Abstract
Motivation: Synonymous codon usage bias has been shown to be correlated with many genomic features among different organisms. However, the biological significance of codon bias with respect to gene function and genome organization remains unclear.
Results: Guanine and cytosine content at the third codon position (GC3) could be used as a good indicator of codon bias. Here, we used relative GC3 bias values to compare the strength of GC3 bias of genes in human and mouse. We reported, for the first time, that GC3-rich and GC3-poor gene products might have distinct sub-cellular spatial distributions. Moreover, we extended the view of genomic gene domains and identified conserved GC3 biased gene domains along chromosomes. Our results indicated that similar GC3 biased genes might be co-translated in specific spatial regions to share local translational machineries, and that GC3 could be involved in the organization of genome architecture.
Availability and implementation: Source code is available upon request from the authors.
Contact: zhaozh@nic.bmi.ac.cn or zany1983@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Due to the redundancy of the genetic code, most amino acids can be translated by multiple codons (called synonymous codons). The frequencies of synonymous codon usage vary among different genes within the same and across different organisms (Hershberg and Petrov, 2008). This phenomenon is termed synonymous codon usage bias (SCUB). Accumulating evidence has suggested different mechanisms for SCUB, including mutational pressure and selection, etc. (Trotta, 2013; Tuller, 2011). Recent bioinformatics and experimental studies have shown strong correlations between SCUB and translation accuracy and speed, mRNA secondary structures and stability, protein folding and function and other factors (Foroughmand-Araabi et al., 2014; Kudla et al., 2009; Novoa and Ribas de Pouplana, 2012; Presnyak et al., 2015; Yang et al., 2014). Although much effort has been made to explain the mechanisms and biological roles of SCUB, whether there are some unexplored biological functions of SCUB, especially with respect to gene function and genome organization, have not yet been elucidated.
Because most synonymous codons differ at only the third nucleotide positions, guanine and cytosine content at the third codon position (GC3) is a good indicator of the extent of SCUB. Previous studies have shown that genes with high GC3 levels may have more sites for DNA methylation, exhibit more variable expression, and accumulate more mutations than those with low GC3 levels (Tatarinova et al., 2010, 2013). In several earlier studies, GC3 has been shown to act as an isochore marker (Aota and Ikemura, 1986; Romiguier et al., 2010), and the relationship between GC3 and the GC content of the flanking regions is still debatable (Clay and Bernardi, 2011; Elhaik et al., 2009). However, the distribution of GC3-biased genes along chromosomes is unknown.
There are numerous genomic features related to GC3 bias. Here, we focused on the genome-wide gene distribution and examined whether genes with different GC3 levels exhibited different functional properties. In order to compare the strengths of GC3 biases among genes within one genome, we developed the ‘relative GC3 bias value’ as a measure. Fisher’s exact tests were performed to remove the compositional bias. We showed that significant GC3-rich and GC3-poor gene products were associated with different cellular components. Additionally, GC3-biased genes were organized in domains along chromosomes. Thus, our results suggested new roles for codon bias and provided insights into the genome arrangement.
2 Methods
2.1 Datasets
Protein-coding sequences from the genome of human and mouse were obtained from the CCDS database (Pruitt et al., 2009). We filtered out sequences using non-natural amino acids.
2.2 Calculation of relative GC3 bias value
Calculations were based on 59 codons, excluding the three stop codons, ATG (Met), and TGG (Trp) since no synonymous codons exist for these. The ‘GC3 bias’ value was defined as the ratio between the frequencies of NNC/Gs to NNA/Ts. The average codon frequency was used for genes with multiple alternatively spliced transcripts. The expected GC3 bias values of individual amino acids were calculated from the overall base frequencies of all genes. For each gene, we calculated the expected GC3 bias value based on its specific amino acid composition. Next, we determined the log2 ‘relative GC3 bias’ value (log2[relative GC3 bias]) by calculating the ratios of GC3 bias values between the observed and expected values. Thus, a positive log2[relative GC3 bias] reflected the higher GC3 level of the gene as compared with the average value. Fisher’s exact tests were performed to assess the significance of differences. To identify ‘GC3-rich’ and ‘GC3-poor’ genes with high stringency, we set the P value < 1e−5 as a significant threshold.
2.3 Definition of GC3-biased gene domains
The log2[relative GC3 bias] values were plotted equidistantly based on the gene positions along the chromosomes. A GC3-rich gene domain was defined as a set of at least N consecutive significant GC3-rich genes. On the contrary, A GC3-poor gene domain was defined as a set of at least N consecutive significant GC3-poor genes. To determine the value of N, we compared domain numbers under different thresholds (N) in human and mouse, and used the control group by randomly shuffling the locations of all genes along chromosomes (1000 times; Supplementary Fig. S2A). N was set as 4 in human, and 3 in mouse to ensure that random domains were less than 15%.
2.4 Comparison between the genomes of human and mouse
Ortholog pairs were identified from HomoloGene (NCBI Resource Coordinators, 2015). We mapped all the GC3-biased gene domains between human and mouse genomes using the UCSC liftOver tool with default parameters (Hinrichs et al., 2006); and then counted the number of orthologous genes which were still identified as GC3-rich or GC3-poor in the other genome (Supplementary Fig. S2B). A domain was considered conserved if at least two orthologous genes were still GC3 biased.
3 Results
3.1 Identification of GC3-biased genes in human and mouse genomes
Previous studies have generally calculated the GC3 percentage of genes to investigate the diversity of GC3 dynamics between species and to determine the correlations between genomic features and GC3 bias. However, this type of measurement is too weak to evaluate the strength of bias between genes within one genome. Here, we used a new index, ‘relative GC3 bias’ which was based on the odds ratio of GC3 occurrences, to identify significant GC3-biased genes.
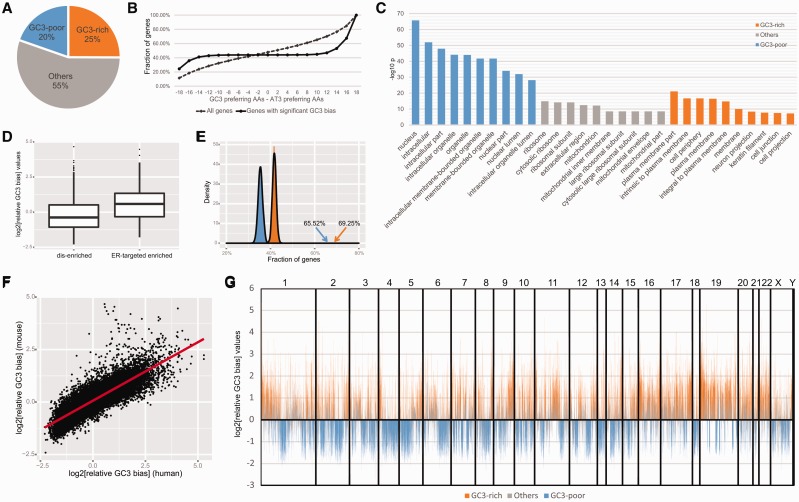
Using P value < 1e−5 (Fisher’s exact test) as a threshold, we found that 20% of genes were GC3-poor and 25% were GC3-rich in the human genome (Fig. 1A, Supplementary Table S1). Moreover, the proportions of GC3-poor and GC3-rich genes were almost twice high as those of the same type in the mouse genome (Supplementary Fig. S1A, Supplementary Table S2). Consistently, the variances of log2[relative GC3 bias] were 1.286 in human and 0.597 in mouse, indicating that GC3 bias was weaker in mouse than in human. Nevertheless, we analyzed the relative GC3 bias of 16 084 ortholog pairs and found that Spearman’s rank correlation rho was as high as 0.87 (P < 1e−15, Fig. 1F), suggesting that GC3 bias was really conserved at least between human and mouse. Scrutinizing into each gene for details, we found that most amino acids of GC3-biased genes used biased codons (Fig. 1B and Supplementary Fig. S1B), suggesting the GC3 bias was not caused by a few given amino acids but rather by overall codon usage.
Fig. 1.
(A) Pie chart summarizing the proportion of genes according to GC3 bias in the human genome. (B) Cumulative curves of differences between GC3 preferring and AT3 preferring amino acids for all genes or genes with significant GC3 bias in the human genome. The two distributions were significantly different (K–S test, P < 1e−15). (C) GO-term (cell component) enrichment analysis. Colors match those in (A). (D) Box plots showing the distributions of log2[relative GC3 bias] values of ER-targeted enriched or dis-enriched genes in human. The two distributions were significantly different (Mann-Whitney test, P < 1e−15). (E) Fractions of GC3-rich/GC3-poor genes having at least one GC3-rich/GC3-poor gene as its neighbor. The observed values were significantly higher than random (P < 1e−15). P values were calculated by drawing 1000 samples (randomly shuffling the locations of all genes along chromosomes) and estimating the distributions. The curves represent random distributions, and arrows represent observed fractions. Colors match those in (A). (F) log2[relative GC3 bias] values of ortholog pairs. The red line shows the linear smoothing. (G) The log2[relative GC3 bias] values of genes along the human genome. Numbers indicate the chromosomes delimited with vertical lines. Colors match those in (A)
3.2 Differential GC3-biased genes may have distinct sub-cellular distributions
To gain insight into the functional relevance of GC3-biased genes, we performed GO analysis using WebGestalt (Zhang et al., 2005) (Supplementary Tables S3 and S4). We found that GC3-rich gene products tended to associate with the plasma membrane and cell periphery, while GC3-poor gene products were more related with nucleus and organelles (Fig. 1C and Supplementary Fig. S1C). Since the cellular component ontology only describes protein locations, we lacked direct evidence to determine whether these gene products were transported after translation or translated at the specific location.
Local translation plays particularly important roles in distal neuronal compartments, and dysregulated RNA localization and translation cause defects in neuronal wiring and survival (Jung et al., 2014). More generally, local translation caused by asymmetric mRNA distribution contributes to the fidelity and sensitivity of spatially localized systems genome-widely (Weatheritt et al., 2014). A recent publication reported a method to characterize localized protein synthesis (Jan et al., 2014), and the authors analyzed translation at the endoplasmic reticulum (ER). To evaluate the above mentioned possibility, we applied our relative GC3 bias values to their results and found that ER-targeted enriched proteins had higher GC3 bias than those dis-enriched proteins (Mann-Whitney test, P < 1e−15; Fig. 1D). The ER can form dynamic contacts with the plasma membrane (Manford et al., 2012), which might be one explanation to our results. It was interesting that different GC3-biased gene products might have distinct sub-cellular spatial distributions. Recently, Gingold et al. (2014) provided direct evidence that alterations of tRNA pools are highly coordinated with changes in mRNA expression in proliferating versus differentiating cells. Furthermore, vaccinia and influenza A virus changed polysome-associated tRNA levels to reflect the codon usage of viral genes, suggesting the existence of local tRNA pools optimized for viral translation (Pavon-Eternod et al., 2012).
These studies have strongly supported the existence of temporal and spatial concomitant mRNA codons and tRNA anticodons pools, thus it might be reasonable to speculate a dynamic and regulated supply-and-demand relationship between translation and localized cellular translational machineries, such as tRNA pools. GC3-rich/GC3-poor genes might be co-translated in specific regions to share local tRNA pools. Methods like MERFISH (Chen et al., 2015) and others (reviewed in Buxbaum et al., 2015) with high resolution to determine mRNA and tRNA subcellular localization should be helpful to further examine this issue.
3.3 GC3-biased genes were organized in domains
Similar GC3-rich/GC3-poor gene products appeared to be clustered in similar sub-cellular distributions via local translation; therefore, we further evaluated whether these genes were also clustered along chromosomes. To this end, we first focused on general clustering of GC3-biased genes in the genome and found that more than 65% of GC3-rich/GC3-poor genes in human (more than 40% in mouse) had at least one GC3-rich/GC3-poor gene as its neighbor (Fig. 1E and Supplementary Fig. S1D); these percentages were significantly higher than what would be expected with random distributions. A very recent study in eukaryotic genomes showed that genes with similar SCUB were close in 3D space, and the functional similarity between genes characterized by SCUB was strongly correlated with their 3D distance (Diament et al., 2014), which was consistent with our findings. Therefore, we then examined the distributions of GC3-biased genes along chromosomes and found that domains composed of neighboring genes sharing similar relative GC3 bias (Fig. 1G and Supplementary Fig. S1E). We defined a GC3-biased domain as a set of consecutive significant GC3-biased genes. Using different gene numbers as thresholds, significantly more domains were consistently found in observed genomes than random distributions (Supplementary Fig. S2A). We then chose moderate thresholds and identified a total of 452 domains in the human genome and 267 domains in the mouse genome, respectively (Supplementary Tables S5 and S6). These domains varied in size (in human: from 20 kb to 10 Mb, median size of 400 kb; in mouse: from 8 kb to 6 Mb, median size of 200 kb). This observation suggested that GC3-biased genes were organized in a specific pattern along the chromosomes and clustered into GC3-rich and GC3-poor domains. Since we only considered consecutive GC3-rich/GC3-poor genes, this strict definition of GC3-biased gene domains might underestimate the number and size of domains. These identified domains could be the core regions for all general GC3-biased gene domains.
To verify whether GC3-biased gene domains were conserved in mammals, we compared between human and mouse, and counted the number of orthologous genes that were still identified as GC3 biased after the domain mapping to the other genome. We found a substantial number of domains containing two or more orthologous GC3-biased genes (an overall assessment is presented in Supplementary Fig. S2B), and such domains were considered conserved. In this way, we observed 59.1% of domains in the human genome and 74.5% of domains in the mouse genome were conserved. Considering GC3 bias in mouse was weaker, and there was a high correlation in GC3 bias between human and mouse, we believe that this phenomenon was conserved between these two mammals. Previous reports have shown that genes with similar SCUB tend to be close to each other on the chromosomes and organized in coherent domains in bacteria (Bailly-Bechet et al., 2006). Therefore, we speculated genes with similar codon usage might cluster along chromosomes in both prokaryotic and eukaryotic genomes. Our data describing this phenomenon provided insights into the compartmentalization of genomes and indicated that there was a strong correlation between gene expression and linear distribution.
4 Conclusions
In this study, we presented a new method for comparing the strength of GC3 bias which might be deduced to other organisms. Using this method, we identified significant GC3-biased genes in human and mouse, and proposed the distinct sub-cellular distributions of GC3-biased genes for the first time. Moreover, we expanded the knowledge of genomic domains of genes, and found GC3-biased genes were organized in domains along the chromosomes. Diament et al. described the high correlation between codon usage and eukaryotic 3D genomic organization, indicating that non-randomly organized codon usage biased genes were linked to genomic organization and protein function. Consistently, our current data demonstrated that these GC3-biased genes were presumably organized in domains along chromosomes, then transcribed and translated in closed subcellular localizations. The identification of conserved GC3-biased gene domains extends the known chromosome architecture domains such as topological associated domains, etc. and will improve our understanding of genome higher-order structures and the functional significance (Sexton and Cavalli, 2015).
It has been well documented since recent years, that functionally related genes are co-transcribed in ‘transcription factories’, and then functionally related mRNA are even subjected to local translation in eukaryotic cells (Jan et al., 2014; Jung et al., 2014; Papantonis and Cook, 2013; Weatheritt et al., 2014). In this report, we found that clustered GC3-biased genes showed enrichment of particular mRNA distributions and protein localization. Based on our observations and results of other studies (Gingold et al., 2014), we speculated that codon usage bias might coordinate multifaceted processes of gene expression and even protein localization.
Further studies are required to provide detailed bioinformatics analysis to identify GC3-rich and GC3-poor genes and domains more concisely. Additionally, the molecular mechanisms mediating the interplay between genomic domains along chromosomes and cellular distributions of gene products are unclear; we believe that there may be a mechanism coordinating these multiple processes from transcription to translational targeting. Further experiments may provide more evidence.
Supplementary Material
Acknowledgements
We thank Jeffrey Chu and members of our laboratory for critical discussions.
Funding
This work was supported by National Basic Research Program of China [2013CB966802], National Natural Science Foundation of China [31370762, 31030026, 31272416, 81372218] and National Key Technology R&D Program of China [2012BAI01B07].
Conflict of Interest: none declared.
References
- Aota S., Ikemura T. (1986) Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res., 14, 6345–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., et al. (2006) Codon usage domains over bacterial chromosomes. PLoS Comput. Biol., 2, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum A.R., et al. (2015) In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol., 16, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.H., et al. (2015) Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay O.K., Bernardi G. (2011) GC3 of genes can be used as a proxy for isochore base composition: a reply to Elhaik et al. Mol. Biol. Evol., 28, 21–23. [DOI] [PubMed] [Google Scholar]
- Diament A., et al. (2014) Three-dimensional eukaryotic genomic organization is strongly correlated with codon usage expression and function. Nat. Commun., 5, 5876–5888. [DOI] [PubMed] [Google Scholar]
- Elhaik E., et al. (2009) Can GC content at third-codon positions be used as a proxy for isochore composition? Mol. Biol. Evol., 26, 1829–1833. [DOI] [PubMed] [Google Scholar]
- Foroughmand-Araabi M.H., et al. (2014) Dependency of codon usage on protein sequence patterns: a statistical study. Theor. Biol. Med. Model., 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H., et al. (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell, 158, 1281–1292. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Petrov D.A. (2008) Selection on codon bias. Annu. Rev. Genet., 42, 287–299. [DOI] [PubMed] [Google Scholar]
- Hinrichs A.S., et al. (2006) The UCSC Genome Browser Database: update 2006. Nucleic Acids Res., 34, D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C.H., et al. (2014) Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science, 346, 1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., et al. (2014) Remote control of gene function by local translation. Cell, 157, 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., et al. (2009) Coding-sequence determinants of gene expression in Escherichia coli. Science, 324, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford A.G., et al. (2012) ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell, 23, 1129–1140. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. (2015) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 43, D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa E.M., Ribas de Pouplana L. (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet., 28, 574–581. [DOI] [PubMed] [Google Scholar]
- Papantonis A., Cook P.R. (2013) Transcription factories: genome organization and gene regulation. Chem. Rev., 113, 8683–8705. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M., et al. (2013) Vaccinia and influenza A viruses select rather than adjust tRNAs to optimize translation. Nucleic Acids Res., 41, 1914–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V., et al. (2015) Codon optimality is a major determinant of mRNA stability. Cell, 160, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., et al. (2009) The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res., 19, 1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romiguier J., et al. (2010) Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res., 20, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Cavalli G. (2015) The role of chromosome domains in shaping the functional genome. Cell, 160, 1049–1059. [DOI] [PubMed] [Google Scholar]
- Tatarinova T., et al. (2010) GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics, 11, 308–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarinova T., et al. (2013) Cross-species analysis of genic GC3 content and DNA methylation patterns. Genome Biol. Evol., 5, 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta E. (2013) Selection on codon bias in yeast: a transcriptional hypothesis. Nucleic Acids Res., 41, 9382–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T. (2011) Codon bias, tRNA pools and horizontal gene transfer. Mob. Genet. Elements, 1, 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatheritt R.J., et al. (2014) Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nat. Struct. Mol. Biol., 21, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.R., et al. (2014) Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS Biol., 12, e1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., et al. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res., 33, W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.