References
- Calikoglu M., Tamer L., Ates Aras N., Karakas S., Ercan B. The association between polymorphic genotypes of glutathione S-transferases and COPD in the Turkish population. Biochem. Genet. 2006;44:307–319. doi: 10.1007/s10528-006-9031-4. [DOI] [PubMed] [Google Scholar]
- Cantlay A.M., Smith C.A., Wallace W.A., Yap P.L., Lamb D., Harrison D.J. Heterogeneous expression and polymorphic genotype of glutathione S-transferases in human lung. Thorax. 1994;49:1010–1014. doi: 10.1136/thx.49.10.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci M., Rossato M., Calzetti F., Tamassia N., Zeminian S., Cassatella M.A., Bazzoni F. IL-10 disrupts the Brd4-docking sites to inhibit LPS-induced CXCL8 and TNF-alpha expression in monocytes: implications for chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2015 doi: 10.1016/j.jaci.2015.04.023. [DOI] [PubMed] [Google Scholar]
- Chan-Yeung M., Ho S.P., Cheung A.H., So L.K., Wong P.C., Chan K.K., Chan J.W., Ip M.S., Mak J.C. Polymorphisms of glutathione S-transferase genes and functional activity in smokers with or without COPD. Int. J. Tuberc. Lung Dis. 2007;11:508–514. [PubMed] [Google Scholar]
- Cheng S.L., Yu C.J., Chen C.J., Yang P.C. Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur. Respir. J. 2004;23:818–824. doi: 10.1183/09031936.04.00104904. [DOI] [PubMed] [Google Scholar]
- Cheng L., Liu J., Li B., Liu S., Li X., Tu H. Cigarette smoke-induced hypermethylation of the GCLC gene is associated with chronic obstructive pulmonary disease. Chest. 2015 doi: 10.1378/chest.14-2309. [DOI] [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Fang L.Z., Zhang J.J., Zhao Z.H., Liu L., Fu W.P. A Study on the Correlation between Exon 5 Polymorph ism of GSTP1 Gene and Clinic Phenotype of COPD. Vol. 2. 2006. pp. 18–22. (Journal of Kunming Medical University). (In Chinese) [Google Scholar]
- Fang X.Y., Xu W.D., Huang Q., Yang X.K., Liu Y.Y., Leng R.X., Pan H.F., Ye D.Q. 5,10-Methylenetetrahydrofolate reductase polymorphisms and colon cancer risk: a meta-analysis. Asian Pac. J. Cancer Prev. 2014;15:8245–8250. doi: 10.7314/apjcp.2014.15.19.8245. [DOI] [PubMed] [Google Scholar]
- Gaspar P., Moreira J., Kvitko K., Torres M., Moreira A., Weimer T. CYP1A1, CYP2E1, GSTM1, GSTT1, GSTP1, and TP53 polymorphisms: do they indicate susceptibility to chronic obstructive pulmonary disease and non-small-cell lung cancer? Genet. Mol. Biol. 2004;27:133–138. [Google Scholar]
- Hanania N.A., Marciniuk D.D. A unified front against COPD: clinical practice guidelines from the American College of Physicians, the American College of Chest Physicians, the American Thoracic Society, and the European Respiratory Society. Chest. 2011;140:565–566. doi: 10.1378/chest.11-1152. [DOI] [PubMed] [Google Scholar]
- Harries L.W., Stubbins M.J., Forman D., Howard G.C., Wolf C.R. Identification of genetic polymorphisms at the glutathione S-transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis. 1997;18:641–644. doi: 10.1093/carcin/18.4.641. [DOI] [PubMed] [Google Scholar]
- He J., Liao X.Y., Zhu J.H., Xue W.Q., Shen G.P., Huang S.Y., Chen W., Jia W.H. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci. Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoidal J.R. Genetics of COPD: present and future. Eur. Respir. J. 2001;18:741–743. doi: 10.1183/09031936.01.00268501. [DOI] [PubMed] [Google Scholar]
- Hu Y.G., Shen Y.F., Ma D. Study on the relationship between GSTP1 gene polymorphism and chronic obstructive pulmonary disease. J. Jianghan Univ. (Nat. Sci.) 2005;33:28–30. (In Chinese) [Google Scholar]
- Hu G., Zhong N., Ran P. Air pollution and COPD in China. J. Thorac. Dis. 2015;7:59–66. doi: 10.3978/j.issn.2072-1439.2014.12.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J.P. Genetic associations: false or true? Trends Mol. Med. 2003;9:135–138. doi: 10.1016/s1471-4914(03)00030-3. [DOI] [PubMed] [Google Scholar]
- Ioannidis J.P., Trikalinos T.A., Khoury M.J. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am. J. Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- Ishii T., Fujishiro M., Masuda M., Nakajima J., Teramoto S., Ouchi Y., Matsuse T. Depletion of glutathione S-transferase P1 induces apoptosis in human lung fibroblasts. Exp. Lung Res. 2003;29:523–536. doi: 10.1080/01902140303777. [DOI] [PubMed] [Google Scholar]
- Jackson D., White I.R., Riley R.D. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat. Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketterer B. Protective role of glutathione and glutathione transferases in mutagenesis and carcinogenesis. Mutat. Res. 1988;202:343–361. doi: 10.1016/0027-5107(88)90197-2. [DOI] [PubMed] [Google Scholar]
- Lakhdar R., Denden S., Knani J., Leban N., Daimi H., Hassine M., Lefranc G., Ben Chibani J., Haj Khelil A. Relationship between glutathione S-transferase P1 polymorphisms and chronic obstructive pulmonary disease in a Tunisian population. Genet. Mol. Res. 2010;9:897–907. doi: 10.4238/vol9-2gmr770. [DOI] [PubMed] [Google Scholar]
- Lu B.B., He Q.Y. Correlation between exon5 polymorphism of glutathione S-transferase P1 gene and susceptibility to chronic obstructive pulmonary disease in northern Chinese population of Han nationality living in Beijing, China. Chin. J. Integr. Med. 2002;41:33–36. (In Chinese) [PubMed] [Google Scholar]
- Ma Z.J., Chen R., Ren H.Z., Guo X., Chen J.G., Chen L.M. Endothelial nitric oxide synthase (eNOS) 4b/a polymorphism and the risk of diabetic nephropathy in type 2 diabetes mellitus: a meta-analysis. Meta Gene. 2014;2:50–62. doi: 10.1016/j.mgene.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannino D.M., Homa D.M., Akinbami L.J., Ford E.S., Redd S.C. Chronic obstructive pulmonary disease surveillance–United States, 1971–2000. Respir. Care. 2002;47:1184–1199. [PubMed] [Google Scholar]
- Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Molfino N.A. Genetics of COPD. Chest. 2004;125:1929–1940. doi: 10.1378/chest.125.5.1929. [DOI] [PubMed] [Google Scholar]
- Murphy T.F., Kirkham C., Jones M.M., Sethi S., Kong Y., Pettigrew M.M. Expression of IgA proteases by Haemophilus influenzae in the respiratory tract of adults with chronic obstructive pulmonary disease. J. Infect. Dis. 2015 doi: 10.1093/infdis/jiv299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.L., Sutton A.J., Jones D.R., Abrams K.R., Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- Restrepo R.D. Year in review 2014: COPD. Respir. Care. 2015;60:1057–1060. doi: 10.4187/respcare.04227. [DOI] [PubMed] [Google Scholar]
- Rodriguez F., de la Roza C., Jardi R., Schaper M., Vidal R., Miravitlles M. Glutathione S-transferase P1 and lung function in patients with alpha1-antitrypsin deficiency and COPD. Chest. 2005;127:1537–1543. doi: 10.1378/chest.127.5.1537. [DOI] [PubMed] [Google Scholar]
- Salvi S. Tobacco smoking and environmental risk factors for chronic obstructive pulmonary disease. Clin. Chest Med. 2014;35:17–27. doi: 10.1016/j.ccm.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Sandford A.J., Joos L., Pare P.D. Genetic risk factors for chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2002;8:87–94. doi: 10.1097/00063198-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Stockley R.A., Mannino D., Barnes P.J. Burden and pathogenesis of chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6:524–526. doi: 10.1513/pats.200904-016DS. [DOI] [PubMed] [Google Scholar]
- Stroup D.F., Berlin J.A., Morton S.C., Olkin I., Williamson G.D., Rennie D., Moher D., Becker B.J., Sipe T.A., Thacker S.B. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- Tian L., Zhang J., Xiao S., Huang J., Zhang Y., Shen J. Impact of polymorphisms of the GGCX gene on maintenance warfarin dose in Chinese populations: systematic review and meta-analysis. Meta Gene. 2015;5:43–54. doi: 10.1016/j.mgene.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestbo J., Hurd S.S., Agusti A.G., Jones P.W., Vogelmeier C., Anzueto A., Barnes P.J., Fabbri L.M., Martinez F.J., Nishimura M., Stockley R.A., Sin D.D., Rodriguez-Roisin R. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013;187:347–365. doi: 10.1164/rccm.201204-0596PP. [DOI] [PubMed] [Google Scholar]
- Vibhuti A., Arif E., Deepak D., Singh B., Qadar Pasha M.A. Genetic polymorphisms of GSTP1 and mEPHX correlate with oxidative stress markers and lung function in COPD. Biochem. Biophys. Res. Commun. 2007;359:136–142. doi: 10.1016/j.bbrc.2007.05.076. [DOI] [PubMed] [Google Scholar]
- Wang W., Hou Z., Wang C., Wei C., Li Y., Jiang L. Association between 5, 10-methylenetetrahydrofolate reductase (MTHFR) polymorphisms and congenital heart disease: a meta-analysis. Meta Gene. 2013;1:109–125. doi: 10.1016/j.mgene.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Zhou H., Yang J., Xiao J., Liang B., Li D., Zhou H., Zeng Q., Fang C., Rao Z., Yu H., Ou X., Feng Y. Association of HHIP polymorphisms with COPD and COPD-related phenotypes in a Chinese Han population. Gene. 2013;531:101–105. doi: 10.1016/j.gene.2013.08.069. [DOI] [PubMed] [Google Scholar]
- Wang W., Li P., Chen Y., Yang J. Association between beta2-adrenergic receptor-16Arg/Gly gene polymorphism and chronic obstructive pulmonary disease risk:systematic review and meta-analysis. Iran J. Public Health. 2014;43:877–888. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li W., Liu W., Cai B., Cheng T., Gao C., Mo L., Yang H., Chang L. GSTM1 and GSTT1 gene polymorphisms as major risk factors for bronchopulmonary dysplasia in a Chinese Han population. Gene. 2014;533:48–51. doi: 10.1016/j.gene.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Wu S.M., Wang F.G., Guo S.J., Ling M. Correlation between exon5 polymorphism of glutathione S-transferase P1 gene and susceptibility to chronic obstructive pulmonary disease in Xin Jiang. J. Xi'an Jiao Tong Univ. (Med. Sci.) 2014;35:509–512. (In Chinese) [Google Scholar]
- Xiao D., Wang C., Du M.J., Pang B.S., Zhang H.Y., Liu J.Z., Weng X.Z., Su L., Christiani D.C. Association between polymorphisms in the gene coding for glutathione S-transferase P1 and chronic obstructive pulmonary disease. Chin. J. Tuberc. Respir. Dis. 2003;26:46–49. (In Chinese) [Google Scholar]
- Xue W.Q., He Y.Q., Zhu J.H., Ma J.Q., He J., Jia W.H. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci. Rep. 2014;4:6791. doi: 10.1038/srep06791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Chen C., Jing J., Li W., Shen H., Wang X. Association between polymorphism of glutathione S-transferase P1 and chronic obstructive pulmonary disease: a meta-analysis. Respir. Med. 2010;104:473–480. doi: 10.1016/j.rmed.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu H. Association between interleukin-18 gene promoter (− 607C/A and − 137G/C) polymorphisms and chronic hepatitis C virus infections: a meta-analysis. Meta Gene. 2015;5:21–31. doi: 10.1016/j.mgene.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Chen J., Wang B., Ding C., Liu H. Association between MTHFR C677T polymorphism and neural tube defect risks: a comprehensive evaluation in three groups of NTD patients, mothers, and fathers. Birth Defects Res. A Clin. Mol. Teratol. 2015;103:488–500. doi: 10.1002/bdra.23361. [DOI] [PubMed] [Google Scholar]
- Yim J.J., Yoo C.G., Lee C.T., Kim Y.W., Han S.K., Shim Y.S. Lack of association between glutathione S-transferase P1 polymorphism and COPD in Koreans. Lung. 2002;180:119–125. doi: 10.1007/s004080000086. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wu Y.M., Liu X., Shi R.F. Correlation between exon5 polymorphism of glutathione S-transferase P1 gene and susceptibility to chronic obstructive pulmonary disease in northern Chinese population of Han. Chin. J. Tuberc. Respir. Dis. 2003;26:57–58. (In Chinese) [Google Scholar]
- Zuntar I., Petlevski R., Dodig S., Popovic-Grle S. GSTP1, GSTM1 and GSTT1 genetic polymorphisms and total serum GST concentration in stable male COPD. Acta Pharma. 2014;64:117–129. doi: 10.2478/acph-2014-0003. [DOI] [PubMed] [Google Scholar]