Abstract
Long non–protein coding RNAs (lncRNAs) are an important class of molecules that help orchestrate key cellular events. Although their functional roles in cells are not well understood, thousands of lncRNAs and a number of possible mechanisms by which they act have been reported. LncRNAs can exert their regulatory function in cells by interacting with epigenetic enzymes. In this study, we developed a tool to study lncRNA-protein interactions for high-throughput screening of small-molecule modulators using AlphaScreen technology. We tested the interaction of two lncRNAs: brain-derived neurotrophic factor antisense (BDNF-AS) and Hox transcript antisense RNA (HOTAIR), with Enhancer of zeste homolog 2 (EZH2), a histone methyltransferase against a phytochemical library, to look for small-molecule inhibitors that can alter the expression of downstream target genes. We identified ellipticine, a compound that up-regulates BDNF transcription. Our study shows the feasibility of using high-throughput screening to identify modulators of lncRNA-protein interactions and paves the road for targeting lncRNAs that are dysregulated in human disorders using small-molecule therapies.
Keywords: long noncoding RNA, brain-derived neurotrophic factor (BDNF), brain-derived neurotrophic factor antisense (BDNFAS), Enhancer of zeste homolog 2 (EZH2), hox transcript antisense RNA (HOTAIR), epigenetic enzyme, RNA protein interaction, AlphaScreen, natural antisense transcript, noncoding RNA
Introduction
Long non–protein coding RNAs (lncRNAs) have been shown to play important functional roles in development1,2 and disease3,4 processes. This large, diverse class of transcripts, which are greater than 200 nucleotides in length, make up the majority of the transcriptional landscape within cells and are expressed in a cell-, tissue-, and development-specific manner.5 Current research has seen a rapid rise in lncRNA research due to an increased appreciation for their functional importance. For example, the non–protein coding antisense transcript to brain-derived neurotrophic factor (BDNF-AS) was reported to regulate the protein-coding gene, BDNF, via an epigenetic mechanism.6 BDNF-AS, through its association with the epigenetic enzyme enhancer of zeste homology 2 (EZH2), is able to guide the histone methyltransferase to the BDNF gene promoter, enabling trimethylation of lysine 27 of histone 3 to repress BDNF transcription (Fig. 1A). BDNF is an important neurotrophin that is critical for early neural development, maintenance, and survival. Furthermore, BDNF has been observed to be down-regulated in a number of neurodegenerative and neuropsychiatric disorders.7 Another well-studied lncRNA, Hox transcript antisense RNA (HOTAIR), has also been shown to interact with EZH28 to transcriptionally suppress several developmental HOXD genes.9 HOTAIR has been implicated in several cancers, making it a potential therapeutic target.4,10,11 Although many reports on dysregulated lncRNAs and their proposed function in disease phenotypes exist, our current options in targeting lncRNAs or their protein partners remain limited.12
Figure 1.
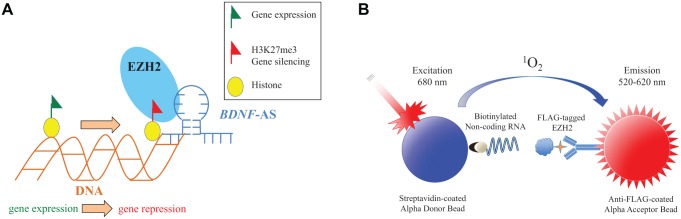
(A) Mechanism of the BDNF-AS–EZH2 interaction. The BDNF-AS transcript interacts with EZH2 (RNA-protein interaction), guiding this ubiquitously expressed epigenetic enzyme to the BDNF locus (RNA-chromatin interaction) where EZH2 is able to epigenetically silence BDNF gene expression. Inhibition of the BDNF-AS–EZH2 interaction can prevent EZH2 recruitment to the BDNF promoter and results in up-regulation of the BDNF gene. (B) Schematic of AlphaScreen adapted to quantify lncRNA-protein interactions. Following the incubation of biotinylated long noncoding RNA BDNF-AS with Flag-tagged EZH2 protein, anti-flag tagged acceptor beads and streptavidin-coated donor beads are added to each well. Upon excitation of the donor beads at 680 nm, ambient oxygen is elevated to an excited state and excites nearby acceptor beads, resulting in a measurable emission at 570 nm that is used to quantify the assay.
Several efforts from pharmaceutical companies have produced EZH2 inhibitors,13 as this enzyme is mutated and up-regulated in several cancers.13,14 However, this approach is not ideal in the nervous system where interactions with this critical epigenetic enzyme are tightly regulated and can have multiple undesirable effects on gene expression. Furthermore, noncoding RNAs do not interact with EZH2 at the C-terminus where their catalytic histone methyltransferase domain is located but rather at a separate and distinct noncoding RNA binding domain.15 As such, our work focused on developing a tool to study the interaction between lncRNAs and an epigenetic enzyme (EZH2) to screen for modulators of these potential drug targets. Although the binding of RNAs to EZH2 has been reported to be nonspecific, some reports indicate that EZH2 can bind specific RNAs with much greater affinity.8,16–18 Many studies have shown that EZH2 does interact with several important long noncoding RNAs to help regulate important cellular processes.19 For example, upon its interaction with Xist, EZH2 helps initiate X-chromosome inactivation, an important event in early fetal development.18 EZH2 also interacts with Kcnq1ot1, an lncRNA that is involved in maternal imprinting of a voltage-gated potassium channel, and has been linked to Beckwith-Wiedemann syndrome.2,20
Several methods exist to study individual RNA-protein interactions21–23; however, these methods require much optimization and importantly are not amenable to large-scale compound screening. To quantify the lncRNA-protein interaction, we adapted PerkinElmer (Waltham, MA) technology to perform an AlphaScreen, which has been previously reported for screening of small molecules targeting other RNA-protein interactions.24 Here, we sought to determine whether the lncRNA-EZH2 interaction can be assayed, quantified, and adapted for high-throughput screening to identify modulators of these interactions. Such modulators can be therapeutically valuable and have the potential to be used in several disease contexts, including neurological disorders and a number of cancers.
Materials and Methods
RNA Preparation and Biotin Labeling
Human BDNF-AS (NR_033315.1) was polymerase chain reaction (PCR) amplified from the pMA-BDNF-AS vector custom produced by Thermo Fisher GeneArt Gene synthesis services (Waltham, MA). Primers for PCR amplification are T7 Forward 5′ TAATACGACTCACTATAG 3′ and SP6 Reverse 5′ ATTTAGGTGACACTATA 3′. Human HOTAIR was PCR amplified from the pcDNA3.1-HOTAIR vector generously provided by Dr. Howard Chang using forward primer 5′ TAA TACGACTCACTATAGGACTCGC 3′ and reverse primer 5′ TTGAAAATGCAT CCAGATAT TAATATATCTACA 3′. The pRL-TK vector containing the Renilla luciferase control transcript (Promega Corp, Madison, WI, cat No. E2241) was linearized with XhoI, run on an agarose gel, and the linearized fragment was excised and purified (Qiagen, Valencia, CA, gel extraction kit cat No. 28704) before performing in vitro transcription. The BDNF-AS and HOTAIR PCR products were used as templates for T7 in vitro transcription using Applied Biosystems (Carlsbad, CA) T7 MEGAScript kit cat No. AM 1333. RNA yield was maximized by incubating overnight at 37 °C in a water bath. The size and purity of RNAs were confirmed using the Agilent RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, cat No. 5067-1511) and run on an Agilent 2100 Bioanalyzer (Agilent Technologies). Prior to analysis, RNA was heated at 70 °C for 2 min and cooled to room temperature to relax RNA secondary structure. RNA was 3′ biotinylated using the Pierce RNA 3′ end biotinylation kit (Thermo Fisher Scientific, cat No. 20160). Biotinylation reactions were extended overnight at 16 °C, and biotinylation efficiency was confirmed using the Pierce Chemiluminescent detection module (Thermo Fisher Scientific, cat No. 89880).
RNA Electrophoretic Mobility Shift Assay
An RNA electrophoretic mobility shift assay (EMSA) was performed using the LightShift Chemiluminescent RNA EMSA kit (Thermo Fisher Scientific, cat No. 20158) as per the manufacturer’s instructions using the 3′ end biotinylated RNA (1 nM). Purified human EZH2 protein (C-terminal Flag/myc tag) was obtained from Origene technologies (Rockville, MD, cat No. TP302054). RNA was incubated in a 37 °C water bath for 30 min to allow the RNA to fold in EMSA binding buffer (10 mM HEPES pH 7.3, 20 mM KCl, 4 mM MgCl2, 1 mM DTT) before EZH2 protein was added to the appropriate samples. The RNA was allowed to interact with EZH2 for 20 min at 37 °C before the samples were run on a 0.5% agarose (Sigma-Aldrich, St. Louis, MO, cat No. A0576) gel for 2 h at 4 °C and 90 V in 0.5 X TBE. Binding reactions were then transferred onto a Biodyne nylon membrane (Thermo Fisher Scientific, cat No. 77016) in 0.5 X TBE for 30 min at 4 °C and 400 mA. RNA was cross-linked to the nylon membrane for 5 min with a UV lamp equipped with 254 nm bulbs. The chemiluminescent detection module (Thermo Fisher Scientific, cat No. 89880) was used per the manufacturer’s instructions to detect biotin-labeled RNA by chemiluminescence on the FluorChem E imager software version 4.1.1 (Bio-Techne, Minneapolis, MN).
Long Noncoding RNA-EZH2 Interaction AlphaScreen Assay
AlphaScreen assays were performed using RNA and protein described above. Optimal concentrations of RNA, EZH2, acceptor, and donor beads were determined using cross-titration of individual components. RNA concentrations (ranging from 1 nM to 1 pM) are as indicated, and the EZH2 concentration was 4 nM in a 40 µL reaction (10 µL per each reaction component: RNA, protein, AlphaScreen acceptor and AlphaScreen donor beads, PerkinElmer, cat No. 6760613) and were plated onto white 384-well OptiPlates (PerkinElmer, cat No. 600790). Prior to performing any experiments, RNA was allowed to fold at 37 °C for 15 min. Following RNA and protein addition diluted in assay buffer—HEPES (30 mM) pH = 7.4, NaCl (100 mM), MgCl2 (2 mM), NP-40 (0.01%), Guanidinum HCl (10 mM), Escherichia coli tRNA (Sigma-Aldrich, cat No. R1753, 50 µg/mL)—the plate was sealed and spun at 1000 rpm and then incubated at 37 °C while shaking at 1000 rpm for 30 min. This assay buffer is a modification of the buffer used in a previous report of RNA-protein interactions measured by AlphaScreen.24 To determine optimal buffer conditions, we titrated individual components of the buffer, including magnesium chloride, guanidinium HCl, E. coli tRNA, bovine serum albumin, as well as a range of buffer pHs, incubation times, and orders of reagent addition. The AlphaScreen Flag-tagged acceptor beads diluted in 1X phosphate-buffered saline (PBS; 10 µg/mL) were added to the plate and incubated for 60 min at room temperature. Subsequently, AlphaScreen streptavidin-coated donor beads diluted in 1X PBS (10 µg/mL) were added to the plate and incubated for 60 min at room temperature. The plate was resealed and spun at 1000 rpm briefly following each reagent addition. Bead additions were performed in the dark room under a green light due to the photosensitivity of the beads. The assay plate was then read on an EnVision 2104 Multilabel Plate Reader (PerkinElmer, Waltham, MA) preprogrammed from the manufacturer with the AlphaScreen module (excitation at 680 nm and Alpha acceptor bead emission was measured at 570 nm).
High-Throughput Screening Assay Using Phytochemical Compound Library
The Prestwick phytochemical compound library (Prestwick Chemical, Illkirch-Graffenstaden, France) was screened at 10 µM compound concentration (0.1% DMSO) in duplicate. RNA (0.3 nM) and EZH2 (4 nM) were then added to the compound plate, and the plate was sealed and spun at 1000 rpm after the final addition. Bead dilutions and additions were performed as described above. Data were plotted using GraphPad Prism software (San Diego, CA).
Cell Culture Drug Treatment
HEK293 cells were used to test the effect of Ellipticine (CAS 519-23-3) in vitro. HEK293 cells express EZH2, BDNF, and BDNF-AS and are an ideal system to test the effect of inhibiting the BDNF-AS–EZH2 interaction. Cells were plated overnight in DMEM supplemented with 10% fetal bovine serum in 6-well plates (500,000 cells per well) in a 37 °C incubator, 5% CO2, and were treated the next day morning with DMSO (0.01%) or Ellipticine (1 µM) for 48 h.
RNA Extraction, cDNA Synthesis, and Real-Time PCR
Cells were washed with 1X PBS and lysed, and RNA was extracted by passing cell lysates through columns supplied in the RNeasy mini kit (Qiagen, Valencia, CA, cat No. 74106) according to manufacturer’s instructions and included on-column DNase treatment. RNA (800 ng) was reverse transcribed using the qScript cDNA SuperMix kit (Quanta Biosciences, Gaithersburg, MD, cat No. 95048). Gene expression was measured by real-time PCR using human BDNF (Life Technologies, Grand Island, NY, cat No. 4331182, assay ID Hs02718934_s1) and human GAPDH (Life Technologies, cat No. 4326317, RefSeq NM_002046.3) Taqman primer-probe assays and Taqman gene expression mastermix (Life Technologies, cat No. 4369016) in a 10 µL reaction. Real-time PCR data were measured using QuantStudio Flex 6 software from Life Technologies (Grand Island, NY), and the delta Ct method was used to determine relative gene expression. Data were analyzed using a two-tailed t test on GraphPad Prism software (San Diego, CA).
Enzyme-Linked Immunosorbent Assay
The media from cells treated with compound (1 mL) were collected for enzyme-linked immunosorbent assay (ELISA) experiments. The ELISA kit for human BDNF from Promega Corporation (cat No. G7611) was used following the manufacturer’s instructions. The average absorbance (measured at 450 nm) in the ellipticine-treated samples was subtracted from the background and normalized to DMSO-treated samples. Experiments were performed with three technical and three biological replicates.
Results
LncRNA BDNF-AS Interacts with EZH2 in a Concentration-Dependent Manner
Although a previous report6 infers that EZH2 and BDNF-AS interact directly at the chromatin level, the physical association has not been shown. To confirm this interaction, we performed RNA EMSA and incubated biotinylated BDNF-AS RNA with increasing concentrations of EZH2, before competing out the interaction with unbiotinylated BDNF-AS. We observed that increasing concentrations of EZH2 prevented the migration of biotinylated BDNF-AS through an agarose gel, an effect that was reversed with an excess of unbiotinylated BDNF-AS (Suppl. Fig. S1). These findings indicate that there is a direct physical interaction between BDNF-AS and EZH2.
Development of AlphaScreen Assay to Quantify Long Noncoding RNA-Protein Interactions
AlphaScreen technology (PerkinElmer) was used to develop a cell-free assay system to measure the interaction of a biotinylated lncRNA with Flag-tagged EZH2 protein. AlphaScreen technology offers a rapid and simple method of quantifying lncRNA-protein interactions using a nonradioactive-amplified luminescent proximity homogeneous bead-based detection method. In our assay, the lncRNA and protein of interest are allowed to interact before AlphaScreen acceptor and donor beads are added to the reaction to measure the association of the binding partners. Upon excitation at 680 nm, the Alpha streptavidin-coated donor beads (that contain the photosensitizer phthalocyanine) convert molecular oxygen to an excited singlet oxygen (not a free radical) with a short (4 µs) half-life. If the RNA and protein are bound, the singlet oxygen can diffuse up to 200 nm to make contact with a thioxene derivative on the Alpha donor bead, resulting in an amplified chemiluminescent emission between 520 and 620 nm25 (Fig. 1B). In the absence of a nearby acceptor bead (when the RNA and protein do not interact), the singlet oxygen falls to the ground state and does not produce a signal. The signal amplification observed with AlphaScreen allows for an assay to be miniaturized with relative ease.
Because of the sensitive nature of the AlphaScreen, great lengths were taken to ensure the quality of protein and RNAs assayed. RNA preparation was optimized to produce pure, high-yield transcripts, and the size and purity were confirmed on the bioanalyzer (Suppl. Fig. S2a). The biotinylation efficiency of RNAs produced was confirmed with dot blot (Suppl. Fig. 2b). Purified Flag/myc-tagged EZH2 (size and purity were confirmed with sodium dodecyl sulfate polyacrylamide gel electrophoresis) was obtained from Origene technologies (Rockville, MD). Several reagent conditions and assay parameters were also optimized, including tRNA and bovine serum albumin concentrations as well as bead and RNA-protein incubation times (Suppl. Figs. 3–6). Optimal RNA and protein concentrations were determined by cross-titration of 1:2 serial dilutions of BDNF-AS into fixed concentrations of EZH2 using bead concentrations of 20 µg/mL (Fig. 2A). To maximize the AlphaScreen signal, all future assays were performed with 4 nM EZH2. AlphaScreen beads were cross-titrated (BDNF-AS [0.3 nM], EZH2 [4 nM]) to determine optimal acceptor and donor bead concentrations (Fig. 2B). Ultimately, 15 µg/mL acceptor and 20 µg/mL donor beads were used to produce maximal assay signal.
Figure 2.
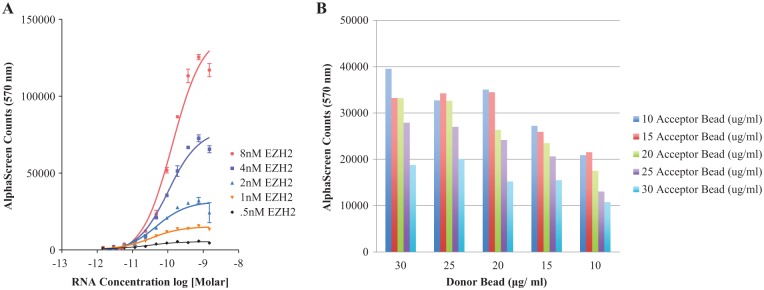
Optimization of AlphaScreen assay conditions. (A) Long noncoding RNA and protein concentrations were optimized by titrating BDNF-AS in fixed concentrations of EZH2 protein. The hook effect was observed at all concentrations of protein, and therefore, the maximal signal or the hook point is graphed. Data represent triplicates of a single experiment that was repeated three independent times. (B) AlphaScreen acceptor and donor beads were cross-titrated to determine optimal bead concentrations to produce maximal assay signal (n = 3).
AlphaScreen Detects Specific Quantifiable Binding between BDNF-AS and EZH2
We performed several experiments to confirm the specificity of the signal detected in the AlphaScreen assay. To show specific binding, we sought to compete out the BDNF-AS-EZH2 signal with increasing concentrations of unbiotinylated transcript. Biotinylated BDNF-AS was competed out with increasing fixed concentrations of unbiotinylated BDNF-AS (1-, 10-, and 100-fold excess; Suppl. Fig. S7). It was observed that a 10-fold increase in unbiotinylated BDNF-AS was able to reduce the assay signal by about 40%, while a 100-fold excess in unlabeled RNA completely quenched the signal.
Next, we determined if a random RNA transcript is able to specifically bind to EZH2 with the same potency as the BDNF-AS transcript. We generated biotinylated RNA using the Renilla luciferase pRL-TK vector. This negative control transcript for EZH2 binding was tested in the assay by titrating Renilla luciferase RNA into 4 nM EZH2. The control Renilla RNA transcript produced a low, concentration-dependent signal in the AlphaScreen (Fig. 3A), indicating some degree of nonspecific binding that was confirmed with RNA EMSA (Fig. 3B). This finding further confirms reports that EZH2 is “promiscuous,” having the ability to bind many RNAs, even nonmammalian transcripts, with low affinity.16,17
Figure 3.
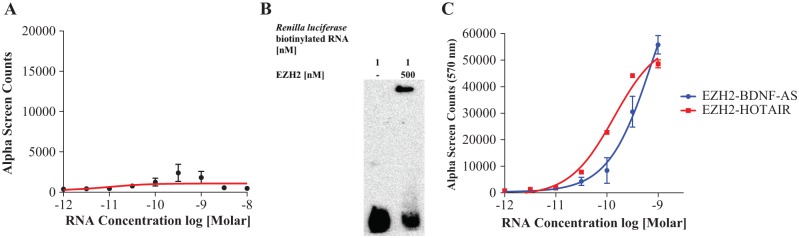
Determining negative and positive RNA controls for EZH2 interactions. EZH2 reportedly binds many RNAs without well-defined protein-binding motifs. As a negative control, a nonrelated Renilla luciferase transcript was tested that did show some affinity for the EZH2 protein in the (A) Alphascreen assay (n = 3) as well as in the (B) RNA EMSA assay. This finding is consistent with reports of EZH2 promiscuity, including the ability of this enzyme to bind nonmammalian transcripts with low affinity.16 (C) The ability of biotinylated lncRNAs BDNF-AS and HOTAIR to interact with EZH2 is measured in the AlphaScreen assay. Both lncRNAs are titrated in EZH2 (4 nM). EZH2 interacts with both lncRNAs in a concentration-dependent manner (n = 3). HOTAIR serves as a positive RNA control and biologically important screen.
AlphaScreen Detects Specific Quantifiable Binding between HOTAIR and EZH2
HOTAIR is a cancer-related lncRNA that has previously been shown to recruit EZH2 to target regions in the genome.9 We applied the same assay conditions as those used for BDNF-AS to investigate the possibility of using our cell-free assay to measure HOTAIR-EZH2 interactions. As expected, increasing concentrations of HOTAIR RNA produced an increase in assay signal, confirming several reports that EZH2 and HOTAIR are indeed binding partners.4,8,17 Binding isotherms for the interaction of BDNF-AS or HOTAIR with EZH2 show an RNA concentration-dependent increase in assay signal when either transcript is incubated with a fixed concentration (4 nM) of EZH2 (Fig. 3C). HOTAIR is an important biological target as it has been implicated in several cancers.4,10,11 Our optimized assay has the potential to be used as a high-throughput screening (HTS) platform to find modulators of HOTAIR-EZH2 binding as well as to study the dynamics of the interaction between this important lncRNA and its protein partner.
Optimization for HTS
To determine the suitability of the assay for screening, several standard parameters were measured. Z-factor is a measure of assay suitability for HTS.26 From the binding isotherms generated (Fig. 3C), a protein concentration of 4 nM and RNA concentration of 0.3 nM produced a signal with Z-factors greater than 0.5, indicating that the assay has little variation at this point. Furthermore, the coefficient of variation was at or below 10%, which is suitable for HTS (Table 1). We generated EC50 values using nonlinear regression and observed consistent EC50 values (0.33 ± 0.065 nM). We also measured day-to-day variation in the Z-factor, coefficient of variation and EC50 to determine whether assay conditions are suitable for HTS (Table 1).
Table 1.
Assay Parameters Measured to Test Day-to-Day Variation in Assay Signal and High-Throughput Screening Suitability.a
| Day 1 | Day 2 | Day 3 | Mean ± SEM | |
|---|---|---|---|---|
| Z-factor | 0.71 | 0.63 | 0.68 | 0.67 ± 0.02 |
| Coefficient of variation (%) | 5.73 | 10.09 | 9.24 | 8.35 ± 1.3 |
| EC50 (nM) | 0.221 | 0.447 | 0.323 | 0.330 ± 0.065 |
Screening optimizations included measuring important assay parameters (Z-factor, coefficient of variation, and in EC50) of the same experiment over several days. This consistency (Z-factor >0.5 and coefficient of variation <10%) suggests the assay is suitable for high-throughput screening at EZH2 (4 nM) and BDNF-AS (0.3 nM).
As most compound libraries are dissolved in DMSO, we measured the stability of our assay in the presence of increasing concentrations of DMSO. We observed that both BDNF-AS–EZH2 and HOTAIR-EZH2 assays were able to tolerate up to 1% of DMSO without significant changes in assay signal (Fig. 4A). Screening was performed at 0.1% DMSO, a concentration that did not affect either the BDNF-AS–EZH2 or HOTAIR-EZH2 interactions. Collectively, our data suggest that both of our lncRNA-EZH2 assays are amendable to HTS and can be used to find small-molecule modulators of such interactions related to various disease phenotypes.
Figure 4.
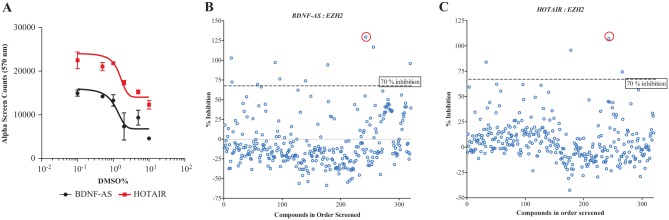
Screening of lncRNA-protein interactions. (A) The tolerance of the assay to DMSO was tested over a 100-fold range (0.1%−10% final DMSO concentration) for both lncRNAs of interest. The lncRNAs, EZH2, and AlphaScreen assay components were found to be stable in DMSO up to 1%, well below the 0.1% DMSO concentration used for screening. Data represent triplicates of a single experiment that was repeated 3 independent times. Scatterplot of Prestwick Phytochemical library (Prestwick Chemical, Illkirch-Graffenstaden, France) screen for (B) BDNF-AS EZH2 (Z-factor = 0.65, 3.1% hit rate) and (C) HOTAIR-EZH2 (Z-factor = 0.85, 1.25% hit rate). A total of 320 compounds (10 µM, 0.1% DMSO) were tested. Data are the average of duplicates expressed as percentage inhibition relative to a positive control (no lncRNA in the reaction, set to 100% inhibition). The negative control (0% signal inhibition) consisted of lncRNA and protein in 0.1% DMSO. Red circles in (B) and (C) indicate biotin, which shows greater than 100% inhibition in the screen, confirming our data and serving as a positive control for future studies.
Primary Screen for Inhibitors of lncRNA-Protein Interactions
Phytochemical compounds have many pharmacophores, a great degree of stereochemistry, and are natural metabolites, making them excellent candidates for the screening of potential bioactive drugs.27 As such, a primary screen was performed using the Prestwick phytochemical library (Prestwick Chemical, Illkirch-Graffenstaden, France) at 10 µM and 0.1% DMSO concentration. The 320-compound natural products library was used to screen for inhibitors of two important lncRNA-protein interactions. The library was screened at a fixed 10 µM concentration against BDNF-AS–EZH2 and HOTAIR-EZH2, and the scatterplots for percentage inhibition of the library were generated (Fig. 4B and C). The data generated from this small-scale screen show the potential of screening larger-compound libraries in our lncRNA-protein interaction assay.
There are no known inhibitors of the RNA-protein interactions; therefore, 100% inhibition represented the absence of RNA in the assay. This value indicates the background signal from interactions between donor and acceptor beads in the absence of a binding partner. The negative control consisted of the RNA and protein of interest in 0.1% DMSO (0% inhibition). Both RNA targets produced quality data. The average Z-factor for the BDNF-AS–EZH2 screen was 0.65 and the hit rate (using a cutoff 70% inhibition as a hit) was 3.1%. For HOTAIR-EZH2, the average Z-factor was 0.85 and the hit rate was 1.25%. We also observed that some compounds enhance the interaction between lncRNAs and EZH2, which are shown as negative values on the graphs. These compounds, although not directly relevant to our work, could have therapeutic value in other disease contexts. As expected, one of the natural compounds present in our library, biotin, produced a hit in both screens (Fig. 4B and C). Free biotin will saturate the streptavidin-coated donor bead, and because free biotin does not interact with EZH2, will inhibit the assay signal. This finding provided us with a compound that can be used as a positive control for inhibition in future screening. Our data suggest that despite using a small library (320 compounds), we are able to identify potential modulators of lncRNA-EZH2 interactions.
Ellipticine Increases BDNF in a Secondary Assay
From our initial screen, 10 putative compound hits for BDNF-AS-EZH2 (Suppl. Table S1) were identified. Interestingly, only one compound (biotin) was identified as a common inhibitor for both transcripts. BDNF-AS–EZH2 compound hits were assayed to establish potency against BDNF-AS–EZH2 (Suppl. Table S1). Validated hits for BDNF-AS–EZH2 inhibition (myricetin, gossypol, ellipticine, and biotin) were tested in secondary cell culture experiments. HEK293 cells were selected as a model to study inhibitors of BDNF-AS–EZH2 as these cells express EZH2, BDNF-AS, and the repressed target of this interaction, BDNF. This cell model was also used in initial studies to study the mechanism of BDNF-AS.6 All validated compounds were tested in a concentration-dependent manner (0.1, 0.3, 1 and 3 µM) to determine toxicity after 48 h of drug treatment. Optimal concentrations for treatment with each drug were determined, and cells were treated for 48 h before changes in target gene expression were measured. One compound, ellipticine, was validated for BDNF-AS–EZH2 in HEK293 cells. Treatment of HEK293 cells with ellipticine (1 µM) was able to up-regulate BDNF transcription (~threefold, p < 0.0001) after 48 h, normalized to GAPDH. The effect of ellipticine treatment on BDNF levels after 24 and 72 h was also measured; however, there was no significant effect on BDNF gene expression. To test if increased BDNF transcription also resulted in increased BDNF protein, an ELISA was performed using the media from HEK293-ellipticine-treated cells because BDNF is a secreted protein. We observed a small (10%) but statistically significant (p < 0.001) up-regulation in BDNF protein (Fig. 5B). Although ellipticine was not able to up-regulate BDNF greatly at the protein level, we do observe an increase in BDNF transcription following drug treatment, indicating that ellipticine treatment does indeed have an effect on the target of BDNF-AS-EZH2.
Figure 5.
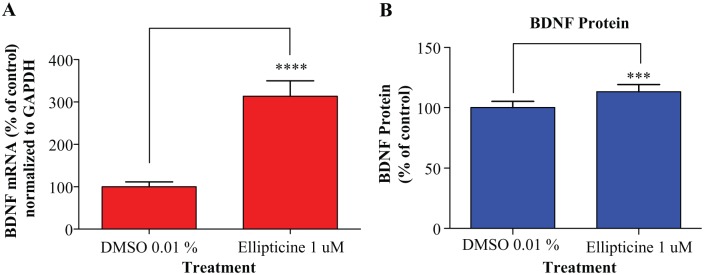
Secondary assays to test the effect of ellipticine on the target of BDNF-AS–EZH2, BDNF, in vitro. (A) HEK293 cells were treated for 48 h with ellipticine (1 µM) before RNA was extracted to measure changes in BDNF gene expression normalized to GAPDH (~threefold up-regulation in BDNF mRNA, p < 0.0001, n = 3). (B) BDNF is a secreted protein; therefore, culture media from HEK293 cells treated with ellipticine in (A) were used to measure changes in BDNF protein with an enzyme-linked immunosorbent assay (10% increase in BDNF, p < 0.001, n = 3). Ellipticine was able to increase BDNF protein modestly, despite the marked increase in BDNF transcription following compound treatment.
Secondary Assays: HOTAIR- EZH2
All four hits for HOTAIR-EZH2 (Suppl. Table S2) were validated. For secondary cell culture experiments, HeLa cells were chosen as a cell model as they express EZH2, HOTAIR, and repressed HOTAIR target gene HOXD11. The validated HOTAIR-EZH2 compounds were tested in a concentration-dependent manner (0.1, 0.3, 1, and 3 µM) to determine toxicity in cells following 48-h drug treatment. Once optimal compound concentrations in HeLa cells were determined, cells were treated for 48 h, and changes in HOXD11 gene expression were measured. These experiments validated one hit, camptothecin, for HOTAIR-EZH2. However, camptothecin also altered the expression of two housekeeping genes, beta actin and GAPDH, and was not further studied.
Discussion
In the past decade, next-generation sequencing platforms and huge multicenter transcriptomics efforts have helped to increase the inventory of functional lncRNAs at an incredible rate. However, our increasing knowledge of the disease relevance of lncRNAs and the observation that they can exert their functions by acting through epigenetic enzymes necessitates a more thorough examination of the structural components of these binding partners.19,21,28 Furthermore, it prompts us to view these interactions as therapeutic targets that have yet to be studied using small-molecule inhibitors. Understanding the nature of RNA-ligand interactions is important and has been highlighted24; however, work in this arena has been slow, partially because of difficulties in targeting RNAs as they do not have a single fixed structure. Several low-throughput assay methods do exist to measure these interactions; however, this is the first report of an assay being used to study lncRNA-protein interactions for high-throughput small-molecule screening purposes. Our assay has the potential to be used in drug discovery as well as to study many different lncRNA-protein binding partners.
This AlphaScreen assay enabled us to study the interaction between specific lncRNAs and an important histone methyltransferase, EZH2, to screen for small-molecule modulators of lncRNA-protein interactions. Z-factors greater than 0.5 and acceptable assay variation (<10%) indicate that this assay could be scaled for screening of much larger compound libraries. Although AlphaScreen is a convenient and commonly used screening technology, one well-known limitation of this assay is the hook effect. At the “hook” or peak assay signal, either donor or acceptor bead is saturated with RNA or protein. Increasing RNA or protein concentration above the hook point results in a decrease in assay signal, producing a bell-shaped curve as opposed to a classical saturation curve.25 Further experimentation showed that reducing concentrations of the protein was not able to overcome the observed hook while maintaining a consistent assay signal. Although the assay is optimal for screening purposes, because of the limitations incurred by the persistence of the hook effect, we were not able to approximate a binding constant for the lncRNA-EZH2 interactions. However, we were able to confirm screening hits in appropriate secondary assays. In this scenario, low-throughput methods, such as RNA EMSA, could be used to determine binding constants for lncRNA-protein interactions, whereas our assay has the distinct advantage of being used for HTS. In the current study, we focused on testing a small natural compounds library (320 compounds) to show the feasibility of compound screening using our assay. We were able to observe one example of a compound (ellipticine) that was able to up-regulate BDNF mRNA and to a lesser degree BDNF protein. Although the identified hits require further development to make them more suitable potential small-molecule leads, this study does serve as a proof of concept that this important class of interactions can be assayed and pharmacologically targeted. Small-molecule therapies that could up-regulate BDNF mRNA, if brain penetrant, could possibly be used to treat a number of neurological diseases in which BDNF is down-regulated (e.g., Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis, and Rett syndrome).7,29 Furthermore, we screened another important biological target, HOTAIR-EZH2. HOTAIR is up-regulated in breast,4 colorectal,10 and pancreatic11 cancers and plays a functional role in the progression of these cancers through its interactions with the Polycomb repressive complex 2, the protein complex containing EZH2. Therefore, compounds that arise from future screening of this target could be potential cancer therapeutics.
This method, compared with previously established methods of measuring lncRNA-protein interactions is faster, easier, and amenable to HTS of many potential lncRNA-protein targets with diverse small-molecule libraries. HTS can lead to two possible types of hits: (1) compounds that directly block the RNA-binding pocket of EZH2 and (2) compounds that bind to RNA and change the secondary or tertiary structure in a way that prevents binding to the intended protein target. In the second scenario, identified hits are highly lncRNA-specific and might act selectively to block the function of EZH2 targets. Nonspecific blocking of EZH2, as previously attempted in cancer, might have mixed and opposing effects on several oncogenes and oncosuppressors. Introducing a new level of specificity by targeting lncRNA-protein interactions might help identify potent and specific cancer therapeutics as lncRNAs are expressed in cell- and development-specific contexts and bind to EZH2 at a specific site separate from the catalytic domain. Although the highly specific targets could be challenging, targeting individual dysregulated lncRNAs and their protein partners could pave the road for precision medicine.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Claude-Henry Volmar and Dr. Shaun Brothers for many helpful insights and discussions. This work was supported by the US NIH NINDS R01NS081208-01A1 awarded to MAF. RPF received funding from American Heart Association Greater Southeast Affiliate.
Footnotes
Author Contributions: RPF, SSU, and MAF designed and performed all experiments. RPF and MAF wrote the manuscript. SSU and CW revised the manuscript.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the US NIH NINDS R01NS081208-01A1 awarded to MAF. RPF received funding from American Heart Association Greater Southeast Affiliate.
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
References
- 1. Brown C. J., Ballabio A., Rupert J. L., et al. A Gene from the Region of the Human X Inactivation Centre Is Expressed Exclusively from the Inactive X Chromosome. Nature 1991, 349, 38–44. [DOI] [PubMed] [Google Scholar]
- 2. Lee M. P., DeBaun M. R., Mitsuya K., et al. Loss of Imprinting of a Paternally Expressed Transcript, with Antisense Orientation to KVLQT1, Occurs Frequently in Beckwith-Wiedemann Syndrome and Is Independent of Insulin-Like Growth Factor II Imprinting. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 5203–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pasmant E., Laurendeau I., Heron D., et al. Characterization of a Germ-Line Deletion, Including the Entire INK4/ARF Locus, in a Melanoma-Neural System Tumor Family: Identification of ANRIL, an Antisense Noncoding RNA Whose Expression Coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [DOI] [PubMed] [Google Scholar]
- 4. Gupta R. A., Shah N., Wang K. C., et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 2010, 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kapranov P., Cheng J., Dike S., et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- 6. Modarresi F., Faghihi M. A., Lopez-Toledano M. A., et al. Inhibition of Natural Antisense Transcripts In Vivo Results in Gene-Specific Transcriptional Upregulation. Nat. Biotechnol. 2012, 30, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagahara A. H., Tuszynski M. H. Potential Therapeutic Uses of BDNF in Neurological and Psychiatric Disorders. Nat. Rev. Drug Discov. 2011, 10, 209–219. [DOI] [PubMed] [Google Scholar]
- 8. Tsai M. C., Manor O., Wan Y., et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rinn J. L., Kertesz M., Wang J. K., et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kogo R., Shimamura T., Mimori K., et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 2011, 71, 6320–6326. [DOI] [PubMed] [Google Scholar]
- 11. Kim K., Jutooru I., Chadalapaka G., et al. HOTAIR Is a Negative Prognostic Factor and Exhibits Pro-Oncogenic Activity in Pancreatic Cancer. Oncogene 2013, 32, 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fatemi R. P., Velmeshev D., Faghihi M. A. De-Repressing LncRNA-Targeted Genes to Upregulate Gene Expression: Focus on Small Molecule Therapeutics. Mol. Ther. Nucleic Acids 2014, 3, e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCabe M. T., Creasy C. L. EZH2 as a Potential Target in Cancer Therapy. Epigenomics 2014, 6, 341–351. [DOI] [PubMed] [Google Scholar]
- 14. McCabe M. T., Graves A. P., Ganji G., et al. Mutation of A677 in Histone Methyltransferase EZH2 in Human B-Cell Lymphoma Promotes Hypertrimethylation of Histone H3 on Lysine 27 (H3K27). Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 2989–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaneko S., Li G., Son J., et al. Phosphorylation of the PRC2 Component Ezh2 Is Cell Cycle-Regulated and Up-Regulates Its Binding to ncRNA. Genes Dev. 2010, 24, 2615–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davidovich C., Zheng L., Goodrich K. J., et al. Promiscuous RNA Binding by Polycomb Repressive Complex 2. Nat. Struct. Mol. Biol. 2013, 20, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davidovich C., Wang X., Cifuentes-Rojas C., et al. Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol. Cell. 2015, 57, 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao J., Sun B. K., Erwin J. A., et al. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science 2008, 322, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brockdorff N. Noncoding RNA and Polycomb Recruitment. RNA 2013, 19, 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pandey R. R., Mondal T., Mohammad F., et al. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Trans-criptional Silencing through Chromatin-Level Regulation. Mol. Cell 2008, 32, 232–246. [DOI] [PubMed] [Google Scholar]
- 21. Rinn J. L., Ule J. ‘Oming in on RNA-Protein Interactions. Genome Biol. 2014, 15, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buenrostro J. D., Araya C. L., Chircus L. M., et al. Quantitative Analysis of RNA-Protein Interactions on A Massively Parallel Array Reveals Biophysical and Evolutionary Landscapes. Nat. Biotechnol. 2014, 32, 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tome J. M., Ozer A., Pagano J. M., et al. Comprehensive Analysis of RNA-Protein Interactions by High-Throughput Sequencing-RNA Affinity Profiling. Nat. Methods 2014, 11, 683–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mills N. L., Shelat A. A., Guy R. K. Assay Optimization and Screening of RNA-Protein Interactions by AlphaScreen. J. Biomol. Screen. 2007, 12, 946–955. [DOI] [PubMed] [Google Scholar]
- 25. Arkin M. R., Glicksman M. A., Fu H., et al. Inhibition of Protein-Protein Interactions: Non-Cellular Assay Formats. In Assay Guidance Manual; Sittampalam G. S., Coussens N. P., Nelson H., et al., Eds. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, 2004. [PubMed] [Google Scholar]
- 26. Zhang J. H., Chung T. D., Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 27. Harvey A. L., Edrada-Ebel R., Quinn R. J. The Re-Emergence of Natural Products for Drug Discovery in the Genomics Era. Nat. Rev. Drug Discov. 2015, 14, 111–129. [DOI] [PubMed] [Google Scholar]
- 28. Shortridge M. D., Varani G. Structure Based Approaches for Targeting Non-Coding RNAs with Small Molecules. Curr. Opin. Struct. Biol. 2015, 30, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W., Pozzo-Miller L. BDNF Deregulation in Rett Syndrome. Neuropharmacology 2014, 76, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


