Abstract
It has been widely reported that the CT60 single-nucleotide polymorphism (SNP), which is in the 3′-untranslated region of the cytotoxic T lymphocyte associated 4 (CTLA4) gene, is strongly correlated with certain immune-mediated diseases. The present case-control study aimed to investigate the genetic association between the CT60 SNP within the CTLA4 gene and Graves' disease (GD). A total of 288 patients with GD and 290 control subjects were recruited for the study. The CT60 SNP of the CTLA4 gene was detected by direct DNA sequencing. The results indicated that the frequencies of the GG genotype and G allele in the case group were evidently higher than that in the control group (P=4×10−6 and P=2.9×10−5, respectively). Furthermore, the G/G genotype of the CT60 SNP was associated with an increased risk for GD (odds ratio=2.223). In conclusion, these results suggested that the CT60 SNP is associated with susceptibility to GD. The frequency of the disease-susceptible G allele of CT60 was significantly associated with an increased risk of GD development.
Keywords: Graves' disease, cytotoxic T lymphocyte-associated 4, CT60, single-nucleotide polymorphism
Introduction
Graves' Disease (GD), also known as toxic diffuse goiter, can increase the level of thyroid hormone, is one of the organ-specific autoimmune diseases and it accounts for 85% of all clinical hyperthyroidism (1,2). The disease often presents in patients aged from 20–40 years old, with a male to female ratio of ~1:8 and a significant familial tendency (3). The clinical performance of GD is not limited to the thyroid, but is a multi-system syndrome, including the high metabolic syndrome group, diffuse goiter, eye symptoms, lesions and thyroid extremity diseases (4–6). Immunologically, GD is characterized by increased circulating antibodies against thyroid-stimulating hormone receptor (TSHR), thyroglobulin (TG) and thyroid peroxidase (TPO). Although the precise pathogenesis involved in the process of GD is not completely understood, certain findings indicate that complex interactions between environmental, genetic, endogenous and local factors are involved in its pathogenesis (7–9). A study on the genetic susceptibility genes of GD has become of interest (10–13).
Previous studies have demonstrated that a number of susceptibility genes are associated with the autoimmune thyroid diseases, including interleukin-21, TSHR, human leukocyte antigen class I and II, cluster of differentiation 40 (CD40) and cytotoxic T lymphocyte associated 4 (CTLA4) (10–15). Among them, CTLA4 as a key negative regulator of the T lymphocyte immune response has attracted an increasing focus on the susceptibility to autoimmune disease.
CTLA4, which is expressed by activate T cells, is a type of transmembrane protein. The CTLA4 gene is located on human chromosome 2q33 (16). The most significant function of the CTLA4 gene is negative regulation of the human immune response. Several polymorphic sites in the CTLA4 gene are associated with thyroid diseases, such as −318C/T promoter and 49A/G exon 1, −224(AT)n dinucleotide repeat sequence single-nucleotide polymorphism (SNP), −1722C/T, −1661A/G, CT60 (rs3087243), CT61 (rs51157131), J031 (rsl1571302) and J027-1 (rsl1571297) (16–22). Several studies also show that the SNPs at the 3′ untranslated region (UTR) are more associated with GD (23–25). CT60 is located in the 3′-UTR of the CTLA4 gene. Increasing attention has previously been paid to the CT60 SNP of CTLA4 in patients with GD (26–28). However, the correlation between CT60 A/G and the development of GD varied among patients from different geographic populations (29). Thus, there are limited reports regarding an association between the CT60 SNP and GD in the Han population of Southern China.
The aim of the present report was to use a case-control study to establish a database of CT60 polymorphisms in the Han population from Southern China and to evaluate CT60 polymorphisms as an indicator of GD susceptibility. The results indicated that the CT60 SNP of the CTLA4 gene was associated with susceptibility to GD in the Han population of Southern China. In addition, the G allele of the CT60 SNP was strongly associated with GD patients.
Materials and methods
GD patients
For the case-control study, a total of 288 unrelated individuals with GD (66 males and 222 females) from Southern China were recruited from the Department of Endocrinology at Sun Yat-Sen Memorial Hospital (Guangzhou, China). The age (mean ± standard deviation) of the GD patients was 37.7±14.2 years (range, 19–80 years). The GD patients were diagnosed from the clinical symptoms and biochemical criteria of thyrotoxicosis, including suppressed serum TSH levels, elevated free thyroxine (fT4) and/or free triiodothyronine (fT3) levels, as well as the presence of TSHR antibodies or antibodies against TG or TPO. All the patients had no evident co-morbidities and provided informed consent.
Data from the patients were compared with those obtained from 290 control subjects without a family history of thyroid diseases or other autoimmune diseases from the Health Care Center at Sun Yat-Sen Memorial Hospital. All the healthy controls were age- and gender-matched with the GD patients and also provided informed consent. The experimental protocol was approved by the Ethics Committee of Sun Yat-Sen Memorial Hospital.
Thyroid function tests
Serum concentrations of fT3, fT4, TSH, anti-TG and anti-TPO were detected with the chemiluminescence immunoassay by the ADVIA Centaur Automated Chemiluminescent Immunoassay Analyzer (Siemens Healthcare, Erlangen, Germany); the normal ranges were 0.82–1.79 ng/ml, 4.5–12.5 µg/dl and 0.40–4.00 µIU/ml, respectively.
CTLA4 3′-UTR CT60 genotyping
Genomic DNA was isolated from whole blood with the Whole Blood Genomic DNA Extraction kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer's instructions. The CT60 SNP was determined by direct DNA sequencing (BigDye DNA Sequencing kit; Applied Biosystems Inc., Norwalk, CT, USA) following polymerase chain reaction (PCR). PCR was performed using the following primers: Forward, 5′-ATAATGCTTCATGAGTCAGCTT-3′ and reverse, 5′-GAG GTGAAGAACCTGTGTTAAA-3′. Amplification of target DNA was performed on the DNA template in a total volume of 50 µl comprising 5.0 µl of 10X PCR buffer, 4.0 µl of deoxyribonucleotide mixture, 0.25 µl of Ex Taq polymerase, 3.0 µl of DNA template (50 ng/µl), 35.75 µl of PCR-Grade water and 1.0 µl of each 20 µmol/l primer. PCR conditions were as follows: An initial denaturation at 94°C for 4 min; 35 cycles of 94°C for 45 sec, 58°C for 45 sec and 72°C for 45 sec; and a final extension for 4 min at 72°C.
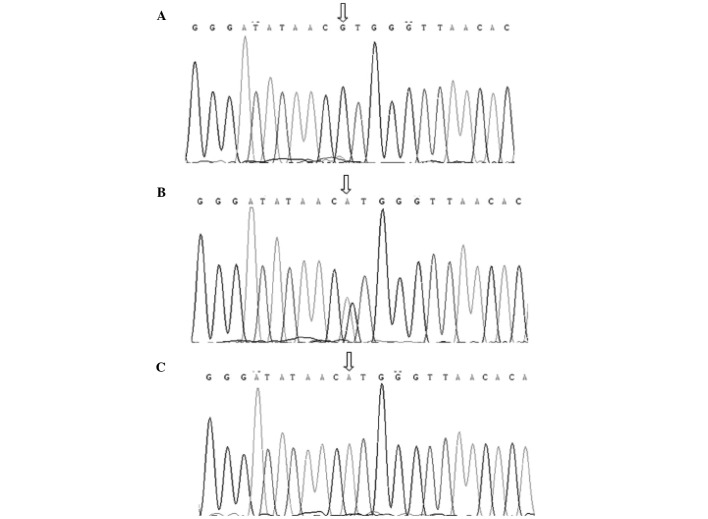
Following amplification, the PCR products were identified by electrophoresis (Power Pac 200; Bio-Rad, Hercules, CA, USA) (Fig. 1) and submitted for DNA sequencing. The sequences are shown in Fig. 2.
Figure 1.
Agarose gel electrophoresis of polymerase chain reaction products of the CT60 single-nucleotide polymorphism.
Figure 2.
Sequencing results of the CT60 single-nucleotide polymorphism in patients of the Han population of Southern China. Sequencing results of genotype (A) GG, (B) AG and (C) AA.
Statistical analysis
The Hardy-Weinberg equilibrium test for the genotyping results in the control group was carried out by Excel (Microsoft Office Excel, Microsoft Corp., Redmond, WA, USA). All the statistical analyses were performed with the SPSS 20.0 software (IBM Corp., Armonk, NY, USA). A χ2 test or the Fisher's exact test was performed to compare the frequencies of genotypes and alleles between the groups. The disease susceptibility of specific genotypes and alleles were estimated with the calculated odds ratio (OR) and 95% confidence interval (CI). P<0.05 was considered to indicate a statistically significant difference.
Results
Hardy-Weinberg equilibrium test
A Hardy-Weinberg equilibrium of the CT60 (rs3087243) genotype distribution was carried out in the control group. As was listed in Table I, χ2 test revealed that the CT60 (rs3087243) loci was in the balance of Hardy-Weinberg (χ2 test=4.216, P=0.122). The CT60 SNP loci can be used as a genetic marker to study the association between the CTLA4 gene and GD.
Table I.
Hardy-Weinberg equilibrium of the genotyping results in the controls.
| Genotype | Observed frequency | Expected frequency | χ2 test | P-value |
|---|---|---|---|---|
| GG | 149 | 157.9 | 4.216 | 0.122 |
| AG | 130 | 112.2 | ||
| AA | 11 | 19.9 |
Genotype distributions of the general population
The population frequencies of genotype and allele of the CT60 SNP in GD patients and controls are listed in Table II. The frequencies of genotype in the cases (AA, 2.4; AG, 27.4; and GG, 70.1%) were significantly different from those in the controls (AA, 3.8; AG, 44.8; and GG, 51.4%) (χ2 test=21.330, P=2.3×10−5). As is shown in Table II, there was an evidently higher frequency of the GG genotype in the cases compared to the controls (P=4.0×10−6; OR, 2.223; 95% CI, 1.579–3.128). Inversely, the frequencies of the AG genotype were significantly lower in the GD group in contrast to the control group (P=1.3×10−5; OR, 0.465; 95% CI, 0.329–0.658). Table II also showed that the frequency of the G allele in the GD group was significantly higher than that in the control group (83.9 vs. 73.8%; χ2 test=17.515, P=2.9×10−5).
Table II.
Genotype and allele frequencies of CT60 in GD patients and controls.
| Genotype/allele | GD, n (%) | Controls, n (%) | χ2 test | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|
| Genotype | ||||||
| GG | 202 (70.1) | 149 (51.4) | 21.322a | 4.0×10−6 | 2.223 | 1.579–3.128 |
| AG | 79 (27.4) | 130 (44.8) | 18.945b | 1.3×10−5 | 0.465 | 0.329–0.658 |
| AA | 7 (2.4) | 11 (3.8) | 0.889c | 0.346 | 0.632 | 0.241–1.653 |
| Allele | ||||||
| G | 483 (83.9) | 428 (73.8) | 17.515 | 2.9×10−5 | 1.844 | 1.382–2.462 |
| A | 93 (16.1) | 152 (26.2) |
Comparison of the frequency of the GG genotype with that of the AG and AA genotype in cases versus controls
comparison of the frequency of the AG genotype with that of the GG and AA genotype in cases versus controls
comparison of the frequency of the AA genotype with that of the GG and AG genotype in cases versus controls. GD, Graves' disease; OR, odds ratio; CI, confidence interval.
Genotype distributions for different gender
All the 578 individuals were divided into two groups according to gender to study the association of the polymorphisms of the CT60 SNP with GD. As is shown in Table III, following subdivision by gender, the frequency distributions of genotype and allele of males and females were similar to those of the general population. Compared to the controls, the frequencies of the GG genotype and the G allele were markedly higher and the frequencies of the AG genotype were evidently lower in the cases. The genotype and allele distributions of the CT60 SNP were also compared between the male and female patients (Table IV). Conversely, no significant differences were found in genotype and allele frequencies between male and female patients.
Table III.
Genotype and allele frequencies of CT60 in male and female GD patients and controls.
| Genotype and allele | GD, n (%) | Controls, n (%) | χ2 test | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|
| Male | ||||||
| Genotype | ||||||
| GG | 51 (77.3) | 49 (52.7) | 9.998 | 1.6×10−3 | 3.053 | 1.508–6.179 |
| AG | 14 (21.2) | 42 (45.2) | 9.705 | 1.8×10−3 | 0.327 | 0.160–0.670 |
| AA | 1 (1.5) | 2 (2.1) | 0.084 | 0.772 | 0.700 | 0.062–7.884 |
| Allele | ||||||
| G | 116 (87.9) | 140 (75.3) | 7.822 | 5.2×10−3 | 2.382 | 1.282–4.427 |
| A | 16 (12.1) | 46 (24.7) | ||||
| Female | ||||||
| Genotype | ||||||
| GG | 151 (68.0) | 100 (50.8) | 12.941 | 3.2×10−4 | 2.063 | 1.387–3.068 |
| AG | 65 (29.3) | 88 (44.7) | 10.665 | 1.1×10−3 | 0.513 | 0.343–0.767 |
| AA | 6 (2.7) | 9 (4.5) | 1.053 | 0.305 | 0.580 | 0.203–1.660 |
| Allele | ||||||
| G | 367 (82.7) | 288 (73.1) | 11.180 | 8.3×10−4 | 1.754 | 1.259–2.444 |
| A | 77 (17.3) | 106 (26.9) |
GD, Graves' disease; OR, odds ratio; CI, confidence interval.
Table IV.
Genotype and allele frequencies of CT60 in male and female GD patients.
| Genotypes and alleles | Males, n (%) | Females, n (%) | χ2 test | P-value |
|---|---|---|---|---|
| Genotype | ||||
| GG | 51 (77.3) | 151 (68.0) | 2.123 | 0.346 |
| AG | 14 (21.2) | 65 (29.3) | ||
| AA | 1 (1.5) | 6 (2.7) | ||
| Allele | ||||
| G | 116 (87.9) | 367 (82.7) | 1.681 | 0.195 |
| A | 16 (12.1) | 77 (17.3) |
GD, Graves' disease.
Genotype distributions of the GD patients
A total of 288 GD patients were grouped according to the family history, ophthalmopathy and relapse history. As is shown in Table V, there was no significant difference in GD patients with and without family history, ophthalmopathy and relapse history.
Table V.
Correlation analyses between the genotypes and alleles of CT60 and clinical features of GD.
| Family history, n (%) | Ophthalmopathy, n (%) | Relapse history, n (%) | |||||
|---|---|---|---|---|---|---|---|
| Characteristics | (+) | (−) | (+) | (−) | (+) | (−) | |
| Genotype | |||||||
| GG | 59 (69.4) | 143 (70.4) | 14 (70.0) | 184 (68.6) | 21 (75.0) | 177 (68.1) | |
| AG | 25 (29.4) | 54 (26.6) | 5 (25.0) | 75 (28.0) | 6 (21.4) | 74 (28.4) | |
| AA | 1 (1.2) | 6 (3.0) | 1 (5.0) | 9 (3.4) | 1 (3.6) | 9 (3.5) | |
| χ2 test | 0.962 | 0.209 | 0.627 | ||||
| P-value | 0.618 | 0.901 | 0.731 | ||||
| Allele | |||||||
| G | 143 (84.1) | 340 (83.7) | 33 (82.5) | 443 (82.6) | 48 (85.7) | 428 (82.3) | |
| A | 27 (15.9) | 66 (16.3) | 7 (17.5) | 93 (17.4) | 8 (14.3) | 92 (17.7) | |
| χ2 test | 0.012 | 0.037 | 0.206 | ||||
| P-value | 0.911 | 0.847 | 0.650 | ||||
GD, Graves' disease.
Discussion
CTLA4 (CD152), as an important immune-regulatory molecule expressed on activated T cells, is able to inhibit a T-cell response while triggered by costimulatory molecules. The CTLA4 gene is located on human chromosome 2q33 and is composed of four exons. It spans ~8.44 kb of genomic DNA and encodes 233 amino acids (30). CTLA4 belongs to the immunoglobulin gene superfamily and is associated with immune tolerance. CTLA4 expresses on the activated CD4+ and CD8+ T cells. It has sequence homology with costimulatory molecules, including CD28, but has an opposing role (31). The main function of the costimulatory molecule CD28 is to stimulate and activate T cells, however, CTLA4 is a negative regulator of T cell response. CTLA4 produces inhibitory signals combining with the relevant molecules in the B7 family, which has an important role in the termination of T-cell activation. The abnormal expression of CTLA4 induces the activation of T cells, breaks down the immune tolerance and results in autoimmune diseases.
The association of CTLA4 gene polymorphisms with the autoimmune diseases, including type 1 diabetes (19,24), GD (20–22,32), abdominal disease (33), systemic lupus erythematous disease (34), rheumatoid arthritis (35), autoimmune pancreatitis (36) and Addison's disease (37), have been investigated. The CTLA4 gene has multiple SNP loci and repeat sequences. In recent years, increasing attention has been paid to the 3′-UTR of the CTLA4 gene. The 3′-UTR is a complex region containing multiple binding sites for microRNA. Therefore, the presence of SNP or base mutation in the 3′-UTR would change the signaling pathway, influence the protein expression and result in diseases.
The CT60 SNP, which has been demonstrated to be most significantly correlated with GD and Hashimoto's thyroiditis (HT) in the Caucasian population (25), is at the 3′-UTR of the CTLA4 gene. The correlation between the CT60 SNP and the autoimmune diseases, which has been confirmed, varied among patients from different ethnicities or regions. Torres et al (34) reported that CT60 (rs3087243) in the CTLA4 gene was associated with systemic lupus erythematosus in the Spanish population. Ban et al (38) suggested that the CT60 SNP in the CTLA4 gene 3′-UTR may have a role in the autoimmune thyroid disease in the Japanese population. Weng et al (28) also found that the G/G genotype and G allele in CT60 were associated with the onset of GD in the Taiwanese population. However, the study of Pastuszak-Lewandoska et al (39) demonstrated that CT60 A/G has no association with HT or GD patients in the Polish population. In the present study, the distributions of the CT60 polymorphisms were also analyzed in GD patients and healthy controls. The GG genotype frequencies of the CT60 SNP in GD patients and healthy controls were 70.2 and 51.4%, respectively (P=4.0×10−6). It is clear that the GD group had a significantly higher GG genotype compared with the control group. Conversely, the AG genotype frequencies were evidently lower in the cases compared with the controls (27.4 vs. 44.8%; P=1.3×10−5). Furthermore, the study demonstrated that the GG genotype was more prevalent in male and female GD patients and fewer male and female patients had the AG genotype, compared with the control subjects. The data provided suggest that the GG genotype of CT60 may be associated with the onset of GD in the Han population of Southern China.
The G allele of CT60 SNP was also revealed to increase the susceptibility to GD. The frequency of the G allele of the control group in the Han population of Southern China was 73.8%, which was higher compared to the Caucasian (25) (53.2%), Swedish (40) (61.3%), Dutch (41) (53.3%) and Italian populations (26) (48%) but similar to the Taiwanese (28) (75.7%) and Japanese populations (38) (72.6%). This may be attributed to the differences of ethnicity, region or sample size. In addition, the frequency of the G allele was associated with GD (83.9 vs. 73.8%, P=0.035) in the Southern Chinese Han population, which was consistent with the research of other ethnic groups (26,28,39,40). These studies, including the present report of the association between G allele and GD, may have increasingly demonstrated our hypothesis that CT60 of CTLA4 appears to be susceptible to GD in the Han population of Southern China.
Additionally, in order to discuss the associations between clinical manifestations with GD, 288 GD patients were subdivided according to the family history, ophthalmopathy and relapse history. However, there was no significant difference identified between GD patients with and without family history, ophthalmopathy and relapse history. Therefore, the CT60 gene polymorphism of CTLA4 may not be associated with the progress of GD. However, more detailed investigations are required to support this hypothesis.
In conclusion, the present findings demonstrated that the CT60 SNP of the CTLA4 gene was significantly associated with GD in the Chinese Han population from Southern China. The G allele of CT60 indicated a positive role in the susceptibility to GD.
Acknowledgements
The authors would like to thank all the patients who consented to take part in the present study, as well as the doctors and nurses for recruitment. The study was supported by Guangdong Medical Science and Technology Research Foundation (grant no. A2010166), Guangdong Province Science and Technology Project (grant no. 2011B031800162) and the Key Project of Guangzhou Science and Technology Project (grant no. 2011J4100114).
References
- 1.Weetman AP. Graves' disease. N Engl J Med. 2000;343:1236–1248. doi: 10.1056/NEJM200010263431707. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson DL, Gange SJ, Rose NR, Graham NM. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–243. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- 3.Falgarone G, Heshmati HM, Cohen R, Reach G. Mechanisms in endocrinology. Role of emotional stress in the pathophysiology of Graves' disease. Eur J Endocrinol. 2013;168:R13–R18. doi: 10.1530/EJE-12-0539. [DOI] [PubMed] [Google Scholar]
- 4.Chaar BT, Kudva GC, Olsen TJ, Silverberg AB, Grossman BJ. Thrombotic thrombocytopenic purpura and Graves disease. Am J Med Sci. 2007;334:133–135. doi: 10.1097/MAJ.0b013e31812e9735. [DOI] [PubMed] [Google Scholar]
- 5.Aydin Y, Berker D, Ustün I, Balcik O, Balli DN, Delibasi T, Peksoy I, Guler S. A very rare cause of aplastic anemia: Graves disease. South Med J. 2008;101:666–667. doi: 10.1097/SMJ.0b013e318172fcb8. [DOI] [PubMed] [Google Scholar]
- 6.Soultati AS, Dourakis SP, Asvesti K, Nezi V, Alexopoulou A, Archimandritis AJ. Autoimmune blistering disease in a patient with Graves disease and vitiligo. Am J Med Sci. 2007;333:309–312. doi: 10.1097/MAJ.0b013e318053d7b3. [DOI] [PubMed] [Google Scholar]
- 7.Brix TH, Christensen K, Holm NV, Harvald B, Hegedüs L. A population-based study of Graves' disease in Danish twins. Clin Endocrinol (Oxf) 1998;48:397–400. doi: 10.1046/j.1365-2265.1998.00450.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomer Y, Barbesino G, Greenberg DA, Concepcion E, Davies TF. Mapping the major susceptibility loci for familial Graves' and Hashimoto's diseases: Evidence for genetic heterogeneity and gene interactions. J Clin Endocrinol Metab. 1999;84:4656–4664. doi: 10.1210/jc.84.12.4656. [DOI] [PubMed] [Google Scholar]
- 9.Brix TH, Kyvik KO, Christensen K, Hegedüs L. Evidence for a major role of heredity in Graves' disease: A population-based study of two Danish twin cohorts. J Clin Endocrinol Metab. 2001;86:930–934. doi: 10.1210/jc.86.2.930. [DOI] [PubMed] [Google Scholar]
- 10.Ban Y, Davies TF, Greenberg DA, et al. Analysis of the CTLA-4, CD28, and inducible costimulator (ICOS) genes in autoimmune thyroid disease. Genes Immun. 2003;4:586–593. doi: 10.1038/sj.gene.6364018. [DOI] [PubMed] [Google Scholar]
- 11.Brand OJ, Barrett JC, Simmonds MJ, et al. Association of the thyroid stimulating hormone receptor gene (TSHR) with Graves' disease. Hum Mol Genet. 2009;18:1704–1713. doi: 10.1093/hmg/ddp087. [DOI] [PubMed] [Google Scholar]
- 12.Heward JM, Allahabadia A, Armitage M, et al. The development of Graves' disease and the CTLA-4 gene on chromosome 2q33. J Clin Endocrinol Metab. 1999;84:2398–2401. doi: 10.1210/jc.84.7.2398. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson EM, Huber A, Tomer Y. The HLA gene complex in thyroid autoimmunity: From epidemiology to etiology. J Autoimmun. 2008;30:58–62. doi: 10.1016/j.jaut.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Sun H, Liu S, et al. CD40 C/T-1 polymorphism plays different roles in Graves' disease and Hashimoto's thyroiditis: A meta-analysis. Endocr J. 2012;59:1041–1050. doi: 10.1507/endocrj.EJ12-0126. [DOI] [PubMed] [Google Scholar]
- 15.Zeng H, Yan H, Zhang Z, Fang W, Ding R, Huang L, Chen M, Zhang J. Association between IL-21 gene rs907715 polymorphisms and Graves' disease in a Southern Chinese population. Exp Ther Med. 2014;8:213–218. doi: 10.3892/etm.2014.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet JF, Denizot F, Luciani MF, RouxDosseto M, Suzan M, Mattei MG, Golstein P. A new member of the immunoglobulin superfamily - CTLA-4. Nature. 1987;328:267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 17.Yanagawa T, Hidaka Y, Guimaraes V, Soliman M, DeGroot LJ. CTLA-4 gene polymorphism associated with Graves' disease in a Caucasian population. J Clin Endocrinol Metab. 1995;80:41–45. doi: 10.1210/jc.80.1.41. [DOI] [PubMed] [Google Scholar]
- 18.Frydecka I, Daroszewski J, Suwalska K, Zołedziowska M, Tutak A, Słowik M, Potoczek S, Dobosz T. CTLA-4 (CD152) gene polymorphism at position 49 in exon 1 in Graves' disease in a Polish population of the Lower Silesian region. Arch Immunol Ther Exp (Warsz) 2004;52:369–374. [PubMed] [Google Scholar]
- 19.Mochizuki M, Amemiya S, Kobayashi K, Kobayashi K, Shimura Y, Ishihara T, Nakagomi Y, Onigata K, Tamai S, Kasuga A, et al. Association of the CTLA-4 gene 49 A/G polymorphism with type 1 diabetes and autoimmune thyroid disease in Japanese children. Diabetes Care. 2003;26:843–847. doi: 10.2337/diacare.26.3.843. [DOI] [PubMed] [Google Scholar]
- 20.Han SZ, Zhang SH, Li R, Zhang WY, Li Y. The common −318C/T polymorphism in the promoter region of CTLA4 gene is associated with reduced risk of ophthalmopathy in Chinese Graves' patients. Int J Immunogenet. 2006;33:281–287. doi: 10.1111/j.1744-313X.2006.00614.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Yang YM, Lv XY. Association of Graves' disease and Graves' ophthalmopathy with the polymorphisms in promoter and exon 1 of cytotoxic T lymphocyte associated antigen-4 gene. J Zhejiang Univ Sci B. 2006;7:887–891. doi: 10.1631/jzus.2006.B0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ban Y, Tomer Y. Susceptibility genes in thyroid autoimmunity. Clin Dev Immunol. 2005;12:47–58. doi: 10.1080/17402520400008897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furugaki K, Shirasawa S, Ishikawa N, Ito K, Ito K, Kubota S, Kuma K, Tamai H, Akamizu T, Hiratani H, et al. Association of the T-cell regulatory gene CTLA4 with Graves' disease and autoimmune thyroid disease in the Japanese. J Hum Genet. 2004;49:166–168. doi: 10.1007/s10038-003-0120-5. [DOI] [PubMed] [Google Scholar]
- 24.Benmansour J, Stayoussef M, AlJenaidi FA, Rajab MH, Rayana CB, Said HB, Mahjoub T, Almawi WY. Association of single nucleotide polymorphisms in cytotoxic T-lymphocyte antigen 4 and susceptibility to autoimmune type 1 diabetes in Tunisians. Clin Vaccine Immunol. 2010;17:1473–1477. doi: 10.1128/CVI.00099-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 26.Petrone A, Giorgi G, Galgani A, Alemanno I, Corsello SM, Signore A, Di Mario U, Nisticò L, Cascino I, Buzzetti R. CT60 single nucleotide polymorphisms of the cytotoxic T-lymphocyte-associated antigen-4 gene region is associated with Graves' disease in an Italian population. Thyroid. 2005;15:232–238. doi: 10.1089/thy.2005.15.232. [DOI] [PubMed] [Google Scholar]
- 27.Daroszewski J, Pawlak E, Karabon L, Frydecka I, Jonkisz A, Slowik M, Bolanowski M. Soluble CTLA-4 receptor an immunological marker of Graves' disease and severity of ophthalmopathy is associated with CTLA-4 Jo31 and CT60 gene polymorphisms. Eur J Endocrinol. 2009;161:787–793. doi: 10.1530/EJE-09-0600. [DOI] [PubMed] [Google Scholar]
- 28.Weng YC, Wu MJ, Lin WS. CT60 single nucleotide polymorphism of the CTLA-4 gene is associated with susceptibility to Graves' disease in the Taiwanese population. Ann Clin Lab Sci. 2005;35:259–264. [PubMed] [Google Scholar]
- 29.Song HD, Liang J, Shi JY, et al. Functional SNPs in the SCGB3A2 promoter are associated with susceptibility to Graves' disease. Hum Mol Genet. 2009;18:1156–1170. doi: 10.1093/hmg/ddn442. [DOI] [PubMed] [Google Scholar]
- 30.Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 31.Kimkong I, Nakkuntod J, SaeNgow S, Snabboon T, Avihingsanon Y, Hirankarn N. Association between CTLA-4 polymorphisms and the susceptibility to systemic lupus erythematosus and Graves' disease in Thai population. Asian Pac J Allergy Immunol. 2011;29:229–235. [PubMed] [Google Scholar]
- 32.Takahashi M, Kimura A. HLA and CTLA4 polymorphisms may confer a synergistic risk in the susceptibility to Graves' disease. J Hum Genet. 2010;55:323–326. doi: 10.1038/jhg.2010.20. [DOI] [PubMed] [Google Scholar]
- 33.Hunt KA, McGovern DP, Kumar PJ, et al. A common CTLA4 haplotype associated with coeliac disease. Eur J Hum Genet. 2005;13:440–444. doi: 10.1038/sj.ejhg.5201357. [DOI] [PubMed] [Google Scholar]
- 34.Torres B, Aguilar F, Franco E, Sánchez E, Sánchez-Román J, Jiménez Alonso J, Núñez-Roldán A, Martín J, González-Escribano MF. Association of the CT60 marker of the CTLA4 gene with systemic lupus erythematosus. Arthritis Rheum. 2004;50:2211–2215. doi: 10.1002/art.20347. [DOI] [PubMed] [Google Scholar]
- 35.Lei C, Dongqing Z, Yeqing S, Oaks MK, Lishan C, Jianzhong J, Jie Q, Fang D, Ningli L, Xinghai H, et al. Association of the CTLA-4 gene with rheumatoid arthritis in Chinese Han population. Eur J Hum Genet. 2005;13:823–828. doi: 10.1038/sj.ejhg.5201423. [DOI] [PubMed] [Google Scholar]
- 36.Chang MC, Chang YT, Tien YW, Liang PC, Jan IS, Wei SC, Wong JM. T-cell regulatory gene CTLA-4 polymorphism/haplotype association with autoimmune pancreatitis. Clin Chem. 2007;53:1700–1705. doi: 10.1373/clinchem.2007.085951. [DOI] [PubMed] [Google Scholar]
- 37.Blomhoff A, Lie BA, Myhre AG, Kemp EH, Weetman AP, Akselsen HE, Huseby ES, Undlien DE. Polymorphisms in the cytotoxic T lymphocyte antigen-4 gene region confer susceptibility to Addison's disease. J Clin Endocrinol Metab. 2004;89:3474–3476. doi: 10.1210/jc.2003-031854. [DOI] [PubMed] [Google Scholar]
- 38.Ban Y, Tozaki T, Taniyama M, Tomita M, Ban Y. Association of a CTLA-4 3′ untranslated region (CT60) single nucleotide polymorphism with autoimmune thyroid disease in the Japanese population. Autoimmunity. 2005;38:151–153. doi: 10.1080/08916930500050319. [DOI] [PubMed] [Google Scholar]
- 39.PastuszakLewandoska D, Sewerynek E, Domańska D, Gładyś A, Skrzypczak R, Brzeziańska E. CTLA-4 gene polymorphisms and their influence on predisposition to autoimmune thyroid diseases (Graves' disease and Hashimoto's thyroiditis) Arch Med Sci. 2012;8:415–421. doi: 10.5114/aoms.2012.28593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayans S, Lackovic K, Nyholm C, Lindgren P, Ruikka K, Eliasson M, Cilio CM, Holmberg D. CT60 genotype does not affect CTLA-4 isoform expression despite association to T1D and AITD in northern Sweden. BMC Med Genet. 2007;8:3. doi: 10.1186/1471-2156-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhernakova A, Eerligh P, Barrera P, et al. CTLA4 is differentially associated with autoimmune diseases in the Dutch population. Hum Genet. 2005;118:58–66. doi: 10.1007/s00439-005-0006-z. [DOI] [PubMed] [Google Scholar]