Abstract
Evidence is mounting that epistasis is widespread among mutations. The cost of carrying two deleterious mutations, or the advantage of acquiring two beneficial alleles, is typically lower that the sum of their individual effects. Much less is known on epistasis between beneficial and deleterious mutations, even though this is key to the amount of genetic hitchhiking that may occur during evolution. This is particularly important in the context of antibiotic resistance: Most resistances are deleterious, but some can be beneficial and remarkably rifampicin resistance can emerge de novo in populations evolving without antibiotics. Here we show pervasive positive pairwise epistasis on Escherichia coli fitness between beneficial mutations, which confer resistance to rifampicin, and deleterious mutations, which confer resistance to streptomycin. We find that 65% of double resistant strains outcompete sensitive bacteria in an environment devoid of antibiotics. Weak beneficial mutations may therefore overcome strong deleterious mutations and can even render double mutants strong competitors.
Keywords: epistasis, resistance, mutation
The effect of a mutation may depend on the genetic background where it occurs, a phenomenon termed epistasis. The existence of pervasive epistasis for mutations that affect fitness-related traits has an important impact on the evolutionary dynamics of a population and the number of paths accessible to it (Weinreich et al. 2005, 2006; Phillips 2008; De Visser et al. 2011; De Visser and Krug 2014). Experimental evolution to novel laboratory environments has been used to determine the importance and type of epistasis underlying evolutionary trajectories (Salverda et al. 2011; Tenaillon et al. 2012; Kryazhimskiy et al. 2014). Recent studies suggest that the fitness of the genetic background is a key factor influencing the ability of microbial populations to adapt or readapt (Khan et al 2011; Sousa et al. 2012; Schenk et al 2013).
Both in bacteria and yeast, low fit clonal populations were found to have a higher capacity to adapt than clonal populations with higher fitness (Kryazhimskiy et al. 2014; Perfeito et al. 2014). Empirical studies have also been performed to directly measure the strength of epistasis (Ɛ). This was done by measuring both the effect of single mutations and pairs of mutations combined onto the same genetic background (De Visser and Krug 2014). Pairs of individually deleterious (Elena and Lenski 1997; Trindade et al. 2009) or beneficial, at the level of a single gene or between loci (Khan et al. 2011; Schenk et al. 2013), were studied. Far less is known about epistasis between beneficial and deleterious mutations, despite its importance to the amount of genetic hitchhiking on the evolution of asexual populations or genomic regions with reduced recombination (Gillespie 2000; Johnson and Barton 2002) and patterns of molecular evolution (Kondrashov DA and Kondrashov FA 2015). An important fitness trait for bacteria is the level of resistance to antibiotics, which can occur in a wide range of concentrations across environments (Andersson and Hughes 2014). Bacterial populations show high levels of polymorphism for resistance alleles and epistasis between resistance alleles is thought to be important in explaining levels of resistance observed in natural populations (Borrell and Gagneux 2011; Müller et al. 2013) and in determining the evolutionary path toward increased resistance to certain antibiotics (Weinreich et al. 2006; MacLean, Hall, et al. 2010; Borrell et al. 2013; De Visser and Krug 2014). Mutations conferring resistance also exhibit strong genotype-by-environment (GxE) interactions. A remarkable example of these types of interactions occurs in alleles that confer rifampicin and streptomycin resistance. Strong epistatic interactions of either positive (alleviating) or negative (increasing) type for the fitness effects of two resistance alleles were found in different species and in different environments (Trindade et al. 2009, 2012; Ward et al. 2009; MacLean, Perron, et al. 2010). Some resistance alleles can even be beneficial in certain environments. For example, Miskinyte and Gordo (2013) found that streptomycin-resistant (StrR) and also rifampicin-resistant (Rif R) resistance mutations can benefit Escherichia coli survival inside macrophages. In this same species, several RifR mutations have also been found to confer a fitness advantage in minimal glucose medium (Trindade et al. 2012). The spontaneous emergence of RifR alleles has even been reported in E. coli evolving in poor medium under high temperature (Rodríguez-Verdugo et al. 2013). Interestingly, in that study the ancestral strain in which rifampicin emerged was streptomycin resistant.
Here we study the fitness effects of rifampicin and streptomycin resistance in poor nutritional medium devoid of antibiotics, where, according to competitive assays, rifampicin mutations are beneficial and streptomycin resistance incurs a fitness cost (Trindade et al. 2012). We ask three questions: How costly is double resistance in this environment? How pervasive is epistasis between beneficial and deleterious alleles? How do the benefits of a single RifR allele vary with the fitness of the genetic background where it emerges?
Results and Discussion
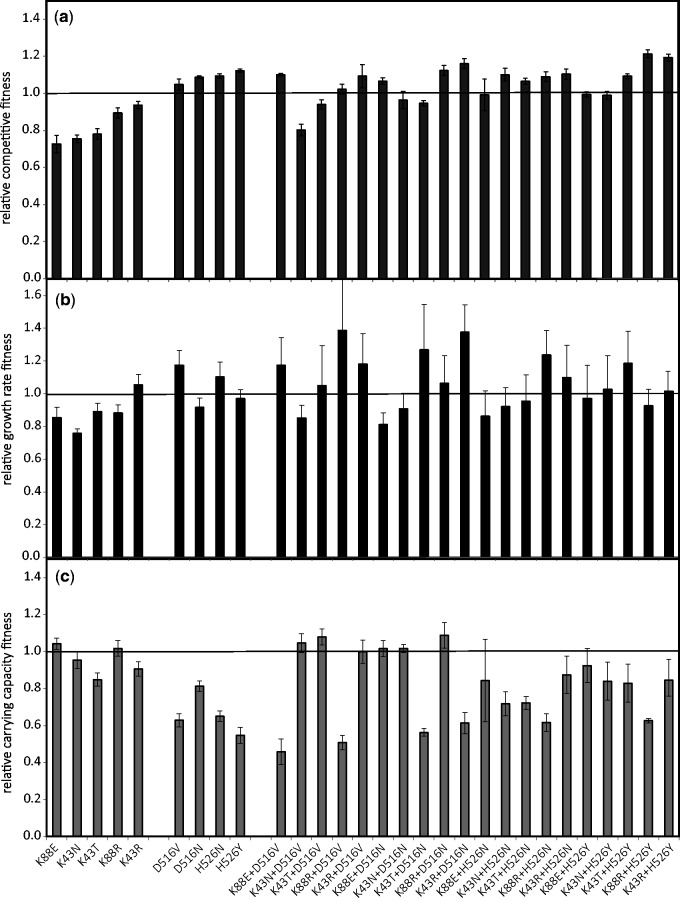
In order to determine the effect on fitness of double resistance, we performed competitive fitness assays between the double resistant and a sensitive strain in minimal media supplemented with glucose. The sensitive strain carries a genetic marker, which is neutral in this environment (Trindade et al. 2012). Figure 1a shows the results of the competitive fitness assays. From the 20 double mutants studied only 3 have a significant fitness cost. We find that 85% of the double resistant strains have no significant cost and therefore are not expected to be eliminated from the population when it grows in poor medium. Hitchhiking of deleterious mutants is observed in the double mutants—K88E+H526N, K43T+H526N, K88R+H526N, K88R+D516V, and K88E+D516N. These represent cases where the deleterious effect of the StrR mutations is not enough to impair fitness below wild type levels (i.e., below 1) and therefore are still expected to outcompete the sensitive strain. Most importantly, 65% of all clones are actually expected to outcompete the sensitive strain even in the absence of any antibiotic. This implies that if single resistance alleles are segregating in populations, double resistance can be a likely end result of natural selection, in environments where only glucose is present and the selective pressure of antibiotics is inexistent.
Fig. 1.
(a) Fitness of each double resistant mutant against a wild-type sensitive strain measured by competitive fitness assays; and resistance trait effects (b) growth rates and (c) carrying capacity. K88R, K43N, K88E, K43T, and K43R are the StrR backgrounds where the effect of the RifR alleles D516V, H526N, H526Y, and D516N was measured. All clones with fitness above one are expected to outcompete the sensitive strain (error bars represent 2SE, n > 4). 65% of the mutants with a combination of the two resistance alleles are beneficial.
For further understanding the consequences of the resistances, we studied their “trait effects”: Growth rate (r) and carrying capacity (K). Each of these traits contributes to the competitive ability of the resistant clones and has different relevance when we consider natural populations, which are likely structured (Hall et al. 2015< Hall et al. 2014>). For example in a metapopulation where extinction and recolonization occurs, the growth rate and carrying capacity will also be important determinants of the maintenance of resistance. Figure 1b indicates that only two RifR mutants are beneficial for r and all show a deleterious effect on their carrying capacity (fig. 1c), despite their competitive superiority. It also shows that many double resistant clones have increased growth rates.
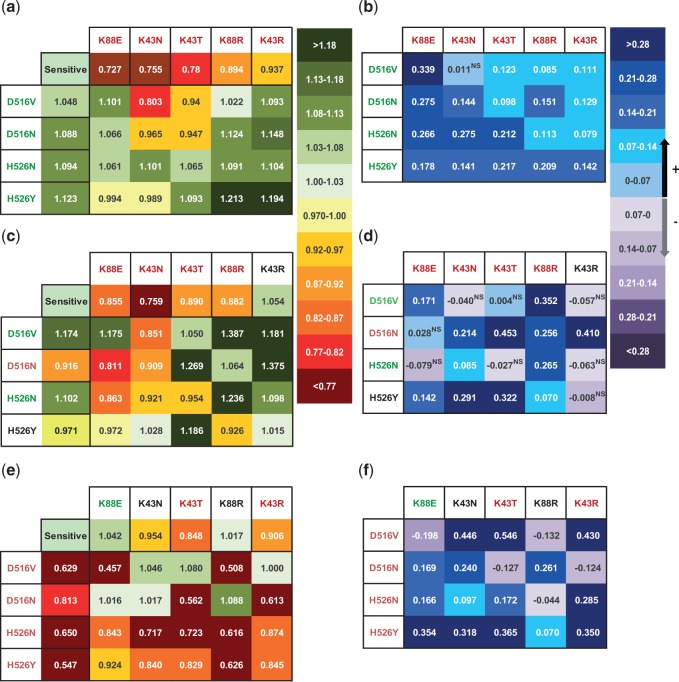
The absence of costs for competitive fitness in the majority of double mutants could be due to an epistatic interaction between the two. To answer whether this was the case, we estimated the level of pairwise epistasis between each pair of mutations. This is measured as the difference between the observed relative fitness of the double mutant (fig. 2a) and the expected fitness, based on the effects of each individual mutation: Ɛ = WRif.Str − WRif*WStr. Figure 2b shows the estimated values of epistasis for competitive fitness. We find that positive epistasis is detected between all pairs of RifR and StrR alleles, which are per se beneficial and deleterious, respectively, when considered individually. In only a single combination of double resistance (D516V/K43N) was the level of epistasis not significant, but still resulted in a positive mean. All other combinations showed a level of Ɛ significantly above 0.
Fig. 2.
Massive positive epistasis between RifR and StrR alleles. (a) Observed competitive fitness of single and double resistant clones. (c) Maximum growth rates and (e) carrying capacity. Level of pairwise epistasis at the level of competitive fitness (b), growth rate (d), and carrying capacity ( f ).
We also studied the pattern of epistasis for the traits r and K of the mutants versus the sensitive strains. Figure 2c–f shows that an overall level of positive epistasis is also observed for both r (mean Ɛ = 0.14, 95% CI [0.06, 0.21], 60% of the cases with significant positive epistasis) and K (mean Ɛ = 0.20, 95% CI [0.10, 0.30], 75% of the cases with positive epistasis). Hence, the costs of double resistance are also smaller than expected when these genotypes grow independently.
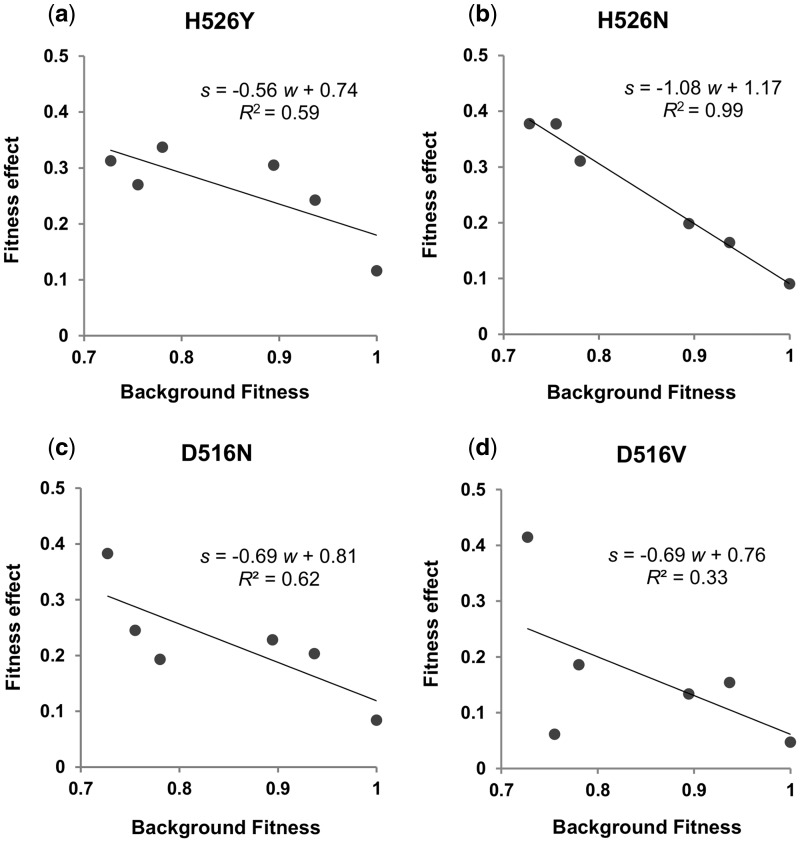
Next, we tested for a correlation between the effect of each beneficial resistance mutation and the fitness of the genetic background where it emerges. Recently, a negative correlation was found between the amount of fitness increase and the initial fitness in bacteria and yeast (Kryazhimskiy et al. 2014; Perfeito et al. 2014; Couce and Tenaillon 2015). Furthermore the effect of specific beneficial mutations was found to negatively correlate with the fitness of the genetic background (Kryazhimskiy et al. 2014), although the data supporting a general pattern are still limited. Figure 3 shows how the beneficial RifR mutations correlate with the background fitness. We observe a strong significant negative correlation between the fitness benefit of H526N mutation with the fitness of the genetic background it arises. For the mutation H526Y, a highly frequent mutation segregating in natural populations of Mycobacterium tuberculosis (Kapur et al. 1994; Yue et al. 2003; Gagneux et al. 2006), its fitness benefit correlates marginally with the fitness of the genetic background. Rodríguez-Verdugo et al. (2013) also found rifampicin alleles conferring higher fitness improvements in more maladapted genetic backgrounds. In the case of mutation D516V, it is always beneficial but its effect does not correlate with the background fitness.
Fig. 3.
Effect of genetic background for each beneficial RifR mutation. Diminishing returns epistasis (Kryazhimskiy et al. 2014) for H526N, that is, the fitness effect of H526N significantly decreases with the fitness of the genetic background (i.e., the cost of streptomycin resistance) (P < 0.001, F = 309.3). No significant correlation for alleles D516V, H526N, and H526Y is detected.
We have previously measured the level of pairwise epistasis for these alleles in rich medium (Luria–Bertani [LB]), an environment where none of these alleles individually was beneficial (Trindade et al. 2009). We can therefore compare how the level and type of epistasis changes with the environment and hence evaluate epistasis by environment interaction (supplementary figs. S2 and S3, Supplementary Material online). Flynn et al. (2013) have recently found that the sign and magnitude of epistasis between beneficial mutations in E. coli can vary with the environment where the cells are grown. For our sample of resistance alleles we observe a significant GxE interaction for the level of epistasis, when comparing poor and rich medium. This is observed when we consider competitive fitness (GxE, P < 0.05, ≈ 9% of the variance) and also the traits r (GxE, P < 0.05, ≈ 35% of the variance) and K (GxE, P < 0.05, ≈ 55% of the variance). Although all resistant clones have a smaller fitness, when competing with the sensitive strains in rich LB medium, some of the mutations confer beneficial effects at the level of r and K (supplementary figs. S2 and S3, Supplementary Material online).
Although the sample is small these results, together with those of previous reports (Remold and Lenski 2004; Lalic and Elena 2012; Zee et al. 2014), suggest that the type of epistasis may change as the environment changes. Our results also hint that epistasis between beneficial and deleterious resistance alleles may be more positive than that between costly resistance alleles.
Materials and Methods
The strains used were E. coli K12 MG1655 wild-type (ara+) and Δara. RifR and StrR clones were the same used in Trindade et al. (2009), which we previously showed to exhibit epistasis in rich medium. Briefly, sets of single spontaneous clones RifR or StrR were obtained by plating in LB agar medium supplemented with the appropriate antibiotic and randomly selecting clones after 24-h incubation at 37 °C. Single resistant clones were exposed to a second antibiotic to select for spontaneous mutants resistant to the two antibiotics. Generalized transduction of the resistance mutations with bacteriophage P1 was performed to eliminate unknown genetic background effects (Trindade et al. 2009). Four RifR clones were chosen for this study based on their superior fitness in minimal medium and five StrR clones were chosen based on their inferior fitness in minimal medium (Trindade et al. 2012). All 20 pairwise combinations giving rise to double resistance were tested. The fitness effects of the double resistance mutations were measured by competitive assays. The double resistant mutants were competed against a reference strain, E. coli K12 MG1655 Δara, in minimal medium supplemented with 0.4% of glucose at 37 °C, in an approximate proportion of 1:1, for 24 h with aeration. Accurate values of each strain initial and final ratios were estimated by plating appropriate dilutions of the mixture in Tetrazolium Agar plates. The fitness effect of each mutant strain—that is, the selection coefficient (s)—was estimated as the per generation difference in Malthusian parameters for the resistant strain and the reference strain: s = ln(Rf/Ri)/t, where t corresponds to the number of generations and Rf and Ri to the final and initial ratios between resistant and reference strains, respectively. In minimal medium, no cost of the Δara marker is detected. Five independent assays were done for each double resistant clone. The competitive fitness values of the double mutants in LB medium presented in supplementary figure S2, Supplementary Material online, are based on Trindade et al. (2009). The traits maximum growth rate (r) and carrying capacity (K) were determined at 37 °C using a 200 µl growth assay in a Bioscreen C Microbiology Reader (Growth Curves Ltd, Finland), after 2 days of acclimation to the growth conditions. Growths were started with 2 × 106 cells and a minimum of four independent assays were done for each single and double resistant clone. The OD600nm of cultures in the Bioscreen was measured every 20 min and the experiments were run for 24 h with continuous shaking (aeration). In these conditions, the growth curves did not display any evidence of death cell. Maximum growth rate was calculated as the maximal slope of the exponential phase using four points corresponding to 1-h time interval. Assuming a logistic growth model, the carrying capacity, or yield, was determined by measuring the final OD600nm after 24 h of growth as commonly used as a proxy for colony-forming units (MacLean and Buckling 2009). For LB, a 1:4 dilution was done before measuring the OD600nm (supplementary fig. S1, Supplementary Material online).
Epistasis (Ɛ) was calculated as in Trindade et al. (2009), that is, Ɛ = WRif.Str − WRif*WStr, where Wi.j is the competitive fitness of the strains, with resistances i and j, against a sensitive strain carrying a neutral marker. The significance of an epistatic interaction was determined by error propagation: The error of the value of Ɛ, σƐ =, as in previous studies (Trindade et al. 2009; Silva et al. 2011; Borrell et al. 2013). Whenever the value of ε was within the error we considered that alleles did not show any significant epistasis.
Supplementary Material
Supplementary figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals. org/).
Acknowledgments
This work was supported by PTDC/BIA-EVF/114622/2009 and PFE-GI-UE-ERC-2010-StG-260421. I.G. acknowledges the salary support of LAO/ITQB & FCT.
References
- Andersson DI, Hughes D. 2014. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 12:465–478. [DOI] [PubMed] [Google Scholar]
- Borrell S, Gagneux S. 2011. Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect. 17:815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell S, Teo Y, Giardina F, Streicher EM, Klopper M, Feldmann J, Muller B, Victor TC, Gagneux S. 2013. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health. 2013:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couce A, Tenaillon OA. 2015. The rule of declining adaptability in microbial evolution experiments. Front Genet. 6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JAGM, Cooper TF, Elena SF. 2011. The causes of epistasis. Proc R Soc Lond B Biol Sci. 278:3617–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JAGM, Krug J. 2014. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet. 15:480–490. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390:395–398. [DOI] [PubMed] [Google Scholar]
- Flynn KM, Cooper TF, Moore FB-G, Cooper VS. 2013. The environment affects epistatic interactions to alter the topology of an empirical fitness landscape. PLoS Genet. 9:e1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944–1946. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. 2000. The neutral theory in an infinite population. Gene 261(1):11-8. [DOI] [PubMed] [Google Scholar]
- Hall AR, Angst DC, Schiessl KT, Ackermann M. 2015. Costs of antibiotic resistance—separating trait effects and selective effects. Evol Appl. 8:261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T, Barton NH. 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162(1):395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V, Li LL, Iordanescu S, Hamrick MR, Wanger A, Kreiswirth BN, Musser JM. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 32(4):1095-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332:1193–1196. [DOI] [PubMed] [Google Scholar]
- Kondrashov DA, Kondrashov FA. 2015. Topological features of rugged fitness landscapes in sequence space. Trends Genet. 31(1):24-33. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344:1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalic J, Elena SF. 2012. Epistasis between mutations is host-dependent for an RNA virus. Biol Lett. 9:20120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, Buckling A. 2009. The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet. 5(3):e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC, Hall AR, Perron GG, Buckling A. 2010. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 11:405–414. [DOI] [PubMed] [Google Scholar]
- MacLean RC, Perron GG, Gardner A. 2010. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186:1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskinyte M, Gordo I. 2013. Increased survival of antibiotic-resistant Escherichia coli inside macrophages. Antimicrob Agents Chemother. 57:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Borrell S, Rose G, Gagneux S. 2013. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet. 29:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Sousa A, Bataillon T, Gordo I. 2014. Rates of fitness decline and rebound suggest pervasive epistasis. Evol Int J Org Evol. 68:150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 9:855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remold SK, Lenski RE. 2004. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet. 36:423–426. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Verdugo A, Gaut BS, Tenaillon O. 2013. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol Biol. 13:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salverda MLM, Dellus E, Gorter FA, Debets AJM, van der Oost J, Hoekstra RF, Tawfik DS, de Visser JAGM. 2011. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7:e1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk MF, Szendro IG, Salverda MLM, Krug J, de Visser JAGM. 2013. Patterns of epistasis between beneficial mutations in an antibiotic resistance gene. Mol Biol Evol. 30:1779–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RF, Mendonça SCM, Carvalho LM, Reis AM, Gordo I, Trindade S, Dionisio F. 2011. Pervasive sign epistasis between conjugative plasmids and drug-resistance chromosomal mutations. PLoS Genet. 7:e1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A, Magalhães S, Gordo I. 2012. Cost of antibiotic resistance and the geometry of adaptation. Mol Biol Evol. 29:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. [DOI] [PubMed] [Google Scholar]
- Trindade S, Sousa A, Gordo I. 2012. Antibiotic resistance and stress in the light of Fisher’s model. Evolution 66:3815–3824. [DOI] [PubMed] [Google Scholar]
- Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. 2009. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5:e1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H, Perron GG, Maclean RC. 2009. The cost of multiple drug resistance in Pseudomonas aeruginosa. J Evol Biol. 22:997–1003. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312:111–114. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Watson RA, Chao L. 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evol Int J Org Evol. 59:1165–1174. [PubMed] [Google Scholar]
- Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. 2003. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Clin Microbiol. 41(5):2209-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee PC, Mendes-Soares H, Yu Y-TN, Kraemer SA, Keller H, Ossowski S, Schneeberger K, Velicer GJ. 2014. A shift from magnitude to sign epistasis during adaptive evolution of a bacterial social trait: brief communication. Evolution 68:2701–2708 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.