Abstract
PKMζ is generated via an alternative transcriptional start site in the atypical protein kinase C (PKC)ζ isoform, which removes N-terminal regulatory elements, including the inhibitory pseudosubstrate domain, consequently rendering the kinase constitutively active. Persistent PKMζ activity has been proposed as a molecular mechanism for the long-term maintenance of synaptic plasticity underlying some forms of memory. Many studies supporting a role for PKMζ in synaptic plasticity and memory have relied on the PKCζ pseudosubstrate-derived ζ-inhibitory peptide (ZIP). However, recent studies have demonstrated that ZIP-induced impairments to synaptic plasticity and memory occur even in the absence of PKCζ, suggesting that ZIP exerts its actions via additional cellular targets. In this study, we demonstrated that ZIP interacts with conventional and novel PKC, in addition to atypical PKC isoforms. Moreover, when brain abundance of each PKC isoform and affinity for ZIP are taken into account, the signaling capacity of ZIP-responsive pools of conventional and novel PKCs may match or exceed that for atypical PKCs. Pseudosubstrate-derived peptides, like ZIP, are thought to exert their cellular action primarily by inhibiting PKC catalytic activity; however, the ZIP-sensitive catalytic core of PKC is known to participate in the enzyme’s subcellular targeting, suggesting an additional mode of ZIP action. Indeed, we have demonstrated that ZIP potently disrupts PKCα interaction with the PKC-targeting protein A-kinase anchoring protein (AKAP) 79 and interferes with ionomycin-induced translocation of conventional PKC to the plasma membrane. Thus, ZIP exhibits broad-spectrum action toward the PKC family of enzymes, and this action may contribute to its unique ability to impair memory.
Introduction
Intramolecular and intermolecular protein interactions control the function of individual signaling enzymes and their recruitment into larger signaling complexes that define the spatiotemporal domains in which cellular signaling operates. Relatively short peptides, corresponding to functional interaction domains of proteins, have been long recognized as useful tools for probing how specific interaction sites shape enzyme function individually or within the context of larger signaling complexes (Souroujon and Mochly-Rosen, 1998). In particular, peptides derived from protein kinase C (PKC) have been exploited to explore mechanisms related to PKC function (House and Kemp, 1987; Souroujon and Mochly-Rosen, 1998; Churchill et al., 2009).
The PKC family of kinases includes 10 isoforms that possess a highly conserved C-terminal catalytic domain, and these isoforms are subdivided into three subfamilies [conventional (cPKC), novel (nPKC), and atypical (aPKC)] based on their second-messenger activator requirements, which arise from structural variations occurring primarily in the N-terminal regulatory region (Steinberg, 2008). The pseudosubstrate domain of each isoform is embedded in this N-terminal regulatory region and holds the kinase in an inactive state until an appropriate activation signal is received, which relieves this pseudosubstrate-mediated autoinhibition (Steinberg, 2008). Peptides derived from this region were initially used to demonstrate the autoinhibitory role of this domain toward PKC activity and made them attractive candidates for use as PKC inhibitors within a cellular context (House and Kemp, 1987; Eichholtz et al., 1993). Because pseudosubstrate domains are naturally occurring and provide large interfaces for multiple points of contact, they are often considered the most specific inhibitors available for a given kinase (Churchill et al., 2009).
Cell-penetrating myristoylated (myr) peptides derived from the pseudosubstrate domain of the aPKC isoform ζ, denoted ZIP, have been extensively used to support the role of PKMζ, the constitutively active splice variant of PKCζ, in synaptic plasticity and memory (Ling et al., 2002; Pastalkova et al., 2006; Kwapis and Helmstetter, 2014); however, recent studies using mice deficient in PKCζ/PKMζ suggest that these isoforms are dispensable for synaptic plasticity in the form of long-term potentiation (LTP) and memory (Lee et al., 2013; Volk et al., 2013). Yet LTP maintenance and several behavioral performance measures used to assess memory are still impaired by ZIP in these mice, suggesting that ZIP exerts its actions via additional cellular targets (Lee et al., 2013; Volk et al., 2013). Because aPKC isoform ι/λ and PKCζ have identical pseudosubstrates, PKCι/λ has been suggested to be a primary candidate for conferring ZIP sensitivity in the absence of PKCζ/PKMζ (Ren et al., 2013; Kwapis and Helmstetter, 2014). However, all PKC pseudosubstrates possess several invariant residues and several well conserved residues (Pears and Parker, 1991; Nishikawa et al., 1997; Wang et al., 2012). In addition, PKC isoforms have overlapping substrate specificities, particularly for substrates derived from modification of their pseudosubstrate domains (Nishikawa et al., 1997), collectively suggesting that ZIP may harbor affinity for all PKC isoforms. Indeed, the PKC targeting protein A-kinase anchoring protein (AKAP) 79 binds the catalytic core PKC of all isoforms by a pseudosubstrate-like mechanism (Faux et al., 1999). In agreement with this hypothesis, we demonstrate that ZIP promiscuously binds all PKC isoforms and interferes with PKC targeting and translocation.
Materials and Methods
ZIP Interaction Assays and PKC Abundance from the Rat Brain.
Adult Sprague-Dawley rats (Harlan, Indianapolis, IN) were sacrificed by an overdose with pentobarbital according to protocols approved by the University of Tennessee Health Science Center Institutional Animal Care and Use Committee. Brains were rapidly removed; in some cases, hippocampi were rapidly microdissected. Once isolated, the tissue was immediately frozen in liquid nitrogen. Frozen brains or hippocampi were pulverized and then Dounce-homogenized in ice-cold lysis buffer [150 mM NaCl, 10 mM HEPES, 5 mM EGTA, 5 mM EDTA, 1% Triton-X-100, and protease inhibitors (Sigma-Aldrich, St. Louis, MO), pH 7.4]. The lysate was clarified by centrifugation at 16,000g for 10 minutes. Protein concentration for the resulting supernatant was determined by Bradford assay (Bio-Rad, Hercules, CA), and the extract was subsequently diluted to 1 mg/ml for all assays. For the ZIP interaction (i.e., pull-down) assays, rat brain, or hippocampal extract (500 μg protein) was combined with biotinylated ZIP (5 μg; Tocris, Bristol, UK) or water control for 2 hours at 4°C with nutation. Streptavidin-coated MyOne-T1 Dynabeads (20 μl packed; Life Technologies, Carlsbad, CA) were added to each sample and incubated for 30 minutes at 4°C with nutation. After four washes with PKC binding solution [150 mM NaCl, 10 mM HEPES, 5 mM EGTA, 5 mM EDTA, 0.1% Tween-20, and protease inhibitors (Sigma-Aldrich) (pH 7.40)], complexes were eluted in 2× Laemmli buffer, and samples were resolved on 4–20% SDS-PAGE gels and transferred to nitrocellulose. For these interaction assays, 5 μg (representing 1.0%) or 1 μg (representing 0.2%) of whole-brain or hippocampal extract, respectively, were run alongside samples and served as a basis for quantitative comparison. For determination of PKC abundance from rat whole brains or hippocampal extracts, 10–20 ng human recombinant PKC isoforms (Life Technologies) were loaded alongside 5-µg extracts onto gels and transferred to nitrocellulose.
Western Blotting.
After sample separation by SDS-PAGE and transfer to nitrocellulose, blots were blocked (1 hour) in phosphate-buffered saline (PBS) + 3% nonfat dry milk + 0.1% Tween-20) and then probed with isoform-specific PKC antibodies. Horseradish peroxidase–conjugated goat-anti rabbit or mouse secondary antibodies (1:10,000 dilution; Millipore, Billerica, MA) were used for detection. Signals were visualized by enhanced chemiluminescence (ThermoFisher Scientific, Waltham, MA). Data were digitally acquired and quantified using a Bio-Rad XRS chemiluminescence documentation system and Quantity One software. The following primary antibodies (and their respective dilutions) were used: mouse monoclonals (BD Bioscience, San Jose, CA) to PKCα (1:1000), PKCβ (1:1000), PKCδ (1:1000), PKCε (1:200–1:1000), PKCθ (1:200–1:1000), and PKCι/λ (1:1000) and rabbit polyclonals (Santa Cruz Biotechnology; Santa Cruz, CA) to PKCγ (1:1000), PKCη (1:200–1:1000), and PKCζ C-terminal (1:200).
PKC Binding and Competition Assays.
Strepavidin-coated dynabeads were incubated with 5 μg biotinylated peptides for 30 minutes at 4°C in 500 µl of PKC binding solution + 0.1% bovine serum albumin (BSA; Sigma-Aldrich). The following biotinylated peptides were used: ZIP (Tocris), AKAP79(31-52), and HT31 (Biomolecules Midwest Inc., Waterloo, IL). Beads were washed four times to remove unbound peptide then incubated with 200 ng of human recombinant PKC (Life Technologies) or human recombinant glutathione transferase/His-tagged-PKMζ (ProQinase, Freiburg, DE) for 2 hours at 4°C. After four washes to remove unbound PKC, complexes were eluted in 2× Laemmli buffer, and samples were separated by SDS-PAGE and analyzed by Western blotting as above. For these experiments, a rabbit polyclonal anti–pan-cPKC antibody (Millipore; 1:1000) was used for the purified conventional isoforms. All other experiments used isoform specific antibodies as above. Bead alone signals were used to correct for nonspecific binding. However, PKCδ and PKCη exhibited a relatively high degree of binding to the beads in the absence of any peptide. As such, these isoforms were corrected for by the signal in the presence of the HT31 peptide. For competition assays, myr-ZIP or myr-scrambled ZIP (SCR; Anaspec, Fremont, CA) were added just prior to addition of PKC and were included in the subsequent washes.
PKC Activity Assays.
In vitro PKCα and PKMζ (20 ng each) kinase reactions (50 μl total reaction volume) were performed in 10 mM HEPES, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/ml BSA and either 10 μM myelin basic protein (residues 4-14) for PKCα or 10 μM PKCε substrate peptide for PKMζ (Anaspec). For PKCα, 0.1 mM CaCl2, 100 μg/ml phosphatidylserine, and 20 μg/ml diacylglycerol were included in the reaction buffer. Myr-ZIP and myr-SCR were added to the reactions 5 minutes before initiation. Reactions were initiated by the addition of ATP to a 10 μM final concentration. After the addition of ATP, samples were incubated 5 minutes (PKCα) or 30 minutes (PKMζ) at 30°C with shaking. Reactions were terminated by the addition of 50 μl Kinase-Glo Plus reagent (Promega, Madison, WI) and allowed to incubate for 10 minutes on a rocker at 20°C. Luminescence was acquired using Bio-Rad XRS chemiluminescence documentation system and Quantity One software. Percent activity was quantified as [(signal without enzyme − signal with enzyme in absence or presence of myr-peptide) / (signal without enzyme − signal with enzyme in absence of myr-peptide)] × 100.
PKC Translocation.
HEK293 cells (American Type Culture Collection, Manassas, VA) were grown on 15-mm round glass coverslips in standard culture media (Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum + penicillin/streptomycin) (Life Technologies), and transfected with AKAP79–green fluorescent protein (GFP) using the Ca2+-phosphate precipitation method. At 1-day post-transfection, cells were washed with PBS (Life Technologies) and then washed in a standard extracellular buffer (150 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 11.1 mM glucose, 10 mM HEPES, pH 7.40). Cells were then washed with buffer alone or with buffer containing either 5 μM myr-ZIP or 5 μM myr-SCR peptides for 10 minutes. After this pretreatment, cells were treated with buffer ± ionomycin (1 μM, 3 minutes; LC laboratories, Woburn, MA) in the continued absence or presence of myr-ZIP or myr-SCR peptides. Cells were rapidly fixed with paraformaldehyde (3.7%, 5 minutes), then washed with PBS, and then permeabilized (PBS + 0.3% Triton-X-100; 5 minutes). Cells were washed with PBS and then incubated with rabbit anti-cPKC (1:500) for 1–2 hours in (PBS + 0.1% Tween-20 and 3% BSA). Cells were subsequently washed and then incubated in AlexaFluor594-coupled goat-anti rabbit IgG antibody (1:500; Life Technologies) for 1 hour and then washed and mounted onto glass slides with Prolong Gold antifade reagent (Life Technologies). Confocal sections from the stained cells were acquired using an Olympus (Center Valley, PA) FV-1000 confocal system assembled on an Olympus IX81 microscope using the 488- and 561-nm laser lines. Images (512 × 512) were acquired using a 60× oil-immersion objective and at 6× digital zoom. No adjustments were made to the settings, and all images shown represent the raw data. Line-scan intensity profiles for green and red channels corresponding to AKAP79-GFP and cPKC were obtained for each cell. The intensity profile for AKAP79-GFP was used to aid in defining the boundary for a ∼2-μm (21 pixels)–wide membrane-localized region for each cell. Sections of the line-intensity profile corresponding to regions outside each cell were used to determine background. After background subtraction, the membrane-to-cytosol ratio for cPKC was derived from the average intensity in these areas.
Statistical Analysis.
Data were subjected to one-way analysis of variance (ANOVA), followed by Student’s t test except for dose-dependent comparisons between myr-ZIP and myr-SCR, which were subjected to two-way ANOVA. Statistical significance is reported as P < 0.05 or P < 0.01.
Results
ZIP Binds to Multiple PKC Isoforms in the Brain.
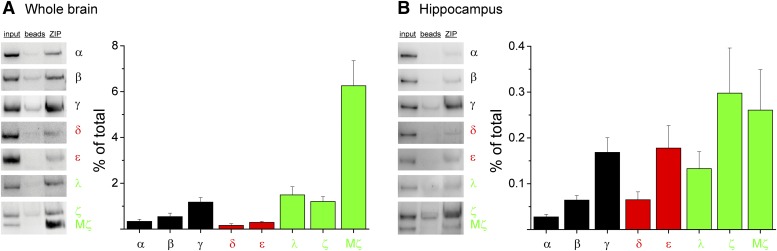
The cell-permeable form of ZIP, myr-ZIP, competitively inhibits PKMζ with IC50 76 nM–2 μM, depending on substrate affinity and concentration, as well as kinase concentration (Lee et al., 2013; Volk et al., 2013; Yao et al., 2013). Myr-ZIP is frequently used in the range of 1–10 μM and is thought to have access to all cellular compartments when applied experimentally to brain tissue; however, the degree to which ZIP binds any PKC isoform in the brain when used in this concentration range is unknown. To address this issue, we performed indirect affinity interaction assays in which rat brain extract was first incubated with a biotinylated form of myr-ZIP (∼4.6 μM) and subsequently precipitated with streptavidin-coupled beads. Samples were subjected to semiquantitative Western blot analysis using a panel of PKC isoform-specific antibodies and compared with input to derive the relative extent to which each isoform bound to ZIP. We found that ZIP-containing precipitates were specifically enriched, compared with beads-alone controls, for all PKC isoforms (Fig. 1), except for the nPKC isoforms θ and η (unpublished data). PKCθ and PKCη have been difficult to reliably detect in the brain (Naik et al., 2000; Lee et al., 2013). Notably, PKMζ coprecipitated with ZIP to the greatest extent, ∼4- to 6-fold greater than PKCζ and PKCι/λ (PKMζ: 6.3 ± 1.1% of total; PKCζ: 1.5 ± 0.4% of total; PKCι/λ: 1.1 ± 0.2% of total). This might be expected given that ZIP must compete with the endogenous pseudosubstrate for the catalytic core in full-length aPKC isoforms, whereas the corresponding region is absent in PKMζ. Overall, ZIP exhibited preferential association with aPKC isoforms relative to cPKC and nPKC isoforms; however, ZIP associated with PKCγ (PKCγ: 1.2 ± 0.2% of total) to a similar extent as the full-length atypical isoforms, suggesting that other isoforms may represent relevant ZIP targets.
Fig. 1.
ZIP binds to multiple PKC isoforms in rat brain. (A) Whole-brain or (B) hippocampal extracts (500 µg protein) were incubated with biotinylated ZIP (5 µg). Streptavidin-coated beads were added to capture biotinylated ZIP-containing complexes, which were separated by SDS-PAGE and subjected to Western blotting using PKC isoform-specific antibodies. Left panels: Representative blots for each isoform are shown. Input lane was loaded with (A) 5 µg or (B) 1 µg of extract. Beads represent bead-alone control. Right panels: Summary graphs quantifying the fraction of each PKC isoform bound to ZIP corrected for nonspecific binding to beads and expressed as percent of total (relative to input). Data are shown as mean + S.E.M., n = 3–8 for each isoform. cPKCs are depicted in black, nPKCs in red, and aPKCs in green.
Each PKC isoform exhibits a unique expression profile across various brain regions (Naik et al., 2000). As ZIP action has been most extensively examined within hippocampus, we performed a parallel analysis of the ability ZIP to interact with each isoform in hippocampal extract. As in whole brain, ZIP bound nearly all PKC isoforms; however, the proportion of each isoform pulled down by ZIP was lower in hippocampal extract (Fig. 1B). Notably, the preferential association of aPKCs, including PKMζ, to ZIP was markedly less pronounced in hippocampus compared with whole brain. This further underscores the promiscuous association of ZIP across the family of PKC isoforms.
Atypical PKCs Exhibit Low Abundance in the Brain.
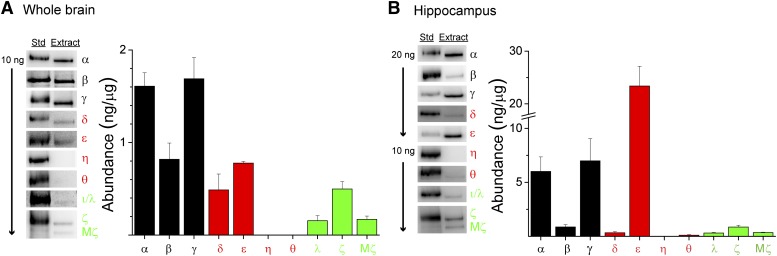
The ability of ZIP to pull down various PKC isoforms from brain extract will be related to the affinity of each isoform for ZIP as well as the abundance of each isoform. Thus, semiquantitative Western blotting was used to determine the abundance of each PKC isoform present in the rat brain extract by comparison against known quantities of recombinant PKC isoforms. cPKC isoforms α, β, and γ had the highest protein expression in whole brain, nPKCs δ and ϵ were present at intermediate levels, and the aPKC isoforms had the lowest protein expression (Fig. 2A). As above, PKCη and PKCθ were not detected in the extracts. Importantly, cPKC isoforms were ∼10-fold more abundant than PKMζ, as well as ∼2- and 10-fold greater than PKCζ and PKCι/λ, respectively. In parallel examination of hippocampal PKC abundance, each PKC isoform (except for PKCδ) was present in substantially greater amounts than in whole brain (Fig. 2B), generally consistent with a previous estimate (Naik et al., 2000). Indeed, even PKCθ was detected at very low levels. Notably, PKCα and PKCγ exhibited ∼4× and PKCϵ exhibited ∼30× greater expression in hippocampus compared with whole brain. In contrast, aPKCs exhibited a more modest ∼2× elevation in expression. Hence, the relative abundance of aPKCs compared with other subfamilies is substantially reduced in hippocampus compared with whole brain. Thus, when considered with the previous pull-down measures, the size of ZIP-interacting pools of cPKC may be similar to, and possibly exceed, that for aPKCs (including PKMζ), the presumptive primary target of ZIP. Moreover, these data suggest that competition resulting from the prevalence of other isoforms contributes to the reduced ability of ZIP to interact with each PKC isoform in hippocampus relative to whole brain (Fig. 1).
Fig. 2.
Atypical PKCs exhibit low abundance in the brain. Relative abundance of PKC isoforms in rat (A) whole-brain or (B) hippocampal extracts were determined by immunoblot. Density of immunoreactive bands at the appropriate molecular weight from brain extract (5 µg) was quantified and compared with purified recombinant PKC input (10 or 20 ng, as indicated). Left panels: Representative blots for each PKC isoform are shown. Right panels: Graphical summary of the relative abundance for each PKC isoform. Data are shown as mean + S.E.M. (n = 3–9). cPKCs are depicted in black, nPKCs in red, and aPKCs in green.
Relative Affinity of ZIP Interactions with PKC In Vitro.
The effective IC50 of ZIP for PKMζ determined by kinase assay is dependent on kinase concentration (Yao et al., 2013). Thus, to establish the relative affinity of various PKC isoforms for ZIP, we performed direct affinity interaction assays by first coupling streptavidin-coated beads with a fixed amount of biotinylated ZIP (5 μg; ∼4.6 μM) and then incubating the ZIP-charged beads with fixed amount (200 ng) of purified recombinant PKC (in the absence of any activators). In contrast to interaction assays using extracts (Fig. 1), where levels of PKC naturally vary widely (Fig. 2), the concentration of each isoform was in a relatively tight range (∼4.6–6.0 nM). A biotinylated peptide corresponding to the PKC binding domain of AKAP79 (residues 31–52) that is known to bind all PKC isoforms was used as a positive control for this assay (Faux et al., 1999), whereas biotinylated-HT31, a peptide of similar size derived from the PKA binding domain of an unrelated AKAP was used as a negative control. As expected, AKAP79(31–52) interacted with all PKC isoforms, whereas HT31 had negligible affinity for any of the PKC isoforms (Fig. 3). ZIP bound to all isoforms except PKCη (Fig. 3), which was similar to the results of the ZIP pull-down assays of PKC from brain (Fig. 1). Although PKCθ was detected only in limited amounts from brain tissue (Fig. 2), it interacts with ZIP when tested in vitro (Fig. 3). Notably, AKAP79(31–52) interacts with all PKC isoforms to a similar or even a greater extent than ZIP (Fig. 3). Unexpectedly, PKCα harbored an apparent affinity for ZIP that matched that of PKCζ and PKMζ. This discrepancy with our assays performed using brain tissue, is likely due not only to the lack of competing PKC isoforms but also to the lack of other PKC- and/or ZIP-interacting proteins that may be endogenously expressed in brain. Given its prevalence in brain, PKCα may represent an additional target by which ZIP exerts its action.
Fig. 3.
Relative affinity of PKC isoforms for ZIP and AKAP79(31-52). In vitro interaction of individual recombinant PKC isoforms (200 ng) with the indicated biotinylated peptides (5 μg) precoupled to streptavidin-coated beads. After PKC incubation and washing, complexes were resolved by SDS-PAGE, and immunoblots were performed using PKC isoform-specific antibodies. Left: Representative blots for each isoform are shown aligned with (Right) their corresponding bars in the graphical summary. Density of immunoreactive bands were corrected for nonspecific binding to beads (or HT31 peptide for δ and η) and normalized to input (50 ng, representing 25% of total). Data are shown as mean + S.E.M. (n = 3–4). cPKCs are depicted in black, nPKCs in red, and aPKCs in green.
ZIP Inhibits Atypical and Conventional PKC.
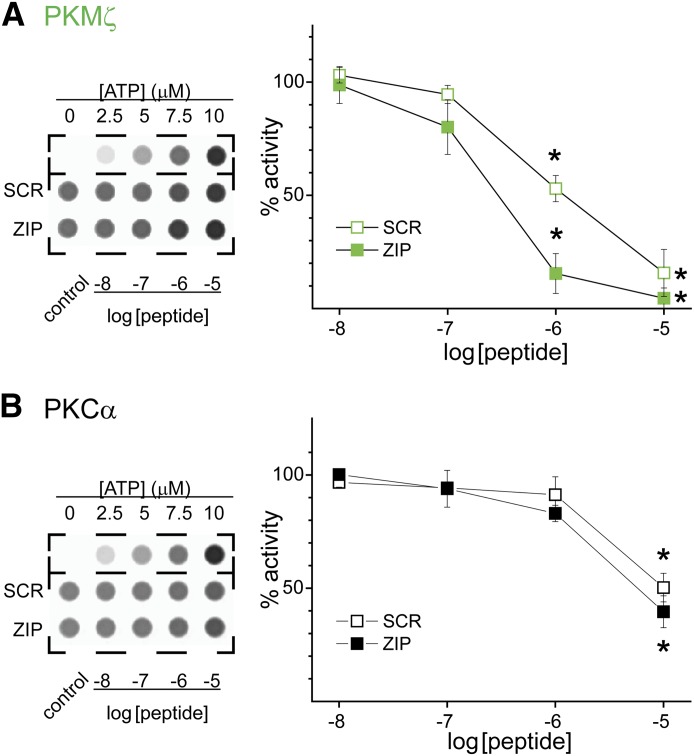
Because PKCα interacts with ZIP to a similar extent as PKMζ, and ZIP inhibits PKMζ, we sought to determine whether ZIP likewise inhibits PKCα activity. Thus, we tested myr-ZIP and its scrambled control peptide myr-SCR for their ability to alter PKC activity. We first assessed the ability of these peptides to inhibit PKMζ to validate our approach. PKMζ activity was blocked completely by 10 µM myr-ZIP with an IC50 of 0.27 µM. Myr-SCR was also quite effective at inhibiting PKMζ with an IC50 = 1.29 µM (Fig. 4A). These results are in agreement with previous studies suggesting that ZIP is ∼3- to 7-fold more potent than its scrambled control (Yao et al., 2008; Lee et al., 2013). Even though PKCα bound to ZIP to a similar extent as PKMζ, PKCα was incompletely inhibited by myr-ZIP (myr-ZIP: 60.4 ± 7.0% inhibition, P < 0.01 compared with control; Fig. 4B). Moreover, myr-SCR inhibited PKCα to a similar extent (myr-SCR: 49.7 ± 6.3% inhibition; P < 0.01 compared with control; Fig. 4B), consistent with other reports that this scrambled control peptide may exhibit actions similar to ZIP (Volk et al., 2013). Although PKCα activity has been previously reported to be less sensitive to ZIP than PKMζ (Ling et al., 2002), it must be emphasized that some degree of PKCα inhibition occurs in response to ZIP within its useful concentration range.
Fig. 4.
ZIP inhibits the activity of atypical and conventional PKC. In vitro kinase activity assays were performed for (A) PKMζ and (B) PKCα. Left panels show representative luminescence signals acquired for the corresponding assays. The ATP standard curve is shown for each assay. In these assays, kinase activity is associated with a decrease in luminescence reflecting ATP consumption. Each assay was initiated by the addition of ATP to 10 μM final concentration. Right panels depict summary graphs of the activity in the presence of SCR or ZIP relative to control. Data are shown as mean ± S.E.M. (n = 3–4). *P < 0.01 compared with their respective control.
ZIP Disrupts PKC Interactions with AKAP79.
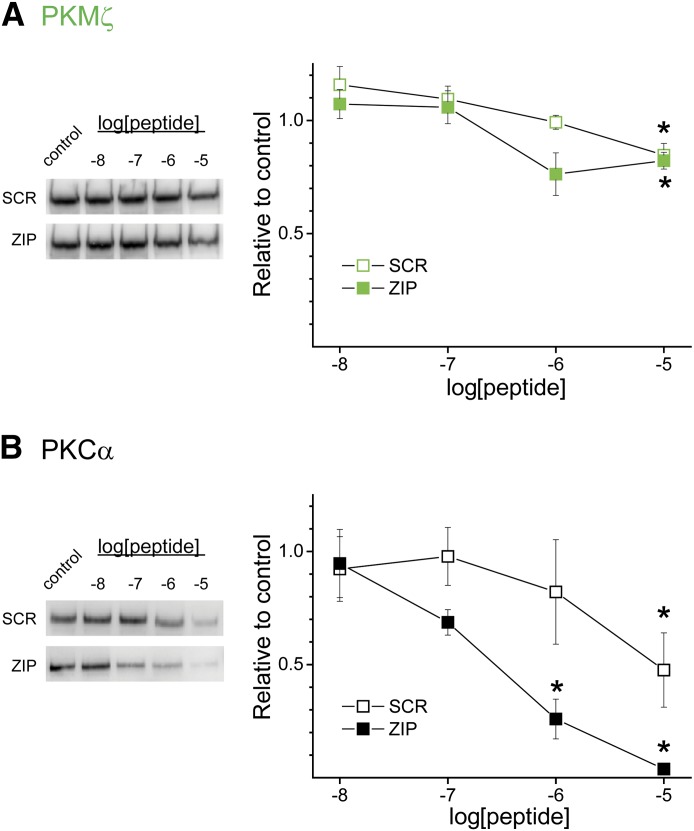
Given the relatively similar apparent affinity of PKCα and PKMζ for ZIP, but the disparate sensitivity of their catalytic activity toward ZIP, we considered that ZIP could potentially interfere with PKCα by a mechanism that is independent of catalytic activity. Indeed, PKCα bound extensively to ZIP in the absence of any activators (Fig. 3). Because PKC, irrespective of its activation state, binds to AKAP79(31–52) by a pseudosubstrate-like mechanism (Faux et al., 1999), we tested the hypothesis that ZIP may competitively displace PKC isoforms from AKAP79(31–52) using an in vitro affinity interaction assay. Streptavidin-coated beads charged with biotinylated-AKAP79(31–52) peptide were incubated with PKMζ or PKCα in the absence or presence of various concentrations of myr-ZIP or myr-SCR. In a remarkable reversal of the effect of these peptides on PKC catalytic activity, myr-ZIP and myr-SCR exhibited relatively limited displacement of PKMζ from AKAP79(31–52) at the highest concentration tested (myr-ZIP: 82.8 ± 3.7% of control; P < 0.01; myr-SCR: 84.7 ± 5.2% of control; P < 0.05; Fig. 5A). In contrast, myr-ZIP potently and completely displaced PKCα from AKAP79(31–52), whereas the scrambled peptide was ∼10-fold weaker (myr-ZIP versus myr-SCR: P < 0.01 evaluated by two-way ANOVA; Fig. 5B). Interestingly, ZIP displaced PKCα from AKAP79(31–52) with an IC50 = 0.27 μM, identical to ZIP-induced PKMζ inhibition, suggesting that displacement of PKCα (and potentially other isoforms) from targeted sites may be an additional mode by which ZIP exerts its effects.
Fig. 5.
ZIP disrupts PKC interactions with AKAP79. Biotinylated-AKAP79(31–52)–charged streptavidin-coated beads were incubated with recombinant PKC (200 ng) in the presence of the indicated concentration of myr-SCR or myr-ZIP. PKC bound to immobilized AKAP79(31–52) was resolved by SDS-PAGE and subjected to immunoblot analysis with isoform-specific PKC antibodies (A) PKMζ and (B) PKCα. Left panels show representative blots for each isoform and treatment. Right panels show the corresponding summary graph based on densitometric analysis. Data are presented as mean ± S.E.M. relative to control (n = 4–5). *P < 0.01 compared with their respective control.
ZIP Disrupts Cellular cPKC Targeting.
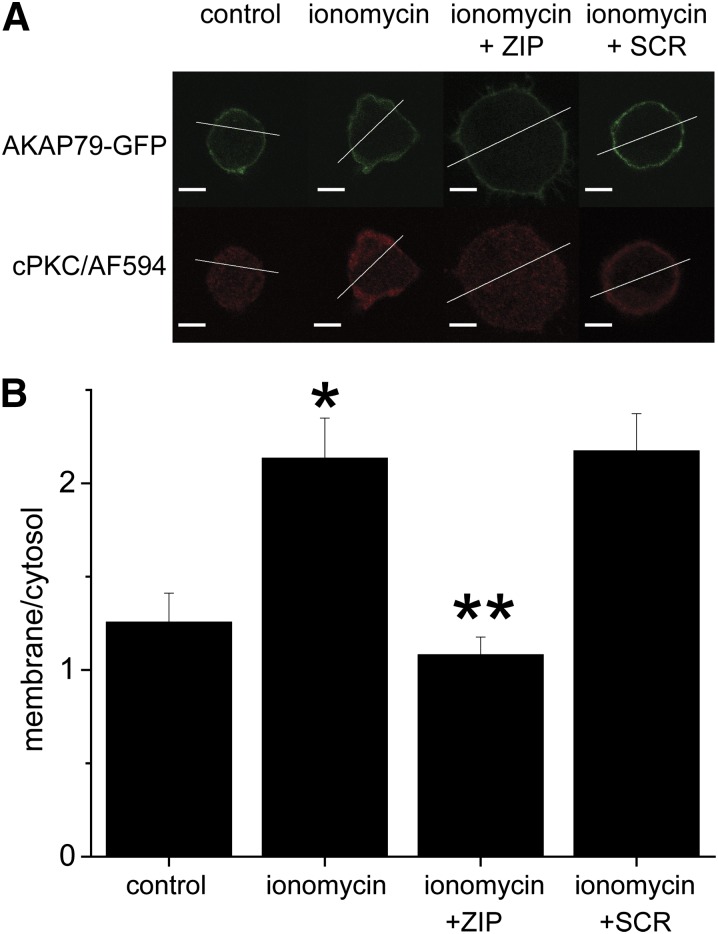
Because ZIP was able to disrupt PKCα/AKAP79(31–52) interactions in vitro, we were interested in determining whether a cellular correlate of this could be resolved. Upon activation, PKC translocates to the plasma membrane (Steinberg, 2008). Because of their affinity for PKC, targeting proteins such as AKAP79 effectively retain PKC near membrane localized substrates, thereby ensuring faithful PKC signaling (Tavalin, 2008). Thus, we anticipated that ZIP could interfere with PKC translocation and/or retention at the plasma membrane. To test this idea, HEK293 cells were first transfected with AKAP79-GFP for use as a marker of the plasma membrane because of its well described polybasic regions that mediate association with plasma membrane resident acidic phospholipids (Dell'Acqua et al., 1998). After transfection, cells were pretreated with vehicle control or myr-ZIP or myr-SCR (5 μM; 10 minutes each). After pretreatment, conventional PKC translocation was induced by brief treatment with ionomycin (1 μM; 3 minutes) in the continued presence or absence of the peptides. Cells were then rapidly fixed, subsequently stained for cPKC using a pan-cPKC antibody and an AlexaFluor594-coupled secondary antibody, and then imaged by confocal microscopy. Line-scan intensity profiles of confocal sections were used to derive membrane/cytosol ratios for cPKC under the various treatment conditions. Our analysis revealed that ionomycin readily induced membrane translocation of cPKC, which was completely blocked by myr-ZIP treatment but unaffected by the scrambled control peptide (Fig. 6). Collectively, these data indicate that ZIP, at the concentrations routinely used, disrupts cPKC targeting and translocation.
Fig. 6.
ZIP disrupts cellular cPKC targeting. HEK293 cells transfected with AKAP79-GFP to delineate the plasma membrane were exposed to standard extracellular buffer alone or buffer containing either 5 μM myr-ZIP or 5 μM myr-SCR peptides for 10 minutes. Cells were then treated with buffer ± ionomycin (1 μM; 3 minutes). Cells were fixed and permeabilized and incubated with anti-cPKC antibody followed by AlexaFluror594-coupled secondary antibody. Line-scan intensity profiles for green and red channels corresponding to AKAP79-GFP and cPKC were obtained for each cell. (A) Representative single confocal plane images with location of line scan for each channel. Scale bar represents 5 μm. (B) After background subtraction, the membrane-to-cytosol ratios for cPKC were derived from the average intensity in these areas and are depicted in the bar graph. Data are shown as the mean + S.E.M. (n = 12–19) and were subjected to one-way ANOVA followed by Student’s t test. *P < 0.01 compared with control; **P < 0.01 compared with ionomycin.
Discussion
Since the initial demonstration of its ability to reverse established LTP, ZIP has achieved widespread use in vivo to implicate PKMζ (and atypical PKCs) as contributing to various forms of memory mediated within different brain regions (Kwapis and Helmstetter, 2014); however, recent studies using mice lacking PKCζ/PKMζ suggest that these PKC isoforms may be dispensable for some forms of learning and memory (Lee et al., 2013; Volk et al., 2013). Moreover, these studies demonstrated that ZIP still had inhibitory effects in the absence of PKCζ/PKMζ, suggesting that additional ZIP targets exist (Lee et al., 2013; Volk et al., 2013). The relevance of these additional sites may be contested, as it is conceivable these may represent latent ZIP targets that only emerge in the absence of PKCζ/PKMζ. Here, we demonstrate that ZIP, at concentrations commonly used, exerts prominent effects on the targeting and translocation of cPKC. Given the prevalence of cPKC isoforms and their previously suggested contributions toward learning and memory based on genetic and inhibitor studies (Abeliovich et al., 1993; Weeber et al., 2000; Bonini et al., 2007), it would appear that the novel actions of ZIP described here likely contribute to the ability of ZIP to disrupt learning and memory, even when PKCζ/PKMζ are present.
Although our data support the idea that ZIP preferentially inhibits PKMζ relative to PKCα (Fig. 4), we found that the opposite was true with regard to the ability of ZIP to competitively displace these PKC isoforms from AKAP79 (31–52) (Fig. 5). The absence and presence of a pseudosubstrate domain in PKMζ and PKCα, respectively, provide a potential explanation for both observations. The lack of a competing pseudosubstrate domain in PKMζ allows ZIP to readily access the kinase’s catalytic site and effectively compete with substrate, whereas ZIP must compete with both substrate and the PKCα pseudosubstrate to access and inhibit PKCα catalytic core. In contrast, ZIP solely competes with AKAP79 (31–52) for binding to the PKMζ catalytic core, whereas both ZIP and the PKCα pseudosubstrate domain compete with AKAP79 (31–52) for binding PKCα. This dual attack by ZIP and the PKCα pseudosubstrate consequently makes PKCα interactions with AKAP79(31-52) more labile. Hence, PKMζ possesses more stable interactions than PKCα with either ZIP or AKAP79 (31–52). As AKAP79 binds all other PKC isoforms by a similar mechanism with similar or lower apparent affinity, it is likely that these interactions are similarly sensitive to ZIP.
PKC translocates to the plasma membrane upon activation but rapidly cycles to and from the membrane (Oancea and Meyer, 1998). Targeting proteins like AKAP79 facilitate PKC signaling by recruiting or retaining PKC near relevant substrates like the GluA1 α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit (Tavalin, 2008; Brooks and Tavalin, 2011). As such, ZIP-mediated competitive displacement of PKC from its targeting interactions effectively solubilizes PKC and would be expected to hinder PKC signaling. Indeed, peptide-mediated displacement of kinases and phosphatases from anchoring proteins is virtually as effective as direct inhibition of these signaling enzymes (Rosenmund et al., 1994; Westphal et al., 1999; Dell’Acqua et al., 2002; Tavalin et al., 2002; Alto et al., 2003). Interestingly, protein phosphatase-1 (PP1) binds to AKAP79 via residues 31–52 and residues 150–250 (Le et al., 2011). However, PKC and PP1 do not appear to compete for binding to AKAP79 (31–52) (Le et al., 2011). As such, AKAP79 should retain PP1 after ZIP-mediated PKC displacement and consequently shift the local balance of kinase to phosphatase activity in favor of the phosphatase. This scenario could represent an ideal mechanism for ZIP to drive restoration of the basal phosphorylation state of critical synaptic proteins that serve to maintain LTP-like potentiated states that are thought to contribute to memory.
One of the prevailing arguments supporting a role for PKMζ in LTP maintenance arises from the apparent parallel pharmacologic sensitivity of both to distinct PKC inhibitors (Ling et al., 2002; Yao et al., 2013). Although LTP induction is sensitive to the broad-spectrum kinase inhibitor staurosporine (which targets most PKC isoforms), LTP maintenance and PKMζ appear to be resistant to this agent (Ling et al., 2002; Yao et al., 2013). Conversely, LTP maintenance and PKMζ are sensitive to ZIP, which acts as a substrate-site competitive inhibitor (Ling et al., 2002; Pastalkova et al., 2006; Yao et al., 2013). Interestingly, AKAP79 alters the inhibitor pharmacology of PKC, rendering it relatively insensitive to ATP-site competitive inhibitors like staurosporine; albeit elevated Ca2+ levels (as would be expected during LTP induction) restore inhibitor sensitivity to AKAP79-anchored PKC (Hoshi et al., 2010; Brooks and Tavalin, 2011). In conjunction with our finding that AKAP79/PKC interactions are targeted by ZIP, this suggests that AKAP79-anchored PKC shares pharmacologic properties with LTP induction and maintenance. Although the studies performed here do not rule out a role for PKMζ in LTP maintenance, it is important to emphasize that our studies suggest that other PKC isoforms cannot be ruled out either based on the use of these inhibitors.
Our data point to AKAP79/PKC interactions as a novel target for ZIP that has potential relevance for LTP based on the known ability of AKAP79 to target PKC, as well as other signaling enzymes to the AMPA receptor GluA1 subunit, and the documented significance of these interactions toward various forms of synaptic plasticity (Dell’Acqua et al., 2002; Tavalin et al., 2002; Lu et al., 2007; Lu et al., 2008; Tavalin, 2008; Jurado et al., 2010; Weisenhaus et al., 2010; Le et al., 2011; Sanderson et al., 2012; Diering et al., 2014); however, it is likely that additional targeting proteins and mechanisms contribute to the ZIP-sensitivity of LTP. For example, p62/ sequestosome 1 (SQSTM1) was suggested to serve as a bridge linking PKCι/λ to GluA1, thereby promoting GluA1 phosphorylation at S818, which drives synaptic AMPA receptor incorporation during early LTP (Boehm et al., 2006; Jiang et al., 2009; Ren et al., 2013). In this case, direct ZIP-induced inhibition of PKCι/λ has been suggested to be responsible (Ren et al., 2013); however, p62/SQSTM1 binds to aPKCs in a region subjacent to the pseudosubstrate domain (Jiang et al., 2009). Consequently, ZIP may be expected to sterically interfere with this interaction. Complementary to this, the AMPA receptor binding domain of p62/SQSTM1 is adjacent to the aPKC binding domain (Jiang et al., 2009) and also could be subject to interference by ZIP. Beyond protein-protein interactions, PKC pseudosubstrate domain–derived peptides, because of their highly basic nature, are known to bind phosphatidylinositol-(3,4,5)-trisphosphate and acidic phospholipids, which impacts cPKC and aPKC activation and subsequent translocation (Mosior and McLaughlin, 1991; Nakanishi et al., 1993; Ivey et al., 2014). Together with our findings, these studies highlight the broad spectrum sensitivity of PKC targeting mechanisms to PKC pseudosubstrate-derived inhibitors. Consequently, our findings suggest the limited utility of ZIP as a means for identifying the isoform dependence of complex phenomena such as LTP, which may rely on the coordinated action of multiple PKC isoforms within each phase. The remarkable ability of ZIP to reverse LTP and disrupt memory performance in various behavioral paradigms, however, suggests that ZIP may still represent a useful agent for dissecting, at the circuit level, the temporal dynamics by which LTP-like mechanisms contribute to the persistence of acquired behaviors.
Abbreviations
- AKAP
A-kinase anchoring protein
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- ANOVA
analysis of variance
- aPKC
atypical protein kinase C
- BSA
bovine serum albumin
- cPKC
conventional protein kinase C
- GFP
green fluorescent protein
- LTP
long-term potentiation
- myr
myristoylated
- nPKC
novel protein kinase C
- PBS
phosphate-buffered saline
- PKC
protein kinase C
Authorship Contributions
Participated in research design: Bogard, Tavalin.
Conducted experiments: Bogard, Tavalin.
Performed data analysis: Bogard, Tavalin.
Wrote or contributed to the writing of the manuscript: Bogard, Tavalin.
Footnotes
This work was supported by the National Institutes of Health National Institute of Neurologic Disorders and Stroke [Grant R01-NS076637].
References
- Abeliovich A, Paylor R, Chen C, Kim JJ, Wehner JM, Tonegawa S. (1993) PKC gamma mutant mice exhibit mild deficits in spatial and contextual learning. Cell 75:1263–1271. [DOI] [PubMed] [Google Scholar]
- Alto NM, Soderling SH, Hoshi N, Langeberg LK, Fayos R, Jennings PA, Scott JD. (2003) Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Natl Acad Sci USA 100:4445–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm J, Kang MG, Johnson RC, Esteban J, Huganir RL, Malinow R. (2006) Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron 51:213–225. [DOI] [PubMed] [Google Scholar]
- Bonini JS, Da Silva WC, Bevilaqua LR, Medina JH, Izquierdo I, Cammarota M. (2007) On the participation of hippocampal PKC in acquisition, consolidation and reconsolidation of spatial memory. Neuroscience 147:37–45. [DOI] [PubMed] [Google Scholar]
- Brooks IM, Tavalin SJ. (2011) Ca2+/calmodulin-dependent protein kinase II inhibitors disrupt AKAP79-dependent PKC signaling to GluA1 AMPA receptors. J Biol Chem 286:6697–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill EN, Qvit N, Mochly-Rosen D. (2009) Rationally designed peptide regulators of protein kinase C. Trends Endocrinol Metab 20:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua ML, Dodge KL, Tavalin SJ, Scott JD. (2002) Mapping the protein phosphatase-2B anchoring site on AKAP79: binding and inhibition of phosphatase activity are mediated by residues 315-360. J Biol Chem 277:48796–48802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Acqua ML, Faux MC, Thorburn J, Thorburn A, Scott JD. (1998) Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. EMBO J 17:2246–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diering GH, Gustina AS, Huganir RL. (2014) PKA-GluA1 coupling via AKAP5 controls AMPA receptor phosphorylation and cell-surface targeting during bidirectional homeostatic plasticity. Neuron 84:790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichholtz T, de Bont DB, de Widt J, Liskamp RM, Ploegh HL. (1993) A myristoylated pseudosubstrate peptide, a novel protein kinase C inhibitor. J Biol Chem 268:1982–1986. [PubMed] [Google Scholar]
- Faux MC, Rollins EN, Edwards AS, Langeberg LK, Newton AC, Scott JD. (1999) Mechanism of A-kinase-anchoring protein 79 (AKAP79) and protein kinase C interaction. Biochem J 343:443–452. [PMC free article] [PubMed] [Google Scholar]
- Hoshi N, Langeberg LK, Gould CM, Newton AC, Scott JD. (2010) Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol Cell 37:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House C, Kemp BE. (1987) Protein kinase C contains a pseudosubstrate prototope in its regulatory domain. Science 238:1726–1728. [DOI] [PubMed] [Google Scholar]
- Ivey RA, Sajan MP, Farese RV. (2014) Requirements for pseudosubstrate arginine residues during autoinhibition and phosphatidylinositol 3,4,5-(PO₄)₃-dependent activation of atypical PKC. J Biol Chem 289:25021–25030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Parameshwaran K, Seibenhener ML, Kang MG, Suppiramaniam V, Huganir RL, Diaz-Meco MT, Wooten MW. (2009) AMPA receptor trafficking and synaptic plasticity require SQSTM1/p62. Hippocampus 19:392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S, Biou V, Malenka RC. (2010) A calcineurin/AKAP complex is required for NMDA receptor-dependent long-term depression. Nat Neurosci 13:1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwapis JL, Helmstetter FJ. (2014) Does PKM(zeta) maintain memory? Brain Res Bull 105:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le AV, Tavalin SJ, Dodge-Kafka KL. (2011) Identification of AKAP79 as a protein phosphatase 1 catalytic binding protein. Biochemistry 50:5279–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AM, Kanter BR, Wang D, Lim JP, Zou ME, Qiu C, McMahon T, Dadgar J, Fischbach-Weiss SC, Messing RO. (2013) Prkcz null mice show normal learning and memory. Nature 493:416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling DS, Benardo LS, Serrano PA, Blace N, Kelly MT, Crary JF, Sacktor TC. (2002) Protein kinase Mzeta is necessary and sufficient for LTP maintenance. Nat Neurosci 5:295–296. [DOI] [PubMed] [Google Scholar]
- Lu Y, Allen M, Halt AR, Weisenhaus M, Dallapiazza RF, Hall DD, Usachev YM, McKnight GS, Hell JW. (2007) Age-dependent requirement of AKAP150-anchored PKA and GluR2-lacking AMPA receptors in LTP. EMBO J 26:4879–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhang M, Lim IA, Hall DD, Allen M, Medvedeva Y, McKnight GS, Usachev YM, Hell JW. (2008) AKAP150-anchored PKA activity is important for LTD during its induction phase. J Physiol 586:4155–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosior M, McLaughlin S. (1991) Peptides that mimic the pseudosubstrate region of protein kinase C bind to acidic lipids in membranes. Biophys J 60:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik MU, Benedikz E, Hernandez I, Libien J, Hrabe J, Valsamis M, Dow-Edwards D, Osman M, Sacktor TC. (2000) Distribution of protein kinase Mzeta and the complete protein kinase C isoform family in rat brain. J Comp Neurol 426:243–258. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Brewer KA, Exton JH. (1993) Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 268:13–16. [PubMed] [Google Scholar]
- Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem 272:952–960. [DOI] [PubMed] [Google Scholar]
- Oancea E, Meyer T. (1998) Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell 95:307–318. [DOI] [PubMed] [Google Scholar]
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. (2006) Storage of spatial information by the maintenance mechanism of LTP. Science 313:1141–1144. [DOI] [PubMed] [Google Scholar]
- Pears CJ, Parker PJ. (1991) Domain interactions in protein kinase C. J Cell Sci 100:683–686. [DOI] [PubMed] [Google Scholar]
- Ren SQ, Yan JZ, Zhang XY, Bu YF, Pan WW, Yao W, Tian T, Lu W. (2013) PKCλ is critical in AMPA receptor phosphorylation and synaptic incorporation during LTP. EMBO J 32:1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Carr DW, Bergeson SE, Nilaver G, Scott JD, Westbrook GL. (1994) Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature 368:853–856. [DOI] [PubMed] [Google Scholar]
- Sanderson JL, Gorski JA, Gibson ES, Lam P, Freund RK, Chick WS, Dell’Acqua ML. (2012) AKAP150-anchored calcineurin regulates synaptic plasticity by limiting synaptic incorporation of Ca2+-permeable AMPA receptors. J Neurosci 32:15036–15052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroujon MC, Mochly-Rosen D. (1998) Peptide modulators of protein-protein interactions in intracellular signaling. Nat Biotechnol 16:919–924. [DOI] [PubMed] [Google Scholar]
- Steinberg SF. (2008) Structural basis of protein kinase C isoform function. Physiol Rev 88:1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalin SJ. (2008) AKAP79 selectively enhances protein kinase C regulation of GluR1 at a Ca2+-calmodulin-dependent protein kinase II/protein kinase C site. J Biol Chem 283:11445–11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalin SJ, Colledge M, Hell JW, Langeberg LK, Huganir RL, Scott JD. (2002) Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J Neurosci 22:3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk LJ, Bachman JL, Johnson R, Yu Y, Huganir RL. (2013) PKM-ζ is not required for hippocampal synaptic plasticity, learning and memory. Nature 493:420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shang Y, Yu J, Zhang M. (2012) Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKCι in complex with a substrate peptide from Par-3. Structure 20:791–801. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Atkins CM, Selcher JC, Varga AW, Mirnikjoo B, Paylor R, Leitges M, Sweatt JD. (2000) A role for the beta isoform of protein kinase C in fear conditioning. J Neurosci 20:5906–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenhaus M, Allen ML, Yang L, Lu Y, Nichols CB, Su T, Hell JW, McKnight GS. (2010) Mutations in AKAP5 disrupt dendritic signaling complexes and lead to electrophysiological and behavioral phenotypes in mice. PLoS One 5:e10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, Sheng M, Scott JD. (1999) Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 285:93–96. [DOI] [PubMed] [Google Scholar]
- Yao Y, Kelly MT, Sajikumar S, Serrano P, Tian D, Bergold PJ, Frey JU, Sacktor TC. (2008) PKM zeta maintains late long-term potentiation by N-ethylmaleimide-sensitive factor/GluR2-dependent trafficking of postsynaptic AMPA receptors. J Neurosci 28:7820–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Shao C, Jothianandan D, Tcherepanov A, Shouval H, Sacktor TC. (2013) Matching biochemical and functional efficacies confirm ZIP as a potent competitive inhibitor of PKMζ in neurons. Neuropharmacology 64:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]