Abstract
The architecture of biogenic structures can be highly influential in determining species contributions to major soil and sediment processes, but detailed 3-D characterisations are rare and descriptors of form and complexity are lacking. Here we provide replicate high-resolution micro-focus computed tomography (μ-CT) data for the complete burrow systems of three co-occurring, but functionally contrasting, sediment-dwelling inter-tidal invertebrates assembled alone, and in combination, in representative model aquaria. These data (≤2,000 raw image slices aquarium−1, isotropic voxel resolution, 81 μm) provide reference models that can be used for the development of novel structural analysis routines that will be of value within the fields of ecology, pedology, geomorphology, palaeobiology, ichnology and mechanical engineering. We also envisage opportunity for those investigating transport networks, vascular systems, plant rooting systems, neuron connectivity patterns, or those developing image analysis or statistics related to pattern or shape recognition. The dataset will allow investigators to develop or test novel methodology and ideas without the need to generate a complete three-dimensional computation of exemplar architecture.
Subject terms: Animal behaviour, Ecosystem ecology, 3-D reconstruction, Biooceanography
Background & Summary
Soils and sediments provide habitat for a wide range of organisms and the vertical exploitation of this ecospace has been important in mediating major ecosystem properties and the diversification of life over geological timescales1–3. Insights about organism-sediment relations, however, have largely been restricted to two dimensions4,5, although important inferences about burrowing mechanics6 and three dimensional architecture7 have been made from burrow castings8 and the use of optically transparent sediment analogues9. Relatively few studies apply non-invasive interrogation of intact sedimentary media10–13, despite significant advances in optical and clinical imaging technology14. High-resolution micro-focus computed tomography (μ-CT) offers a way of not only imaging the organisms themselves15,16 but also visualising the structure of a whole sediment core in three dimensions to allow quantitative examination of organismal burrowing17.
Experimental details are given in Hale et al.18. Briefly, surficial sediment (less than 3 cm depth; mean particle size, 54.80 μm; mud content, 55.93%) and three co-occurring functionally contrasting inter-tidal invertebrates (the polychaete Hediste diversicolor, the gastropod Hydrobia ulvae and mud shrimp Corophium volutator) were collected from the mid-shore at Breydon water, Great Yarmouth, UK (N52° 37.030′, E01° 41.390′) and returned to the Biodiversity and Ecosystem Futures Facility at the University of Southampton to acclimatise to laboratory conditions (5 days). Sediment was sieved (500 μm mesh) in a seawater (sand filtered, UV sterilized and salinity 33 practical salinity units) bath to remove macrofauna and allowed to settle for 48 h to retain the fine fraction (less than 63 μm). Circular aquaria (internal diameter=10 cm, 15 cm tall, n=20) were filled to a depth of 8 cm with sediment homogenate overlain by 4 cm of seawater.
Overlying seawater was replaced after 24 h to remove excess nutrients associated with assembly. Aquaria were aerated and maintained at 12±0.1 °C under a 12:12 h light (Aqualine T5 Reef White 10 K fluorescent light tubes, Aqua Medic) cycle. Fauna were not added until the lower regions of the sediment cores showed evidence of reducing conditions (visible anoxic microniche formation). Replicate (n=5) invertebrate communities (1 g wet weight aquaria−1; ~127 g m−2) were assembled in monoculture (Hediste diversicolor, HD; Hydrobia ulvae, HU; or Corophium volutator, CV) and in equal mixture (Mix).
These μ-CT sediment scans can provide reference models which may be of use in a range of connected fields, such as for the development of novel structural analysis routines and computer models in ecology17,19, pedology20, geomorphology, ichnology21, palaeobiology, and mechanical engineering22. We envisage those investigating transport networks23, vascular systems, plant rooting systems24, neuron connectivity patterns25,26, or developing image analysis or statistics related to pattern or shape recognition will find these data of interest. We have made this dataset available to allow investigators to develop or test novel methodology and ideas without the need to generate a complete three-dimensional computation of exemplar architecture.
Methods
Reconstruction of biogenic structures in the aquaria was achieved using a 225/450 kVp Nikon/Metris custom designed micro-focus computed tomography scanner housed within the μ-VIS X-ray Imaging Centre, University of Southampton. As the system used to acquire the scan data requires the cores to be held vertically batches of 5 aquaria were stacked and secured in a custom-made holding brace to ensure stability and prevent sediment or seawater leakage during rotation and scanning (Fig. 1). During each acquisition, the aquaria were rotated through 360° whilst collecting 3,142 projections averaging over 8 frames per 250 ms projection (for a total of 2 s per projection, ca. 105 min per acquisition). Ring artifact reduction was enabled. X-ray conditions were set to 300 kVp and 326 μA with a 3 mm Cu filter, and an XRD 1621 CN3 H5 PerkinElmer flat panel detector (CsI scintillator) was used to collect the images. In the resulting reconstructed images, levels of grey scale reflect the level of X-ray attenuation caused by variation in bulk density3. Hence, brighter pixels represent denser material (sediment) and darker pixels represent less dense material (burrow voids). Raw image slices (n=2,000 aquarium−1, voxel resolution=81 μm) were processed as follows: First, the projection data was reconstructed using CTPro3D (v. XT 2.2 service pack 10, Nikon Metrology, UK) and CTAgent (v. XT 2.2 service pack 10, Nikon Metrology, UK). The reconstructed volumes were converted to 8 bit format using FIJI27 (Version 1.49a) to reduce file sizes and computational loading. Finally, these images were opened as a 3D project in VGStudio Max (v. 2.1 Volume Graphics GmbH, Germany) and an edge-preserving 3D 5 pixel non-linear digital median filter was applied to reduce noise in the images.
Figure 1. Five aquaria stacked in the holding brace in the micro-focus computed tomography scanner housed within the μ-VIS X-ray Imaging Centre, University of Southampton.

Three types of images were produced. Whole core scans of 16-bit quality (Core_Volume_01_16bit to Core_Volume_20_16bit: Data Citation 1), processed image whole core scans of 8-bit quality with a 3D 5 pixel non-linear digital median filter applied (Core_Volume_01 to Core_Volume_20: Data Citation 1), an example slice of which is shown in Fig. 2, and processed burrow images (Burrow_Volume_01 to Burrow_Volume_20: Data Citation 1). To produce the burrow images the three-dimensional image captured of the aquaria and the holding brace was discarded to leave the central sediment core volume. Within the sediment core, regions of interest, the low density burrows, were segmented using a threshold based seed point growing algorithm that identified three-dimensional areas of similar low densities to produce a three-dimensional image of the burrow network (Fig. 3) called the burrow volume.
Figure 2. A representative transverse core slice from the Core Volumes image set showing distinct low density burrows (dark grey) through the (light grey) higher density sediment.
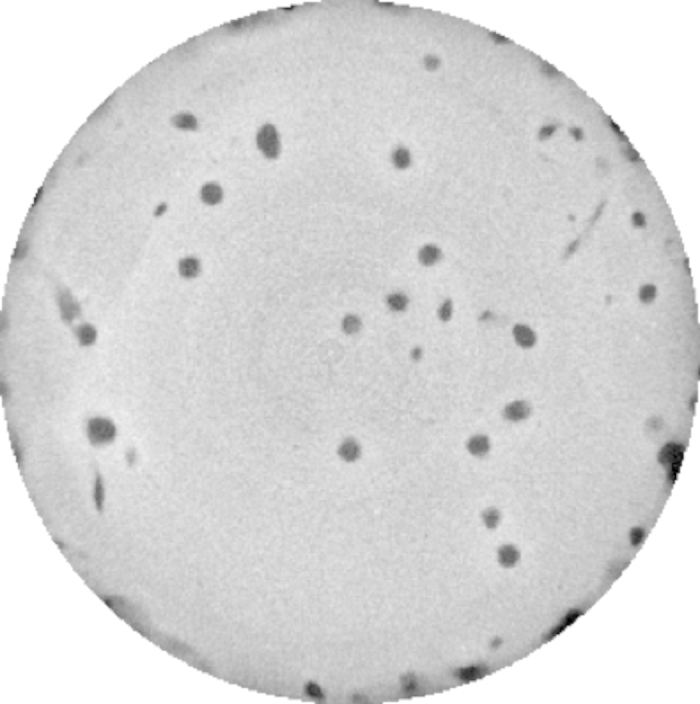
A Core Volume image set consists of number images that are sequentially stacked to create the three-dimensional core volume image. The central sediment core is 10 cm in diameter.
Figure 3. Representative example reconstructed three-dimensional burrow models for (a) Hediste diversicolor, (b) Hydrobia ulvae, (c) Corophium volutator, and (d) the three species in mixture created from the stacked Burrow Volumes images in VG Studio.
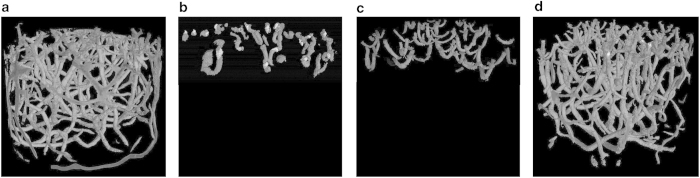
The sediment cores containing the burrows are 10 cm in diameter.
Data Records
All data records listed in this section are available at the Harvard Dataverse (Data Citation 1). Details of supplementary experimental procedures and additional materials, including videos of the three dimensional burrow structures are available from Hale et al.18. Computed tomography three-dimensional files have been converted to stacked tagged image file format (TIFF) images with associated dimension data (image width, image breadth, stack height) to enable access by multiple processing programs. There are three sets of images (n=20). Sediment core volume images for each replicate in 16- bit (Core_Volume_01_16bit to Core_Volume_20_16bit) and 8- bit (Core_Volume_01 to Core_Volume_20) and burrow volume images for each replicate (Burrow_Volume_01 to Burrow_Volume_20).
Technical Validation
The system geometry at the μ-VIS X-ray Imaging Centre, University of Southampton, is checked and validated periodically using a 3 ruby sphere reference object that has been measured using optical profilometry (Xyris 4000 CL Surface Profiler, Taicaan technologies Europe). The centroid distances (threshold independent) of these ruby spheres when measured using CT are in agreement with the optical profilometry measurements to within 0.2%. For the presented scans, measurement validation was carried out post-scan by ensuring reference distances were accurately represented in the final images (within 1%).
Usage Notes
The TIFF images provided should be imported as a three dimensional image sequence. The starting image is 0. The number of images and dimensions of each stack for each sediment core or burrow volume is provided in Tables 1,2,3 (available online only). When importing, image names should be sorted numerically.
Table 1. Stacked image data for the 16-bit quality unfiltered sediment core volumes (n=20).
| Core_ID | Scan_ID | Replicate_ID | Species_ID | dimension_X | dimension_y | dimension_Z | number_of_images | species_id_folder | stack_folder | imagestack_fileprefix | first_image | last_image |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 34 | 1 | Rep_01 | Corophium volutator | 2000 | 2000 | 2000 | 2000 | Species_Corophium | Core_Volume_01_CV01_16bit | Core_Volume_01_CV01_16bit_2000_2000_2000_ | Core_Volume_01_CV01_16bit_2000_2000_2000_0000.tif | Core_Volume_01_CV01_16bit_2000_2000_2000_1999.tif |
| 24 | 2 | Rep_01 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_02_HD01_16bit | Core_Volume_02_HD01_16bit_2000_2000_2000_ | Core_Volume_02_HD01_16bit_2000_2000_2000_0000.tif | Core_Volume_02_HD01_16bit_2000_2000_2000_1999.tif |
| 39 | 3 | Rep_01 | Mixed species | 2000 | 2000 | 2000 | 2000 | Species_Mixed | Core_Volume_03_Mx01_16bit | Core_Volume_03_Mx01_16bit_2000_2000_2000_ | Core_Volume_03_Mx01_16bit_2000_2000_2000_0000.tif | Core_Volume_03_Mx01_16bit_2000_2000_2000_1999.tif |
| 29 | 4 | Rep_01 | Hydrobia ulvae | 2000 | 2000 | 2000 | 2000 | Species_Hydrobia | Core_Volume_04_HU01_16bit | Core_Volume_04_HU01_16bit_2000_2000_2000_ | Core_Volume_04_HU01_16bit_2000_2000_2000_0000.tif | Core_Volume_04_HU01_16bit_2000_2000_2000_1999.tif |
| 40 | 5 | Rep_02 | Mixed species | 2000 | 2000 | 2000 | 2000 | Species_Mixed | Core_Volume_05_Mx02_16bit | Core_Volume_05_Mx02_16bit_2000_2000_2000_ | Core_Volume_05_Mx02_16bit_2000_2000_2000_0000.tif | Core_Volume_05_Mx02_16bit_2000_2000_2000_1999.tif |
| 23 | 6 | Rep_02 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_06_HD02_16bit | Core_Volume_06_HD02_16bit_2000_2000_2000_ | Core_Volume_06_HD02_16bit_2000_2000_2000_0000.tif | Core_Volume_06_HD02_16bit_2000_2000_2000_1999.tif |
| 36 | 7 | Rep_03 | Mixed species | 2000 | 2000 | 2000 | 2000 | Species_Mixed | Core_Volume_07_Mx03_16bit | Core_Volume_07_Mx03_16bit_2000_2000_2000_ | Core_Volume_07_Mx03_16bit_2000_2000_2000_0000.tif | Core_Volume_07_Mx03_16bit_2000_2000_2000_1999.tif |
| 31 | 8 | Rep_02 | Corophium volutator | 2000 | 2000 | 2000 | 2000 | Species_Corophium | Core_Volume_08_CV02_16bit | Core_Volume_08_CV02_16bit_2000_2000_2000_ | Core_Volume_08_CV02_16bit_2000_2000_2000_0000.tif | Core_Volume_08_CV02_16bit_2000_2000_2000_1999.tif |
| 30 | 9 | Rep_02 | Hydrobia ulvae | 2000 | 2000 | 2000 | 2000 | Species_Hydrobia | Core_Volume_09_HU02_16bit | Core_Volume_09_HU02_16bit_2000_2000_2000_ | Core_Volume_09_HU02_16bit_2000_2000_2000_0000.tif | Core_Volume_09_HU02_16bit_2000_2000_2000_1999.tif |
| 25 | 10 | Rep_03 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_10_HD03_16bit | Core_Volume_10_HD03_16bit_2000_2000_2000_ | Core_Volume_10_HD03_16bit_2000_2000_2000_0000.tif | Core_Volume_10_HD03_16bit_2000_2000_2000_1999.tif |
| 28 | 11 | Rep_03 | Hydrobia ulvae | 2000 | 2000 | 2000 | 2000 | Species_Hydrobia | Core_Volume_11_HU03_16bit | Core_Volume_11_HU03_16bit_2000_2000_2000_ | Core_Volume_11_HU03_16bit_2000_2000_2000_0000.tif | Core_Volume_11_HU03_16bit_2000_2000_2000_1999.tif |
| 38 | 12 | Rep_04 | Mixed species | 2000 | 2000 | 2000 | 2000 | Species_Mixed | Core_Volume_12_Mx04_16bit | Core_Volume_12_Mx04_16bit_2000_2000_2000_ | Core_Volume_12_Mx04_16bit_2000_2000_2000_0000.tif | Core_Volume_12_Mx04_16bit_2000_2000_2000_1999.tif |
| 22 | 13 | Rep_04 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_13_HD04_16bit | Core_Volume_13_HD04_16bit_2000_2000_2000_ | Core_Volume_13_HD04_16bit_2000_2000_2000_0000.tif | Core_Volume_13_HD04_16bit_2000_2000_2000_1999.tif |
| 27 | 14 | Rep_04 | Hydrobia ulvae | 2000 | 2000 | 2000 | 2000 | Species_Hydrobia | Core_Volume_14_HU04_16bit | Core_Volume_14_HU04_16bit_2000_2000_2000_ | Core_Volume_14_HU04_16bit_2000_2000_2000_0000.tif | Core_Volume_14_HU04_16bit_2000_2000_2000_1999.tif |
| 35 | 15 | Rep_03 | Corophium volutator | 2000 | 2000 | 2000 | 2000 | Species_Corophium | Core_Volume_15_CV03_16bit | Core_Volume_15_CV03_16bit_2000_2000_2000_ | Core_Volume_15_CV03_16bit_2000_2000_2000_0000.tif | Core_Volume_15_CV03_16bit_2000_2000_2000_1999.tif |
| 32 | 16 | Rep_04 | Corophium volutator | 2000 | 2000 | 2000 | 2000 | Species_Corophium | Core_Volume_16_CV04_16bit | Core_Volume_16_CV04_16bit_2000_2000_2000_ | Core_Volume_16_CV04_16bit_2000_2000_2000_0000.tif | Core_Volume_16_CV04_16bit_2000_2000_2000_1999.tif |
| 37 | 17 | Rep_05 | Mixed species | 2000 | 2000 | 2000 | 2000 | Species_Mixed | Core_Volume_17_Mx05_16bit | Core_Volume_17_Mx05_16bit_2000_2000_2000_ | Core_Volume_17_Mx05_16bit_2000_2000_2000_0000.tif | Core_Volume_17_Mx05_16bit_2000_2000_2000_1999.tif |
| 33 | 18 | Rep_05 | Corophium volutator | 2000 | 2000 | 2000 | 2000 | Species_Corophium | Core_Volume_18_CV05_16bit | Core_Volume_18_CV05_16bit_2000_2000_2000_ | Core_Volume_18_CV05_16bit_2000_2000_2000_0000.tif | Core_Volume_18_CV05_16bit_2000_2000_2000_1999.tif |
| 26 | 19 | Rep_05 | Hydrobia ulvae | 2000 | 2000 | 2000 | 2000 | Species_Hydrobia | Core_Volume_19_HU05_16bit | Core_Volume_19_HU05_16bit_2000_2000_2000_ | Core_Volume_19_HU05_16bit_2000_2000_2000_0000.tif | Core_Volume_19_HU05_16bit_2000_2000_2000_1999.tif |
| 21 | 20 | Rep_05 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_20_HD05_16bit | Core_Volume_20_HD05_16bit_2000_2000_2000_ | Core_Volume_20_HD05_16bit_2000_2000_2000_0000.tif | Core_Volume_20_HD05_16bit_2000_2000_2000_1999.tif |
Table 2. Stacked image data for the 8-bit quality sediment core volumes with 3D 5 pixel non-linear digital median filtered applied (n=20).
| Core_ID | Scan_ID | Replicate_ID | Species_ID | dimension_X | dimension_y | dimension_Z | number_of_images | species_id_folder | stack_folder | imagestack_fileprefix | first_image | last_image |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 34 | 1 | Rep_01 | Corophium volutator | 1230 | 1230 | 700 | 700 | Species_Corophium | Core_Volume_01_CV01 | Core_Volume_01_CV01_1230_1230_700_ | Core_Volume_01_CV01_1230_1230_700_0000.tif | Core_Volume_01_CV01_1230_1230_700_0699.tif |
| 24 | 2 | Rep_01 | Hediste diversicolor | 1701 | 1401 | 928 | 928 | Species_Hediste | Core_Volume_02_HD01 | Core_Volume_02_HD01_1701_1401_928_ | Core_Volume_02_HD01_1701_1401_928_0000.tif | Core_Volume_02_HD01_1701_1401_928_0927.tif |
| 39 | 3 | Rep_01 | Mixed species | 1401 | 1401 | 892 | 892 | Species_Mixed | Core_Volume_03_Mx01 | Core_Volume_03_Mx01_1401_1401_892_ | Core_Volume_03_Mx01_1401_1401_892_0000.tif | Core_Volume_03_Mx01_1401_1401_829_0828.tif |
| 29 | 4 | Rep_01 | Hydrobia ulvae | 1501 | 1501 | 1201 | 1201 | Species_Hydrobia | Core_Volume_04_HU01 | Core_Volume_04_HU01_1501_1501_1201_ | Core_Volume_04_HU01_1501_1501_1201_0000.tif | Core_Volume_04_HU01_1501_1501_1201_1200.tif |
| 40 | 5 | Rep_02 | Mixed species | 1207 | 1217 | 923 | 923 | Species_Mixed | Core_Volume_05_Mx02 | Core_Volume_05_Mx02_1207_1217_923_ | Core_Volume_05_Mx02_1207_1217_923_0000.tif | Core_Volume_05_Mx02_1207_1217_923_0922.tif |
| 23 | 6 | Rep_02 | Hediste diversicolor | 1601 | 1601 | 1401 | 1401 | Species_Hediste | Core_Volume_06_HD02 | Core_Volume_06_HD02_1601_1601_1401_ | Core_Volume_06_HD02_1601_1601_1401_0000.tif | Core_Volume_06_HD02_1601_1601_1401_1400.tif |
| 36 | 7 | Rep_03 | Mixed species | 1501 | 1501 | 1101 | 1101 | Species_Mixed | Core_Volume_07_Mx03 | Core_Volume_07_Mx03_1501_1501_1101_ | Core_Volume_07_Mx03_1501_1501_1101_0000.tif | Core_Volume_07_Mx03_1501_1501_1101_1100.tif |
| 31 | 8 | Rep_02 | Corophium volutator | 1501 | 1501 | 1401 | 1401 | Species_Corophium | Core_Volume_08_CV02 | Core_Volume_08_CV02_1501_1501_1401_ | Core_Volume_08_CV02_1501_1501_1401_0000.tif | Core_Volume_08_CV02_1501_1501_1401_1400.tif |
| 30 | 9 | Rep_02 | Hydrobia ulvae | 1501 | 1501 | 801 | 801 | Species_Hydrobia | Core_Volume_09_HU02 | Core_Volume_09_HU02_1501_1501_801_ | Core_Volume_09_HU02_1501_1501_801_0000.tif | Core_Volume_09_HU02_1501_1501_801_0800.tif |
| 25 | 10 | Rep_03 | Hediste diversicolor | 2000 | 2000 | 2000 | 2000 | Species_Hediste | Core_Volume_10_HD03 | Core_Volume_10_HD03_2000_2000_2000_ | Core_Volume_10_HD03_2000_2000_2000_0000.tif | Core_Volume_10_HD03_2000_2000_2000_1999.tif |
| 28 | 11 | Rep_03 | Hydrobia ulvae | 1501 | 1501 | 1001 | 1001 | Species_Hydrobia | Core_Volume_11_HU03 | Core_Volume_11_HU03_1501_1501_1001_ | Core_Volume_11_HU03_1501_1501_1001_0000.tif | Core_Volume_11_HU03_1501_1501_1001_1000.tif |
| 38 | 12 | Rep_04 | Mixed species | 1601 | 1601 | 1301 | 1301 | Species_Mixed | Core_Volume_12_Mx04 | Core_Volume_12_Mx04_1601_1601_1301_ | Core_Volume_12_Mx04_1601_1601_1301_0000.tif | Core_Volume_12_Mx04_1601_1601_1301_1300.tif |
| 22 | 13 | Rep_04 | Hediste diversicolor | 1601 | 1601 | 1301 | 1301 | Species_Hediste | Core_Volume_13_HD04 | Core_Volume_13_HD04_1601_1601_1301_ | Core_Volume_13_HD04_1601_1601_1301_0000.tif | Core_Volume_13_HD04_1601_1601_1301_1300.tif |
| 27 | 14 | Rep_04 | Hydrobia ulvae | 1601 | 1601 | 951 | 951 | Species_Hydrobia | Core_Volume_14_HU04 | Core_Volume_14_HU04_1601_1601_951_ | Core_Volume_14_HU04_1601_1601_951_0000.tif | Core_Volume_14_HU04_1601_1601_951_0950.tif |
| 35 | 15 | Rep_03 | Corophium volutator | 1601 | 1601 | 1201 | 1201 | Species_Corophium | Core_Volume_15_CV03 | Core_Volume_15_CV03_1601_1601_1201_ | Core_Volume_15_CV03_1601_1601_1201_0000.tif | Core_Volume_15_CV03_1601_1601_1201_1200.tif |
| 32 | 16 | Rep_04 | Corophium volutator | 1601 | 1601 | 1001 | 1001 | Species_Corophium | Core_Volume_16_CV04 | Core_Volume_16_CV04_1601_1601_1001_ | Core_Volume_16_CV04_1601_1601_1001_0000.tif | Core_Volume_16_CV04_1601_1601_1001_1000.tif |
| 37 | 17 | Rep_05 | Mixed species | 1601 | 1601 | 1501 | 1501 | Species_Mixed | Core_Volume_17_Mx05 | Core_Volume_17_Mx05_1601_1601_1501_ | Core_Volume_17_Mx05_1601_1601_1501_0000.tif | Core_Volume_17_Mx05_1601_1601_1501_1500.tif |
| 33 | 18 | Rep_05 | Corophium volutator | 1601 | 1601 | 1201 | 1201 | Species_Corophium | Core_Volume_18_CV05 | Core_Volume_18_CV05_1601_1601_1201_ | Core_Volume_18_CV05_1601_1601_1201_0000.tif | Core_Volume_18_CV05_1601_1601_1201_1200.tif |
| 26 | 19 | Rep_05 | Hydrobia ulvae | 1601 | 1601 | 1101 | 1101 | Species_Hydrobia | Core_Volume_19_HU05 | Core_Volume_19_HU05_1601_1601_1101_ | Core_Volume_19_HU05_1601_1601_1101_0000.tif | Core_Volume_19_HU05_1601_1601_1101_1100.tif |
| 21 | 20 | Rep_05 | Hediste diversicolor | 1601 | 1601 | 1401 | 1401 | Species_Hediste | Core_Volume_20_HD05 | Core_Volume_20_HD05_1601_1601_1401_ | Core_Volume_20_HD05_1601_1601_1401_0000.tif | Core_Volume_20_HD05_1601_1601_1401_1400.tif |
Table 3. Stacked image data for the burrow volumes (n=20).
| Core_ID | Scan_ID | Replicate_ID | Species_ID | dimension_X | dimension_y | dimension_Z | number_of_images | species_id_folder | stack_folder | imagestack_fileprefix | first_image | last_image |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 34 | 1 | Rep_01 | Corophium volutator | 1229 | 1184 | 265 | 265 | Species_Corophium | Burrow_Volume_01_CV01 | Burrow_Volume_01_CV01_1229_1184_265_ | Burrow_Volume_01_CV01_1229_1184_265_000.tif | Burrow_Volume_01_CV01_1229_1184_265_264.tif |
| 24 | 2 | Rep_01 | Hediste diversicolor | 1207 | 1219 | 909 | 909 | Species_Hediste | Burrow_Volume_02_HD01 | Burrow_Volume_02_HD01_1207_1219_909_ | Burrow_Volume_02_HD01_1207_1219_909_000.tif | Burrow_Volume_02_HD01_1207_1219_909_908.tif |
| 39 | 3 | Rep_01 | Mixed species | 1219 | 1236 | 872 | 872 | Species_Mixed | Burrow_Volume_03_Mx01 | Burrow_Volume_03_Mx01_1219_1236_872_ | Burrow_Volume_03_Mx01_1219_1236_872_000.tif | Burrow_Volume_03_Mx01_1219_1236_872_871.tif |
| 29 | 4 | Rep_01 | Hydrobia ulvae | 1220 | 1133 | 411 | 411 | Species_Hydrobia | Burrow_Volume_04_HU01 | Burrow_Volume_04_HU01_1220_1133_411_ | Burrow_Volume_04_HU01_1220_1133_411_000.tif | Burrow_Volume_04_HU01_1220_1133_411_410.tif |
| 40 | 5 | Rep_02 | Mixed species | 1205 | 1215 | 900 | 900 | Species_Mixed | Burrow_Volume_05_Mx02 | Burrow_Volume_05_Mx02_1205_1215_900_ | Burrow_Volume_05_Mx02_1205_1215_900_000.tif | Burrow_Volume_05_Mx02_1205_1215_900_899.tif |
| 23 | 6 | Rep_02 | Hediste diversicolor | 1228 | 1224 | 906 | 906 | Species_Hediste | Burrow_Volume_06_HD02 | Burrow_Volume_06_HD02_1228_1224_906_ | Burrow_Volume_06_HD02_1228_1224_906_000.tif | Burrow_Volume_06_HD02_1228_1224_906_905.tif |
| 36 | 7 | Rep_03 | Mixed species | 1228 | 1223 | 898 | 898 | Species_Mixed | Burrow_Volume_07_Mx03 | Burrow_Volume_07_Mx03_1228_1223_898_ | Burrow_Volume_07_Mx03_1228_1223_898_000.tif | Burrow_Volume_07_Mx03_1228_1223_898_897.tif |
| 31 | 8 | Rep_02 | Corophium volutator | 1123 | 1121 | 220 | 220 | Species_Corophium | Burrow_Volume_08_CV02 | Burrow_Volume_08_CV02_1123_1121_220_ | Burrow_Volume_08_CV02_1123_1121_220_000.tif | Burrow_Volume_08_CV02_1123_1121_220_219.tif |
| 30 | 9 | Rep_02 | Hydrobia ulvae | 900 | 1212 | 313 | 313 | Species_Hydrobia | Burrow_Volume_09_HU02 | Burrow_Volume_09_HU02_900_1212_313_ | Burrow_Volume_09_HU02_900_1212_313_000.tif | Burrow_Volume_09_HU02_900_1212_313_312.tif |
| 25 | 10 | Rep_03 | Hediste diversicolor | 1220 | 1235 | 854 | 854 | Species_Hediste | Burrow_Volume_10_HD03 | Burrow_Volume_10_HD03_1220_1235_854_ | Burrow_Volume_10_HD03_1220_1235_854_000.tif | Burrow_Volume_10_HD03_1220_1235_854_853.tif |
| 28 | 11 | Rep_03 | Hydrobia ulvae | 1129 | 1188 | 332 | 332 | Species_Hydrobia | Burrow_Volume_11_HU03 | Burrow_Volume_11_HU03_1129_1188_332_ | Burrow_Volume_11_HU03_1129_1188_332_000.tif | Burrow_Volume_11_HU03_1129_1188_332_331.tif |
| 38 | 12 | Rep_04 | Mixed species | 1228 | 1223 | 879 | 879 | Species_Mixed | Burrow_Volume_12_Mx04 | Burrow_Volume_12_Mx04_1228_1223_879_ | Burrow_Volume_12_Mx04_1228_1223_879_000.tif | Burrow_Volume_12_Mx04_1228_1223_879_878.tif |
| 22 | 13 | Rep_04 | Hediste diversicolor | 1236 | 1222 | 943 | 943 | Species_Hediste | Burrow_Volume_13_HD04 | Burrow_Volume_13_HD04_1236_1222_943_ | Burrow_Volume_13_HD04_1236_1222_943_000.tif | Burrow_Volume_13_HD04_1236_1222_943_942.tif |
| 27 | 14 | Rep_04 | Hydrobia ulvae | 1204 | 1151 | 305 | 305 | Species_Hydrobia | Burrow_Volume_14_HU04 | Burrow_Volume_14_HU04_1204_1151_305_ | Burrow_Volume_14_HU04_1204_1151_305_000.tif | Burrow_Volume_14_HU04_1204_1151_305_304.tif |
| 35 | 15 | Rep_03 | Corophium volutator | 1204 | 1191 | 252 | 252 | Species_Corophium | Burrow_Volume_15_CV03 | Burrow_Volume_15_CV03_1204_1191_252_ | Burrow_Volume_15_CV03_1204_1191_252_000.tif | Burrow_Volume_15_CV03_1204_1191_252_251.tif |
| 32 | 16 | Rep_04 | Corophium volutator | 1201 | 1168 | 274 | 274 | Species_Corophium | Burrow_Volume_16_CV04 | Burrow_Volume_16_CV04_1201_1168_274_ | Burrow_Volume_16_CV04_1201_1168_274_000.tif | Burrow_Volume_16_CV04_1201_1168_274_273.tif |
| 37 | 17 | Rep_05 | Mixed species | 1228 | 1244 | 981 | 981 | Species_Mixed | Burrow_Volume_17_Mx05 | Burrow_Volume_17_Mx05_1228_1244_981_ | Burrow_Volume_17_Mx05_1228_1244_981_000.tif | Burrow_Volume_17_Mx05_1228_1244_981_980.tif |
| 33 | 18 | Rep_05 | Corophium volutator | 1179 | 1195 | 332 | 332 | Species_Corophium | Burrow_Volume_18_CV05 | Burrow_Volume_18_CV05_1179_1195_332_ | Burrow_Volume_18_CV05_1179_1195_332_000.tif | Burrow_Volume_18_CV05_1179_1195_332_331.tif |
| 26 | 19 | Rep_05 | Hydrobia ulvae | 1207 | 1140 | 335 | 335 | Species_Hydrobia | Burrow_Volume_19_HU05 | Burrow_Volume_19_HU05_1207_1140_335_ | Burrow_Volume_19_HU05_1207_1140_335_000.tif | Burrow_Volume_19_HU05_1207_1140_335_334.tif |
| 21 | 20 | Rep_05 | Hediste diversicolor | 1235 | 1218 | 859 | 859 | Species_Hediste | Burrow_Volume_20_HD05 | Burrow_Volume_20_HD05_1235_1218_859_ | Burrow_Volume_20_HD05_1235_1218_859_000.tif | Burrow_Volume_20_HD05_1235_1218_859_858.tif |
There are no limitations on data use.
Additional Information
Tables 1,2,3 are only available in the online version of this paper.
How to cite this article: Hale, R. et al. High-resolution computed tomography reconstructions of invertebrate burrow systems. Sci. Data 2:150052 doi: 10.1038/sdata.2015.52 (2015).
Supplementary Material
Acknowledgments
This work was undertaken while R.H. was in receipt of a University of East Anglia Funded PhD studentship. R.H. thanks the University of East Anglia for additional financial support for travel and subsistence while in Southampton. Financial support for μ-CT was provided by the University of Southampton. We thank Dr Louise Darroch at the British Oceanographic Data Centre for her assistance with metadata organisation. We thank the Harvard Dataverse for hosting our data and Sonia Barbosa, Kevin Condon and Dwayne Liburd for their technical assistance. This work was partially supported by NERC grant NE/K001906/1.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Hale R., Boardman R., Mavrogordato M. N., Sinclair I., Tolhurst T. J., Solan M. 2015. Harvard Dataverse. http://dx.doi.org/10.7910/DVN/4XNRE3 [DOI] [PMC free article] [PubMed]
References
- Buatois L. A. et al. Colonization of the brackish-water systems through time: Evidence from the trace-fossil record. Palaois 20, 321–347 (2005). [Google Scholar]
- Martin R. E., Quigg A. & Podkovyrov V. Marine biodiversification in response to evolving phytoplankton stoichiometry. Palaeogeogr. Palaeoclimatol. Palaeoecol 258, 277–291 (2008). [Google Scholar]
- Seilacher A. & Pflüger F. in Biostabilization of Sediments (eds Krumbein W. E., Peterson D. M. & Stal L. J. ) 97–105 (Bibliotheks-und Informationssystem der Carl von Ossietzky Universität Odenburg, 1994). [Google Scholar]
- Germano J. D. Sediment profile imaging: a rapid seafloor impact assessment tool for oil spills. Arctic and Marine Oil Spill Technical Seminar 18, 1271–1279 (1995). [Google Scholar]
- Solan M. & Kennedy R. Observation and quantification or in-situ animal sediment relations using time-lapse sediment profile imagery (t-SPI). Mar. Ecol. Prog, Ser. 228, 179–191 (2002). [Google Scholar]
- Dorgan K. M., Jumars P. A., Johnson B., Boudreau B. P. & Landis E. Burrow extension by crack propagation. Nature 433, 475 (2005). [DOI] [PubMed] [Google Scholar]
- Griffis R. B. & Suchanek T. H. A model of burrow architecture and trophic modes in thalassinidean shrimp (Decapoda: Thalassinidea). Mar. Ecol. Prog. Ser. 79, 171–183 (1991). [Google Scholar]
- Nickell L. A. & Atkinson R. J. A. Functional morphology of burrows and trophic modes of three thalassinidean shrimp species, and a new approach to the classification of thalassinidean burrow morphology. Mar. Ecol. Prog. Ser. 128, 181–197 (1995). [Google Scholar]
- Downie H. et al. Transparent soil for imaging the rhizosphere. PLoS ONE 7, e44276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel O., Kretzschmar A., Capowiez Y., Kohli L. & Zeyer. J. Computer-assisted tomography of macroporosity and its application to study the activity of the earthworm Aporrectodea nocturna . Eur. J. Soil Sci. 48, 727–737 (1997). [Google Scholar]
- Joschko M. et al. A nondestructive method for the morphological assessment of earthworm burrow systems in 3 dimensions by X-ray computed-tomography. Biol. Fert. Soils 11, 88–92 (1991). [Google Scholar]
- Mazik K., Curtis N., Fagan M. J., Taft S. & Elliott M. Accurate quantification of the influence of benthic macro- and meio-fauna on the geometric properties of estuarine muds by micro computer tomography. J. Exp. Mar. Biol. Ecol 354, 192–201 (2008). [Google Scholar]
- Perez K. T. et al. Application of computer-aided tomography (CT) to the study of estuarine benthic communities. Ecol. Appl. 9, 1050–1058 (1999). [Google Scholar]
- Weiddleder R. & Pittet M. J. Imaging in the era of molecular oncology. Nature 452, 580–589 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boistel R. et al. The future of three-dimensional microscopic imaging in marine biology. Mar. Ecol. 32, 1–15 (2011). [Google Scholar]
- Dinley J. et al. Micro-computed X-ray tomography: a new non-destructive method of assessing sectional, fly-through and 3D imaging of a soft-bodied marine worm. J. Microsc. 238, 123–133 (2010). [DOI] [PubMed] [Google Scholar]
- Rosenberg R., Gremare A., Duchene J. C., Davey E. & Frank M. Visualization and quantification of marine benthic biogenic structures and particle transport utilizing computer-aided tomography. Mar. Ecol. Prog. Ser. 363, 171–182 (2008). [Google Scholar]
- Hale R., Mavrogordato M. N., Tolhurst T. J. & Solan M. Characterizations of how species mediate ecosystem properties require more comprehensive functional effect descriptors. Sci. Rep. 4, e6463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R., Davey E., Gunnarsson J., Norling K. & Frank M. Application of computer-aided tomography to visualize and quantify biogenic structures in marine sediments. Mar. Ecol. Prog. Ser. 331, 23–34 (2007). [Google Scholar]
- Brimhall G. H. et al. Deformational mass transport and invasive processes in soil evolution. Science 255, 695–702 (1992). [DOI] [PubMed] [Google Scholar]
- La Croix A., Gingras M., Dashtgard S. E. & Pemberton G. Computer modelling bioturbation: The creation of porous and permeable fluid-flow pathways. AAPG Bull. 96, 545–556 (2012). [Google Scholar]
- Quinn G. W., Quinn J. B., Mecholsky J. J. & Quinn G. D. in Mechanical Properties and Performance of Engineering Ceramics and Composites: Ceramic Engineering and Science Proceedings (ed. Lara-Curzio E. ) 26, Number 2, 77–84 (John Wiley & Sons, Inc., 2005). [Google Scholar]
- Bebber D. P., Hynes J., Darrah P. R., Boddy L. & Fricker M. D. Biological solutions to transport network design. Proc. R. Soc. B 274, 2307–2315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer S. et al. Recovering complete plant root system architectures from soil via X-ray μ-computed tomography. Plant Methods 9, 8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E. & Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198 (2009). [DOI] [PubMed] [Google Scholar]
- Ramnani N., Behrens T. E., Penny W. & Matthews P. M. New approaches for exploring anatomical and functional connectivity in the human brain. Biol. Psychiat. 56, 613–619 (2004). [DOI] [PubMed] [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hale R., Boardman R., Mavrogordato M. N., Sinclair I., Tolhurst T. J., Solan M. 2015. Harvard Dataverse. http://dx.doi.org/10.7910/DVN/4XNRE3 [DOI] [PMC free article] [PubMed]


