Abstract
Molecular chaperones fold many proteins and their mutated versions in a cell and can sometimes buffer the phenotypic effect of mutations that affect protein folding. Unanswered questions about this buffering include the nature of its mechanism, its influence on the genetic variation of a population, the fitness trade-offs constraining this mechanism, and its role in expediting evolution. Answering these questions is fundamental to understand the contribution of buffering to increase genetic variation and ecological diversification. Here, we performed experimental evolution, genome resequencing, and computational analyses to determine the trade-offs and evolutionary trajectories of Escherichia coli expressing high levels of the essential chaperonin GroEL. GroEL is abundantly present in bacteria, particularly in bacteria with large loads of deleterious mutations, suggesting its role in mutational buffering. We show that groEL overexpression is costly to large populations evolving in the laboratory, leading to groE expression decline within 66 generations. In contrast, populations evolving under the strong genetic drift characteristic of endosymbiotic bacteria avoid extinction or can be rescued in the presence of abundant GroEL. Genomes resequenced from cells evolved under strong genetic drift exhibited significantly higher tolerance to deleterious mutations at high GroEL levels than at native levels, revealing that GroEL is buffering mutations in these cells. GroEL buffered mutations in a highly diverse set of proteins that interact with the environment, including substrate and ion membrane transporters, hinting at its role in ecological diversification. Our results reveal the fitness trade-offs of mutational buffering and how genetic variation is maintained in populations.
Keywords: GroEL, mutational buffering, experimental evolution, Escherichia coli
Introduction
A population may harbor genetic variants that are adaptive, neutral or slightly deleterious in a given environment. Some neutral variants do not have any phenotypic manifestation and are referred to as phenotypically silent or cryptic variants. A number of experiments have demonstrated that these variants bear preadaptive mutations that can spark innovations in new environments (Rutherford and Lindquist 1998; Keys et al. 1999; Zakany and Duboule 1999; Queitsch et al. 2002; True and Carroll 2002; Amitai et al. 2007; Romero and Arnold 2009; Tokuriki and Tawfik 2009; Hayden et al. 2011; Hayden and Wagner 2012; Tokuriki et al. 2012; Rohner et al. 2013). Many of these variants are silent thanks to mechanisms of mutational robustness that buffer their slightly deleterious effects on fitness and facilitate their persistence in a population. However, even though characterizing mechanisms of mutational buffering is a fundamental aim of evolutionary biology (de Visser et al. 2003), it remains unclear how these mechanisms exactly act, what their metabolic cost is, and under what conditions could they fuel evolution.
A known mechanism of mutational robustness is provided by molecular chaperones. These proteins assist the folding of many other proteins and prevent the formation of nonspecific protein aggregates in the cell through noncovalent interactions (Hartl and Hayer-Hartl 2009). They help fold proteins regardless of whether—or even because—these proteins bear destabilizing mutations. This means that through the correct folding of mutated proteins, molecular chaperones can allow the persistence of destabilizing mutations in a population. They are generally considered modulators of the relationship between a protein’s primary sequence and its structure—that is, between genotype and phenotype. Pioneering work in this area demonstrated that impairment of the folding activity of Hsp90, an essential chaperone in signal transduction pathways in eukaryotes, can unveil cryptic genetic variation causing the emergence of a large number of aberrant phenotypes in populations of Drosophila melanogaster (Rutherford and Lindquist 1998). Similar observations were made in the plant Arabidopsis thaliana (Queitsch et al. 2002). Moreover, in duplicated kinases, a protein that requires Hsp90 for folding evolves faster than a closely related protein that does not require Hsp90 and is encoded by a duplicate gene (Lachowiec et al. 2015). A link between the ability of Hsp90 to increase morphological variation and the emergence of novel adaptations was also revealed in natural surface populations of the fish Astyanax mexicanus (Rohner et al. 2013). Impairing Hsp90 in this species leads to the phenotypic manifestation of developmental variants (e.g., eyeless phenotypes) that are better adapted to the dark environment of a cave.
In bacteria, GroEL is an essential molecular chaperone that promotes the evolution of its client proteins, which are those proteins requiring GroEL’s assistance for folding (Bogumil and Dagan 2010; Williams and Fares 2010). GroEL is a member of the class of chaperones known as chaperonins, which are large double-ring complexes that enclose client proteins for folding. Specifically, GroEL has heptameric rings and cooperates with the cochaperonin GroES, which forms the lid of a folding cage (Hartl et al. 2011). GroEL seems to play a key role in the mutualistic symbiosis of bacteria and insects by buffering the effects of deleterious mutations accumulated during the bottlenecks that the bacterial populations experience in every transfer between host generations (Moran 1996; Fares et al. 2005). Most of these mutations are protein-destabilizing mutations (van Ham et al. 2003). Interestingly, groE, the operon encoding both GroEL and its cochaperone GroES, is highly expressed in all endosymbiotic bacteria of insects known to date, probably to satisfy the demand for folding activity in cells possessing a large load of deleterious mutations. In support of this hypothesis, groE overexpression rescues bacterial cells that have declined in their fitness after having been experimentally evolved under the effect of strong genetic drift (Moran 1996; Fares, Barrio, et al. 2002; Fares, Ruiz-Gonzalez, et al. 2002; Fares et al. 2004). Despite the apparent ability of GroEL in rescuing deleterious phenotypes, whether GroEL buffers mutations in a particular type of proteins and not others and whether such buffering has a cost for the cell remain largely unexplored.
Evidence that mutational buffering is enhanced in certain organisms (e.g., endosymbiotic bacteria) but not others (e.g., free-living bacteria) suggests that a cost is associated with increasing the activity responsible of the buffering. In this study we have conducted experiments of laboratory evolution followed by genome resequencing and comparative genomics in Escherichia coli to determine the cost of overexpressing groE, as well as GroEL's role in allowing the survival of mutations.
Results
Experimental Evolution of E. coli with Enhanced groE Expression
To evaluate the fitness cost associated with groE overexpression, we performed an evolution experiment under two conditions: 1) Populations evolving under very strong genetic drift imposed by frequent single-colony bottlenecks, and 2) populations evolving under mild genetic drift imposed by serial transfers (fig. 1). All populations were initiated from a single hypermutable clone of E. coli that lacks the DNA mismatch repair gene mutS (ΔmutS) and has a mutation rate 3 orders of magnitude higher than the wild type. This ancestral strain was transformed with a 15-copy plasmid containing the operon groE, which encodes both GroES and GroEL under the control of a promoter inducible by l-arabinose (pGro7). We refer to this plasmid-bearing strain as pgroE. When l-arabinose is present in the growth medium, groE is expressed at very high levels, whereas in the absence of l-arabinose groE is basally expressed at a higher level than that of the wild type owing to the presence in the cell of the 15-copy plasmid (fig. 2). We used as a control the same strain of E. coli transformed with a plasmid that lacked the operon groE but was otherwise identical to the plasmid pGro7. We refer to this strain as pΔgroE. We performed all evolution experiments under a temperature of 37 °C (see Materials and Methods).
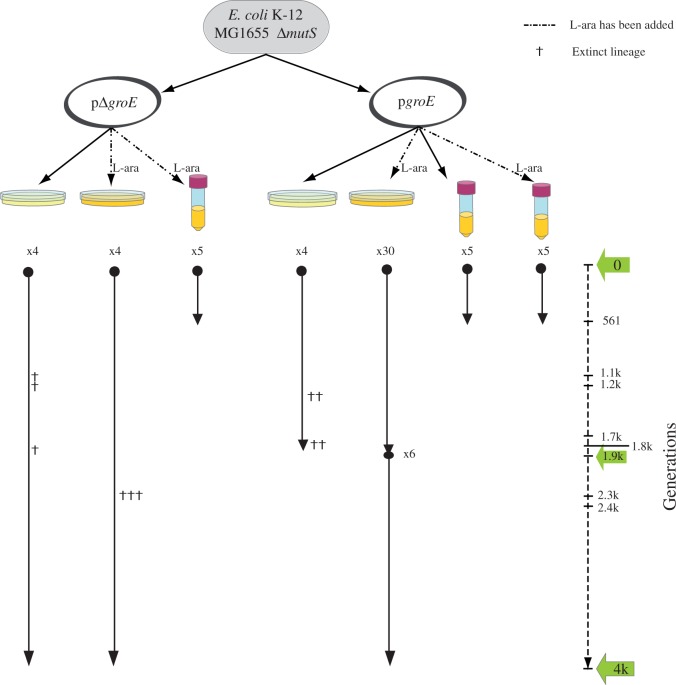
Fig. 1.
Evolutionary history of populations evolved in this study. A single ancestor Escherichia coli K-12 MG1655 population lacking the repair gene mutS was evolved for hundreds to thousands of generations. Two evolution lines were derived from the ancestral population. One line included a 15-copy plasmid containing the operon groE under the control of the l-arabinose inducible promoter (pgroE) and the other line (referred to as the control), which contained the plasmid without the operon groE (pΔgroE). Each of the populations evolved under the effect of weak-to-strong genetic drift at a temperature of 37 °C. The number of independently evolved lines is indicated with an x followed by a number. The right-most dashed line represents the length (in generations) of the evolution experiment, which took about 1,900 (1.9K) and 4K generations of E. coli for populations evolving under strong genetic drift, and 561 generations in the case of populations evolving under mild genetic drift. Green arrows indicate the time points at which bacterial fitness was assayed.
Fig. 2.
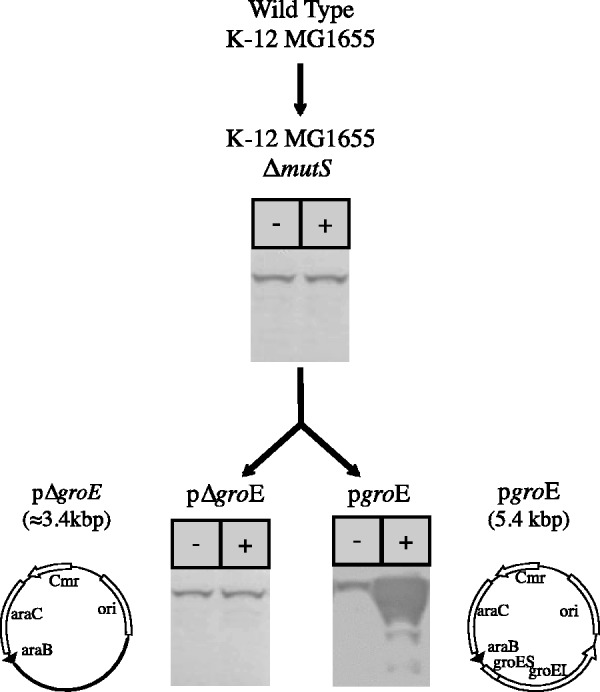
Outline of the Escherichia coli variant strains built in this study and the expression of the groE operon in each of the strains under different conditions. K-12 MG1655 ΔmutS is a hypermutable strain lacking the DNA repair gene mutS obtained from K-12 MG1655. The plasmids pGro7 (Takara Inc. # 3340) and pΔgroE (modified from pGro7) were introduced in K-12 MG1655 ΔmutS, obtaining the strains pgroE and pΔgroE. The strain pgroE presents a higher level of GroEL than the wild type, corresponding to the 15-plasmid copies containing groE operon and basally expressed, when grown in media without inducer (−). In the presence of the inducer l-arabinose (+), groE is overexpressed several orders of magnitude above the constitutive level. The control lines pΔgroE contain the plasmid lacking the operon groE, otherwise is identical to all other strains. Western images correspond to lines grown at 37 °C. GroEL is stained using monoclonal antibodies.
For the evolution through single-colony bottlenecks, and thus under strong genetic drift, a single pgroE colony was passaged daily to a fresh plate. Thirty independent replicate lines were evolved in the presence of l-arabinose and four replicate lines in its absence. Growing pgroE in the absence of l-arabinose controls for the effect of this sugar. At the same time, it allowed us to evaluate the incidence of mutations that silence the plasmid’s groE operon in populations that evolve in the absence of the inducer. Likewise, we evolved eight replicate lines of the control strain pΔgroE: Four lines evolved in the presence of l-arabinose and the other four in its absence. For evolution under weak genetic drift, we serially transferred ten populations of pgroE by 100-fold dilution into a fresh flask. Five of the ten populations evolved in the presence of the inducer and the other five in its absence. Five populations of the control strain pΔgroE were evolved in this same manner in the presence of the inducer (fig. 1).
We evolved the bottlenecked populations for up to 180 passages, corresponding to 4,000 (4K) generations. Importantly, not all populations survived for the entire length of the experiment, and extinctions (symbolized by a cross on the corresponding lineages in fig. 1) affected the evolving lines differently. Specifically, 10 of the 12 bottlenecked populations evolving without the overexpression of groE—eight pΔgroE and four pgroE without inducer—went extinct around 1.8K and 2.3K generations. In contrast, all 30 lineages with groE overexpression—pgroE with inducer—continued growing up to 1.9K generations. To ensure that overexpression of groE was providing significantly larger chances to survive, we continued the evolution of 6 of the 30 surviving bottlenecked pgroE populations in groE overexpression conditions for 2K additional generations, with all 6 surviving for 4K generations of the evolution experiment.
We also evolved ten large populations for 85 days or passages, corresponding to approximately 561 generations of E. coli (log2(100) = 6.6 generations per passage) (fig. 1). In contrast to the observed dynamics of bottlenecked populations, all ten populations evolving under the effect of mild genetic drift survived until the end of the experiment. Taken together, these observations suggest that when the effect of genetic drift is strong, those populations with high GroEL levels survive preferentially, because GroEL is present in sufficient amounts to buffer the deleterious effects of mutations. In contrast, when genetic drift is weaker, most deleterious mutations are purified by selection regardless of groE expression. We next explore the possibility that the overexpression of groE in large populations imposes a fitness cost that is not compensated by the benefits of its buffering capacity.
Enhancing GroEL Mutational Buffering Has a High Fitness Cost
Increasing groE expression may enhance the buffering of the deleterious effects of protein-destabilizing mutations. However, with the exception of endosymbiotic bacteria of insects, bacteria do not increase groE expression under normal conditions, possibly due to a large fitness cost associated with high-levels of groE expression. Here, we investigated the fitness cost associated with groE overexpression and whether this cost could be counterbalanced under conditions where mutational buffering is highly needed in the cell. To this end, we examined groE expression levels in the ten evolving pgroE populations and the four control populations of pΔgroE cells. Of the ten populations of pgroE cells, five evolved in the presence of l-arabinose and five in its absence (fig. 1). As explained earlier, the effect of genetic drift in these ten populations was lower than in the bottlenecked populations, and therefore the cost of groE overexpression should be higher in them.
At the start of the experiment (generation 0), the pgroE populations that were growing in the presence of the inducer l-arabinose showed higher levels of groE expression than pgroE populations growing in the absence of l-arabinose. The latter populations, in turn, presented higher groE expression than the control pΔgroE populations (left gel of fig. 3a). However, as populations continued evolving (e.g., at generations 66 and 561), groE expression in the presence of l-arabinose declined dramatically, returning to levels comparable to those of the control lines in all five populations (left gel of fig. 3a and b). This result strongly points to a high cost of groE overexpression, which may have resulted in the rapid loss of groE overexpression and the accumulation of cells with substantially lower groE levels than in the initial pgroE population. The rapid loss of groE overexpression suggests that the ability of groE to compensate the effects of deleterious mutations in the hypermutable strains did not counterbalance the fitness costs of high GroEL levels.
Fig. 3.
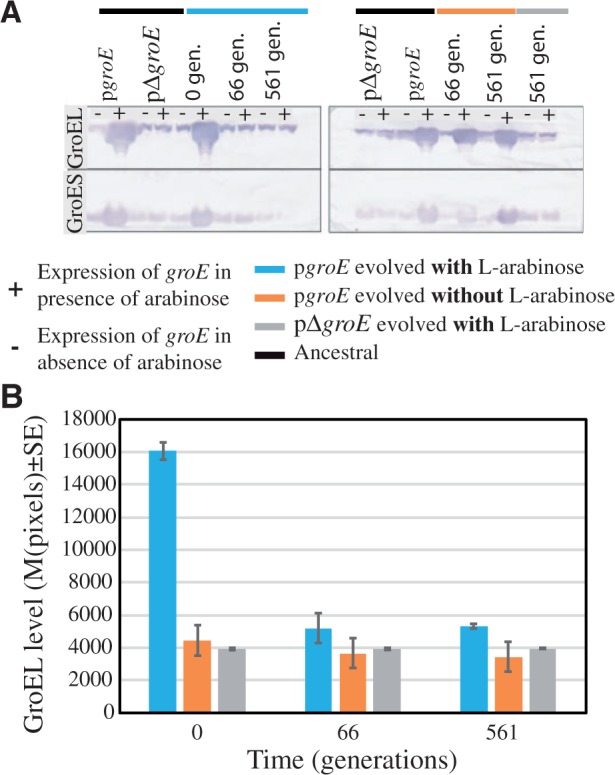
Reduction of GroEL levels in Escherichia coli MG1655 ΔmutS populations evolving under mild genetic drift effects. (A) GroEL detection by quantification of protein dot-blot hybridized with monoclonal GroEL antibody (ABCam #ab905022 at 1:1,000). The left panel corresponds to groE expression in the presence of l-arabinose (pgroE) and its control pΔgroE at the start of the evolution experiment (0 generations), 10 passages (66 generations), and 85 passages (561 generations). The right panel corresponds to the same experiment as in the left panel but without the presence of the inducer l-arabinose (groE). (B) Expression of groE quantified using the mean pixels ± SE from the Western blots in (A) at different time points expressed as generations. The different lines examined were labeled in blue (pgroE growing in the presence of l-arabinose), orange (pgroE growing in the absence of l-arabinose), and gray (pΔgroE).
If groE overexpression is costly and this cost is reduced by mutations, then cells that have evolved in the absence of the inducer l-arabinose should retain their ability to overexpress groE when induced with l-arabinose. Indeed, pgroE populations that were evolved in the absence of l-arabinose for 1.8K generations overexpressed groE when they were induced with l-arabinose at generations 66 and 561 (right gel in fig. 3a). These observations again point to a large cost of groE overexpression that led to selection against cells with high GroEL levels. Taken together, these results demonstrate that overexpression of groE is costly and deleterious, although not lethal, in large microbial populations.
To quantify the fitness cost of groE overexpression, we competed head-to-head the ancestral pgroE cells against the ancestral pΔgroE cells in the presence of the inducer l-arabinose (see Materials and Methods). We observed that the fitness of pgroE declined significantly compared with pΔgroE (Wrel = 0.67 ± 0.05) varying between 0.62 and 0.72. Therefore, overexpression of groE had an average fitness cost between 38% and 28%.
Overexpression of groE Is Tolerated under Strong Genetic Drift and Buffers the Effects of Mutations
In the evolution experiment through single-colony transfers (fig. 1), we noticed that all bottlenecked populations that were overexpressing groE (i.e., pgroE cells growing in the presence of l-arabinose) persisted under strong genetic drift for over 4K generations. In contrast, bottlenecked populations that evolved without the overexpression of groE (i.e., pgroE populations evolving in the absence of l-arabinose, and pΔgroE populations) became extinct after 1.8K and 2.3K generations. In fact, the majority of the control lines (10 of the 12 lines: 6 of the 8 pΔgroE lines and 4 of the 4 pgroE lines evolving in the absence of arabinose) could not be transferred after 1.8K or 2.3K generations (fig. 1).
To ascertain that the pgroE lines evolving in the presence of l-arabinose had not silenced groE overexpression, we compared groE expression levels in the pgroE lines evolved with l-arabinose for 1.9K generations with that of the control lines (pΔgroE) at different time points of the evolution experiment (fig. 4). Although groE overexpression diminished during evolution relative to the ancestor of the pgroE lines, it remained significantly higher than in the control lines, and this trend continued until the end of the experiment (fig. 4). The extinction of populations growing without groE overexpression and the persistence of those populations evolving with groE overexpression suggest that overexpression of groE is required, at a relatively early stage of the evolution experiment (i.e., during the first 1.8K generations of evolution), when the strength of genetic drift is high, possibly to buffer the effects of deleterious mutations.
Fig. 4.
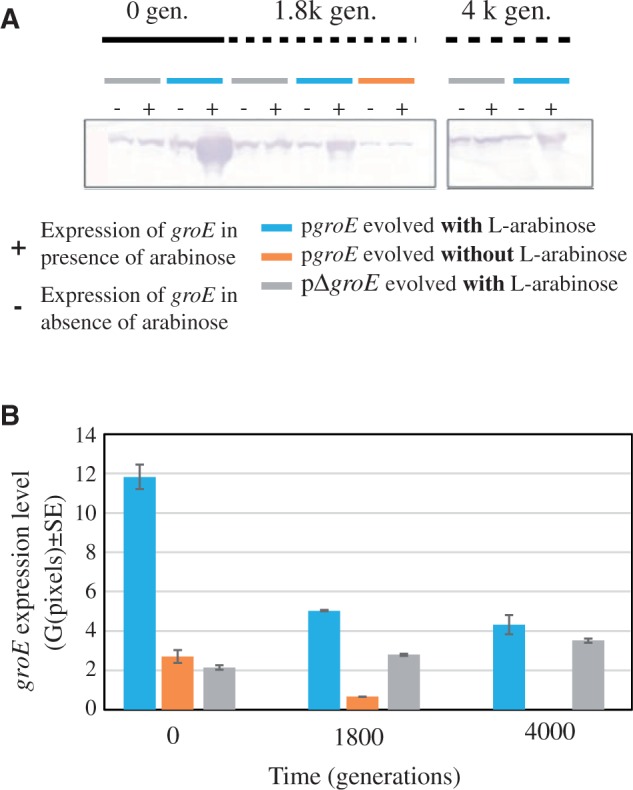
High expression levels of groE are tolerated in populations evolving with strong genetic drift effects. (A) Expression of groE was measured at three points of the evolution of bottlenecked populations: Generation 0, generation 1.8K, and generation 4K. We compared the expression of three lineages, the control lineage pΔgroE (gray horizontal bar), the lineage with groE under the control of l-arabinose inducible promoter (pgroE) evolved in the presence of l-arabinose (blue bar), and the same lineage pgroE evolved in the absence of l-arabinose (orange bar). (B) We quantified the expression level as the mean number of pixels (±SE) by Western blotting using hybridization with monoclonal GroEL antibody (ABCam #ab905022 at 1:1,000) at different time points expressed as generations. Bars correspond to lines evolved at 37 °C. Bottlenecked lines were rescued at these time points from glycerol stocks and grown simultaneously under arabinose induction conditions (+) or in media without the inducer arabinose (−).
The Extinction of Cell Lines without groE Overexpression Evolving under Strong Genetic Drift Is due to the Deleterious Effects of Accumulated Mutations
Most of the cell lines evolving under strong genetic drift and without groE overexpression were extinct before 2.4K generations. To ascertain that declining fitness caused these extinctions, we compared the fitness of the evolved strains isolated one passage before their extinction with that of their ancestors (see Materials and Methods). Two principal scenarios could explain their extinction: 1) These lines accumulated mutations with deleterious effects that led to a gradual decline in fitness during evolution and 2) the extinction of these lines was due to the lethal effect produced by new mutations emerging in an already highly deleterious genomic background.
We revived the four extinct pgroE cell lines that had evolved in the absence of l-arabinose from the frozen record at a passage predating their extinction, and reinitiated their evolution in two conditions: 1) In the presence of l-arabinose (i.e., under groE overexpression), and 2) in the absence of l-arabinose. None of the four revived lines could be passaged into further plates without l-arabinose—they became extinct despite several attempts to revive them. However, when we grew these lines in the presence of l-arabinose, they persisted until we stopped the evolution experiment more than 220 generations of evolution beyond their extinction point. These results support the notion that cells were close to the threshold of the “error catastrophe” (Eigen 1971, 2002) at which novel mutations would lead to their extinction. Rescue of these cells in the presence of abundant GroEL levels indicates that the mutational buffering provided by GroEL is important and that GroEL may have buffered a large set of mutations that had accumulated in the cell in an uncontrolled way.
To determine the changes in the fitness of the evolved strains with and without groE overexpression in bottlenecked populations, we compared 1) the fitness of the evolved strains overexpressing groE (pgroE) with that of their ancestral strain, and 2) the fitness of the evolved control lines without groE overexpression (pΔgroE) against that of their ancestral strain. We performed these competitions experiments at generations 1.8K (for which we had 2 surviving pΔgroE lines and all 30 pgroE lines) and 4K (for which we had one surviving lineage from pΔgroE, evolved in the presence of l-arabinose, another surviving lineage from pΔgroE evolved in the absence of l-arabinose, and all six pgroE lineages). All pgroE populations that evolved in the absence of l-arabinose became extinct before 1.8K generations and we could therefore not use them for fitness calculations.
We observed that the fitness of pgroE lines (overexpressing groE) decreased more than that of pΔgroE cells during the first 1.8K generations (fig. 5). However, pΔgroE lines declined in fitness to a significantly greater extent than pgroE lines. The single surviving pΔgroE population approached extinction at 4K generations, whereas the pgroE populations increased their fitness during their evolution from 1.8K to 4K generations (fig. 5, supplementary table S1, Supplementary Material online). As bottlenecked populations are subject to Muller’s ratchet (the irreversible accumulation of deleterious mutations in small clonal populations), this result suggests that GroEL expression may have helped the spreading of mutations that compensate the effects of deleterious mutations, but this requires further investigation.
Fig. 5.
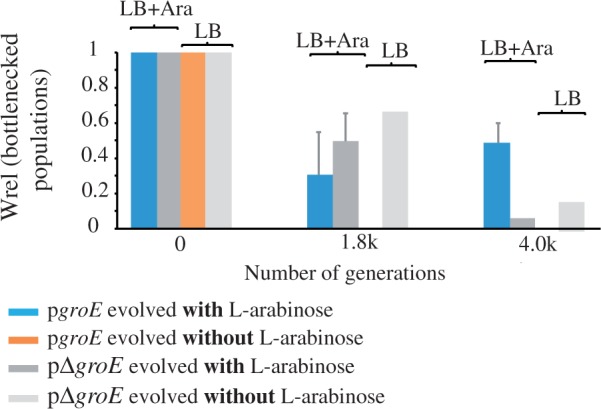
Relative fitness of bottlenecked populations. Average relative fitness (Wrel ± SE) of the evolved bottlenecked lines measured at 0, 1,870 (1.8K), and 3,960 (4K) generations of their evolution. Wrel data were obtained by head-to-head competitions of the evolved lines against their ancestors. Populations were grown for the Wrel quantification in the same kind of media used for their evolution, LB supplemented with the inducer arabinose and LB media without the inducer. Blue bars correspond to pgroE lines evolved with l-arabinose, dark gray to pΔgroE lines evolved with l-arabinose, light gray to pΔgroE lines evolved without l-arabinose, and orange to pgroE evolved without l-arabinose. Data to build this graph are presented in supplementary table S1, Supplementary Material online.
The Mutational Spectrum of the Evolving E. coli Cells
All the analyses performed at this point demonstrate that groE overexpression is taxing and only tolerated when cells evolve under strong genetic drift caused by strong population bottlenecks. Such populations mostly persisted if groE was overexpressed, suggesting that high levels of GroEL are required to buffer the effects of deleterious mutations. To study the impact of groE overexpression on genome evolution, we resequenced the genomes of a clone with groE overexpression (e.g., pgroE) and a control clone (pΔgroE) evolved through single-colony transfer, and did so at two time points (1.8K and 4K generations). Then, we determined and compared the mutational spectra of these clones (supplementary tables S2–S5, Supplementary Material online).
Contrary to what one might expect, the pgroE clone had accumulated fewer nonsynonymous mutations (i.e., those nucleotide substitutions that cause amino acid replacements in the encoded protein) than the pΔgroE clone, despite the mutational buffering that GroEL might provide to pgroE populations (table 1). Specifically, the control clone that evolved in the absence of groE overexpression exhibited more than twice as many nonsynonymous mutations as the clone overexpressing groE (table 1). This observation could have three principal causes. The first is a lack of mutational buffering by GroEL, which seems unlikely given that pgroE can protect against population extinction. The second potential cause is that the fewer mutations observed when overexpressing groE might have a larger fitness effect.
Table 1.
Mutational Spectra of Bacteria Evolved with Enhanced GroEL Levels.
| Generation | Lineages | Coding Regions |
Intergenic Spacers | |||
|---|---|---|---|---|---|---|
| NSNPs | SSNPs | Insertions | Deletions | |||
| 1.8K | pgroE | 133 | 73 | 17 | 30 | 64 |
| pΔgroE | 291 | 84 | 22 | 4 | 70 | |
| 4K | pgroE | 179 | 109 | 20 | 37 | 79 |
| pΔgroE | 636 | 117 | 18 | 12 | 137 | |
Note.— NSNPs, nonsynonymous single nucleotide polymorphisms; SSNPs, synonymous single nucleotide polymorphisms.
In support of the latter hypothesis, the number of strongly protein disruptive mutations, which were caused by single nucleotide insertions and deletions (indels), was larger in the pgroE line (47 indels, corresponding to 26.11% of all coding-disrupting mutations) than in the pΔgroE line (26 indels, corresponding to 8.2% of all disruptive mutations), a difference that is significant (Fisher’s exact test: Odds ratio F = 3.94, P = 1.41 × 10−7).
In a related analysis, we studied the kinds of amino acid replacements that occurred in our evolved lines in terms of three physicochemical amino acid features: 1) Polarity, 2) charge, and 3) polarity and volume. We considered an amino acid substitution “radical”—and thus likely to have a deleterious effect—if it occurred between two amino acids belonging to two different physicochemical groups (e.g., if it changed the neutral amino acid glycine into the positively charged amino acid arginine). In this way, we found that 91% of all changes in the pgroE line (163 of 180 changes) were radical. In contrast, only 80% of changes were radical in the pΔgroE control line (254–317 changes). The proportion of radical changes in the groE line was significantly higher (Fisher’s exact test: Odd’s ratio F = 2.374, P = 0.002). Analyses of the genomes of pgroE and pΔgroE evolved for 4K generations yielded similar results: The number of indels was greater in the pgroE line than it was in the pΔgroE line (24.15% and 4.55% of all changes were indels in the pgroE and pΔgroE, respectively. Fisher’s exact test: F = 6.73, P = 3.86 × 10−22). In addition, the proportion of radical amino acid substitutions was significantly higher in the pgroE line than in the pΔgroE line (88% and 74% radical changes, respectively; Fisher’s exact test: F = 2.48, P = 1.08 × 10−5).
The third potential cause of the lower number of mutations under groE overexpression regards primarily those proteins that do not interact with GroEL. If the cost of groE overexpression interacts synergistically with amino acid changes in these proteins, increasing their deleterious effects, then selection may preferentially eliminate such changes, which might explain their lower numbers. However, an analysis focusing on mutations in proteins known to interact with GroEL (Fujiwara et al. 2010), did not provide sufficient statistical power to test this hypothesis, because the number of such proteins with nonsynonymous mutations was very low.
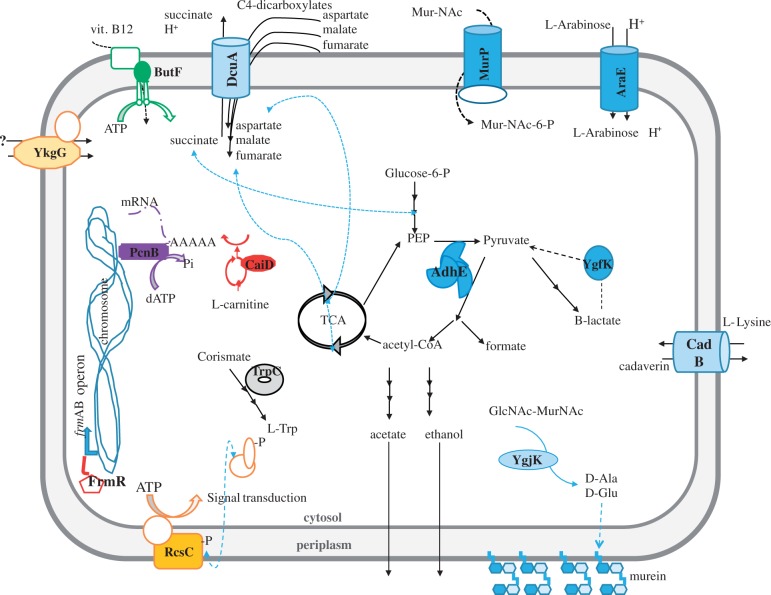
We also examined the distribution of genes with nonsynonymous mutations and indels among the different gene ontology (GO) categories of gene functions (Harris et al. 2004). If GroEL buffers a subset of mutations, then some GO categories for mutated genes might be enriched in the presence of groE overexpression (pgroE) but not in its absence (pΔgroE). To find out, we compared the distribution of mutated proteins under these two conditions using GO terms as provided by AmiGO2 (http://amigo.geneontology.org), using the three GO levels of biological processes, molecular functions, and cellular components (Carbon et al. 2009). The control line pΔgroE showed no enrichment for mutated genes in any of the GO categories. In contrast, the pgroE line was substantially enriched for mutated genes in several GO categories for all three levels of GO functions (table 2). Most affected categories were related to energy production and transmembrane transport of ions and substrates (fig. 6). Examples include genes encoding the proteins DcuA, involved in dicarboxylate uptake, MurP, a component of the N-acetilmuramic acid PTS (Phosphotransferase protein) permease, and CadB, a lysine-cadaverin antiporter (fig. 6).
Table 2.
Biased Distribution of Genes with Nonsynonymous Mutations in Bacteria Evolved with Enhanced GroEL Levels (pgroE).
| Generation | GO Classificationa | Enriched GO Categoryb | Probabilityc |
|---|---|---|---|
| 1.8K | Biological processes | • Single organism process | 1.25 × 10−4 |
| • Single organism cellular process | 2.39 × 10−2 | ||
| • Energy derivation by oxidation | 7.35 × 10−3 | ||
| • Cellular respiration | 2.53 × 10−3 | ||
| Molecular functions | • Cation transmembrane transporter | 6.54 × 10−3 | |
| • Anion activity | 1.15 × 10−2 | ||
| • Ion activity | 2.45 × 10−2 | ||
| • Substrate specific transmembrane transporter activity | 4.18 × 10−2 | ||
| • Carbohydrate derivative activity | 5 × 10−2 | ||
| Cellular components | • Cellular component | 4.61 × 10−3 | |
| • Membrane | 8.74 × 10−3 | ||
| • Plasma membrane | 1.17 × 10−2 | ||
| • Cell part | 1.60 × 10−2 | ||
| • Cell | 1.60 × 10−2 | ||
| • Cell periphery | 1.97 × 10−2 | ||
| 4K | Biological processes | • Single organism process | 3.65 × 10−5 |
| • Single organism cellular process | 1.86 × 10−2 | ||
| • Cellular respiration | 1.68 × 10−2 | ||
| • Energy derivation by oxidation | 4.82 × 10−2 | ||
| Molecular functions | • Ion binding | 1.99 × 10−3 | |
| • Catalytic activity | 7.34 × 10−3 | ||
| • Anion binding | 2.02 × 10−2 | ||
| • Molecular function | 3.63 × 10−2 | ||
| Cellular components | • Cellular component | 3.92 × 10−3 | |
| • Membrane | 1.33 × 10−2 | ||
| • Plasma membrane | 3.48 × 10−3 | ||
| • Cell part | 7.57 × 10−3 | ||
| • Cell | 7.57 × 10−3 | ||
| • Cell periphery | 1.17 × 10−2 | ||
| • Oxidoreductase complex | 4.53 × 10−2 |
aGO major categories.
bGO categories enriched for genes with nonsynonymous SNPs.
cProbability of enrichment of GO categories under a hypergeometric test corrected for multiple testing using Bonferroni correction.
Fig. 6.
Subcellular localization of proteins mutated in the evolved populations under variable GroEL conditions. A cell-like structure with periplasmic space between inner- and outermembranes has been used to highlight the cellular position and affected pathways detected in evolving populations. The mutated proteins in the presence of groE overexpression are labeled in bold and indicated with solid color forms. When the affected protein belongs to a complex (e.g., the protein MurP), the other subunits are represented as empty forms with the same color lines. Colors are grouped by GO terms, so that all proteins belonging to the same molecular function are color-coded equally.
Taken together, our results support the hypothesis that groE overexpression buffers the accumulation of strongly deleterious mutations in a subset of proteins. However, this enhanced buffering does have a fitness cost. In addition, it may constrain the fixation of deleterious mutations in proteins that do not interact with GroEL.
Discussion
We show that the overexpression of the chaperonin system groEL–groES, known to increase the resistance of phenotypes to mutational insults in bacteria (Moran 1996; Fares, Barrio, et al. 2002; Fares, Ruiz-Gonzalez, et al. 2002), can only be tolerated in populations evolving under strong genetic drift. The reason is the fitness cost of overexpressing groE. Enhancing the production of this large molecular complex has a great energetic cost for the cell. The translation of a single amino acid costs four ATP molecules, and the production of GroEL/S amounts to the translation of 8,351 amino acids: 7 GroES subunits (7 × 97 amino acids) and 14 GroEL subunits (14 × 458 amino acids) (Bogumil and Dagan 2012). Besides, GroEL-mediated folding consumes more ATP: Seven molecules in each encapsulation cycle (Hartl et al. 2011). Therefore, when a population’s effective size is large and selection is more efficient (e.g., in the populations under mild genetic drift, the effective population size Ne ≈ 106, calculated from the CFUs = 2.3 × 108 and 4.8 × 108 when diluted by 100 in the passages) cells expressing groE at a much higher level than the wild type are quickly purged from the population. In contrast, such levels of groE expression can be maintained for thousands of generations in periodically bottlenecked populations, where the effect of genetic drift is large. High levels of GroEL are likely maintained in these populations because overexpression of groE compensates the deleterious effects of mutations that are not purged by selection. Consistent with this hypothesis is our observation that low expression of groE in cell lines under strong genetic drift and large mutational loads leads to massive population extinctions.
Previous studies have shown that GroEL can rescue bottlenecked populations with declining fitness (Fares, Ruiz-Gonzalez, et al. 2002; Maisnier-Patin et al. 2005) and allow the fixation of adaptive mutations in proteins that require GroEL for folding (Bogumil and Dagan 2010; Williams and Fares 2010). In this study, we also characterize the cost–benefit trade-off of this mutational buffering system.
In addition, we show by genome sequencing that GroEL’s mechanism of buffering may be more complex than thought before, and that the set of proteins susceptible to this mechanism may be larger than anticipated. Relevant in this regard is our observation that populations overexpressing groE and evolving under strong genetic drift accumulate disruptive mutations in only some functional classes of proteins, those involved in transport of ions and substrates across membrane and enzymes. Most of the diversification enabled by GroEL in our experiment concerns proteins that are at the cell–environment interface and that may enable adaptations to other substrates or conditions, hinting at their role in ecological diversification. In contrast, populations with wild type groE expression levels do not show this bias, which is likely caused by the fact that GroEL only buffers a subset of all proteins in the cell.
Only a small fraction of proteins (a maximum of 11 and minimum of 4 proteins in total) known to require GroEL for folding (Fujiwara et al. 2010) undergo amino acid changes, probably because most GroEL clients are essential proteins and their mutation can lead to lethal phenotypes (Williams and Fares 2010). However, the list of known GroEL clients is likely to miss many proteins that interact with GroEL in a transient manner. Because GroEL helps fold partially folded proteins through nonspecific interactions with their hydrophobic exposed regions, it is likely that proteins not previously reported as GroEL clients can become so once destabilized by mutations. We hypothesize therefore that the list of GroEL clients may be substantially longer than thought before and that being a GroEL client may depend on the environment, the genotype, and the environment-by-genotype interactions. But this deserves further investigation.
Systems such as GroEL that increase the robustness of proteins to destabilizing mutations, including functionally innovative mutations (Wang et al. 2002; Tokuriki et al. 2008), can increase a population’s mutational “reservoir” in protein-coding genes, expediting their rates of evolution. Our results show a link between a cell’s GroEL level and its ability to survive despite the presence of deleterious mutations. Accordingly, we show that high GroEL levels allow buffering strong protein disrupting mutations.
Overexpression of groE has also been proposed to play a key role in stabilizing proteins with mutations in natural biological systems like endosymbiotic bacteria of insects that evolve under strong genetic drift and where genome mutational loads increase under a Muller’s ratchet dynamic (Muller 1964), a process by which the genomes of asexual populations accumulate deleterious mutations in an irreversible manner (Moran 1996; Fares, Barrio, et al. 2002). Accordingly, groE is highly overexpressed in the primary endosymbiont Sodalis pierantonius str. SOPE of the weevil Sitophilus oryzae (Oakeson et al. 2014), and in Buchnera and Blochmannia, primary endosymbiotic bacteria of aphids and ants, respectively (Wilcox et al. 2003; Stoll et al. 2009; Fan et al. 2013). Even in extremely reduced genomes, such as those of Sulcia muelleri and other symbiotic bacteria, groE is expressed at a very high level (McCutcheon et al. 2009; McCutcheon and Moran 2012; Bennett and Moran 2013). The universal overexpression of groE in endosymbiotic bacteria may help stabilize partially misfolded mutated protein as well as unstable protein intermediates (Bershtein et al. 2013), and allow the persistence of symbiotic bacteria (Moran 1996; Fares, Barrio, et al. 2002; Fares, Ruiz-Gonzalez, et al. 2002; Fares et al. 2004; Maisnier-Patin et al. 2005). Therefore, increasing the expression of groE allows satisfying the need for folding unstable proteins in cells evolving under strong genetic drift.
Our observations raise the possibility that mutualistic symbioses between bacteria and insects may have evolved through the kinds of modest genetic changes that occur in our experiment. The infection of an insect host by a free-living bacterium may have increased the expression of groE as a physiological response to the initially stressful environment of the host (Gahan et al. 2001). Once the host has been infected, the vertical transmission of the trapped bacterium across host generations, or its horizontal transmission between individuals of the same population may minimize the negative effects of groE overexpression due to genetic drift, whereas this overexpression in turn may allow the persistence of the symbiotic association by mitigating the effects of deleterious mutations. The overexpression of groE was probably simultaneous with the establishment of the mutualistic symbiosis. It is very unlikely that the overexpression originated after the symbiosis because we observe that most cell lineages evolving under strong genetic drift without groE overexpression go extinct. It is also unlikely that the overexpression predated the establishment of the intracellular symbiosis because such overexpression should have been deleterious in a free-living bacterial population with a large effective population size. Instead, our data support the notion that the overexpression of groE was concomitant with the infectious nature of the symbiotic bacterial ancestor, being an illustrative example of exaptation.
Experimental evolution shows great potential for testing hypotheses about the mechanisms underlying certain evolutionary events. Such experiments make it possible to observe evolution while it is unfolding, and thus allow inquiring about the consequences of variation in selective forces, such as shifts in selection-drift balance (Tenaillon et al. 2012; Dragosits and Mattanovich 2013; Wielgoss et al. 2013; Wiser et al. 2013; Cheng et al. 2014). Collectively, our results demonstrate the importance of mutational buffering mechanisms in allowing the persistence of otherwise deleterious genetic variation in populations, which may influence the tempo and mode of the emergence of key biological innovations.
Materials and Methods
Bacterial Lines
Escherichia coli K-12 substr. MG1655 ΔmutS was obtained from I. Matic (INSERM U571, Paris, France) through J. Blazquez’s group at Centro Nacional de Biotecnología (CSIC, Madrid, Spain). This mutS deletion strain is deficient in MutS (a component of the mismatch repair system of E. coli that recognizes and binds to mispaired nucleotides allowing for their removal by further action of MutL and MutH) and has a predicted mutation rate that is 1,000-fold higher than wild type (Bjedov et al. 2007; Turrientes et al. 2013). This strain was transformed with pGro7 (Takara Inc. # 3340) (Nishihara et al. 1998), to obtain a cohort of lineages with capacity to overexpress groES and groEL by induction with l-arabinose (pgroE). A second cohort was generated with plasmid pΔgroE, which was obtained by removal of groES and groEL from the pGro7 plasmid through a restriction digestion with BamHI and HindIII followed by religation, after manufacturer licensing to do so. This cohort was used as a control for our evolution experiment. Cells were grown routinely in Luria–Bertani (LB) media (Pronadisa #1551; 1% bacto-tryptone, 1% NaCl, 0.5% yeast extract) supplemented with 20 µg/ml of chloramphenicol (Sigma #C0378) if containing any of the two plasmids.
Evolution Experiment
Evolution was performed according to two different propagation procedures: 1) Through single-cell bottlenecks in solid media (agar plates; 1.5% Bacto-agar European grade; Pronadisa #1800), or 2) through serial transfer of large bacterial populations in liquid LB media. A total of 30 bottlenecked and 10 large population lineages were established with plasmid pGro7. Half of these populations were evolved with 0.2% l-arabinose (Acros Organics, #104985000) in the growth medium, in order to induce groES/EL overexpression, and the other half without it. We used the same E. coli strain transformed with the plasmid pΔgroE as a control. In this case, we propagated replicates for bottlenecked and large populations, growing in the same conditions as pgroE. Controls consisted of the same series without the inducer (fig. 1). We passaged each lineage serially every 24 h by streaking a single colony, in the case of bottlenecked populations, or by sampling 0.1 ml of the large population into 10 ml of fresh media. We passaged all lineages until their extinction or otherwise for 180 days, corresponding to approximately 4K generations of the bacteria (∼22 generations per day for bottlenecked bacterial lineages and 6.6 generations per day for large populations). A glycerol stock of each lineage was prepared every ten passages and stored at −80 °C to establish a “bacterial fossil record.”
Deep Sequencing and DNAseq Data Analysis
Whole-genome sequencing (WGS) was performed using paired-end Illumina sequencing at two different sequencing facilities. The extraction of genomic DNA was performed with the QIAmp DNA mini kit (Qiagen, Venlo [Pays Bas], Germany) for a QiaCube automatic extractor using bacterial pellets obtained from approximately 10 ml cultures (from frozen stock or sampled directly from the most recent bacterial culture) of two lineages from bottlenecked populations of pgroE and pΔgroE evolved for approximately 1.8K and 4K generations. Clonal DNAseq libraries were constructed using the TrueSeq DNA polymerase chain reaction-free HT sample preparation kit (Illumina) and labeled with individual indices to allow for running in a single lane. Quality and quantity of clonal DNAseq libraries were assessed using a 2100 Bioanalyzer (Agilent). All clonal DNAseq libraries were paired-end sequenced on an Illumina HiSeq2000 platform using a 2 × 100 cycles configuration (this service was provided by LifeSequencing SL., Valencia, Spain).
After removing a sample from the culture after passage 85 for freezing, the remaining culture was pelleted. Genomic DNA was extracted from the frozen bacterial cell pellets with the Illustra bacteria genomicPrep mini spin kit (GE Healthcare). DNAseq libraries were constructed with a Nextera XT DNA sample preparation kit (Illumina) and labeled with individual indices to allow for multiplexed running in a single lane. A sequencing run was performed in a MiSeq Benchtop Sequencer using 2 × 150 bp with 300 cycles pair-end reads configuration. MiSeq sequencing was performed at Valgenetics SL sequencing facility (Valencia, Spain).
WGS of the ancestral strain, E. coli K-12 substr. MG1655 ΔmutS, was performed using paired-end and shotgun 454 sequencing (Roche/454 Life Sciences, Branford, CT). A de novo assembly using the GS De Novo Assembler version 2.6 (Roche) was performed. This assembly was compared with E. coli K-12 substr. MG1655 (NC_000913.2) using Mauve (Darling et al. 2010) in order to detect potentially novel sequences within the ancestral genome. One such region was observed at position 806 kb, where 3.5 kb had been inserted and contained two genes (cI and tetA, both from Ec plasmid pGBG1). Lost regions were detected by mapping against MG1655 using ssaha v2.5.4 (Ning et al. 2001). Only two lost regions were observed, one of 4.4 kb in size and containing araABC (located at 67 kb), and another of 2.5 kb in size and containing mutS (located at 2.8 Mb). The mapped and de novo assemblies were combined into one using in-house scripts. We examined reads to confirm indels using ssaha and breseq v0.24rc4 (Deatherage and Barrick 2014). Annotations from MG1655 and pGBG1 were transferred using RATT (Otto et al. 2011).
Sequencing reads were converted from Illumina quality scores into Sanger quality scores. We then used the breseq v 0.24rc4 (version 4) pipeline (Deatherage and Barrick 2014) for aligning the Illumina reads to our E. coli parental genome and for identifying single nucleotide polymorphisms (SNPs) and indels (using bowtie2 [Langmead and Salzberg 2012]). Individual runs of breseq, with junction prediction disabled but otherwise default parameters, were performed for the ancestral sequence, as well as for each of the evolved lines. Finally, in house scripts were run to create tables containing all SNPs and indels for each lineage (including the ancestral) (supplementary tables S2–S5, Supplementary Material online). Files containing reads for the sequences used in this study can be accessed through the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under the accession number (SRP058803).
GroEL Measurement and Quantification
Crude protein extraction was performed with cultures from all the frozen stocks for the bottlenecked and large evolved populations, grown in liquid LB media with or without the inducer (l-arabinose). Total protein content was quantified by the Bradford method. Normalized crude extracts (the same total amount of protein per sample) were used for SDS-PAGE (12.5%) followed by Western blotting to assess and quantify groEL overexpression. Westerns were conducted as described elsewhere, using Rabbit polyclonal primary antibody to E. coli GroEL and GroES at 1:1,000 and 1:5,000 dilutions, respectively (from ABCam, #ab905022 and #ab69823, respectively). Control samples were reloaded to allow for comparisons between western blots. Relative GroEL quantification in each sample was performed (from western blot or dot blot membranes) using ImageJ software (http://rsb.info.nih.gov/ij; Schneider et al. 2012).
Growth Rate and Fitness Measurements
Growth rate and fitness characterization were conducted at endpoint of both the bottlenecked and the large population lines. Growth parameters for bottlenecked lines were evaluated using the Bioscreen C plate-reader system (Oy Growth Curves Ab Ltd, Helsinki, Finland). Each population was precultured overnight at the corresponding temperature and media, and used to inoculate 200 µl of fresh medium (LB diluted 1:8; Malouin et al. 1991) to an OD595 of 0.06–0.07, distributed in 100-well Honeycomb plates. Populations from each time point were grown at least in triplicate in the same plate with and without inducer. Each experimental run was conducted including negative control (i.e., blank fresh media) and positive control (i.e., ancestral lines), and using at least three replicates per 100-well Honeycomb plate. Each run consisted of two Honeycomb plates that allowed testing up to 200 cultures. Positive controls were used to allow for interplate and interrun comparison. Plates were incubated at 37 °C with continuous shaking (medium force) in the Bioscreen C. Growth was monitored for a period of 24–48 h taking OD595 measurement every 15 min. Growth rate was determined after OD595 normalization following a previous formula (Warringer and Blomberg 2003). Growth rates were averaged across replicated cultures with as many as 12 replicates for some time points, and determined with the best-fit model (Baty and Delignette-Muller 2004). From these data, relative fitness of the clonal lines was determined as one minus log 2 of the ratio of evolved versus the ancestor lineage (Lenski 1991).
To quantitatively determine small fitness variations throughout the evolution experiment, the mean fitness of each population lineage was determined by head-to-head competitions between evolved and ancestral strains, and between ancestral lineages containing different groE expression levels. Competitions were performed under the same temperature conditions as in the evolution experiment, but using a poor medium (LB diluted 1:8). Competitors were distinguished by the presence of pAmCyan (encoded on their plasmids Clontech #632440) fluorescent proteins in one competitor (e.g., ancestral population) and its absence in the other competitor (e.g., evolved population) and vice versa. Competition assays were performed in triplicate for each lineage. Data were obtained as CFUs (colony forming units), and then log-transformed for normalization. From these data, the net growth rate of each competitor was measured, and relative fitness was determined as the ratio of the evolved versus the ancestral lineage (Lenski 1991).
Quantification of the Fitness Cost of groE Overexpression
To quantify the fitness trade-off of groE overexpression, we competed head-to-head the ancestral pgroE cells against the ancestral pΔgroE cells in the presence of the inducer l-arabinose. We conducted these competitions by growing equal amounts of cells of each type in the same flask for 24 h. Then, we plated dilutions of these cultures to count the number of colonies of each cell type. To differentiate the cells of each competing type, we used as a marker a plasmid (pAmCyan) expressing a fluorescent protein. We transformed pΔgroE cells with this plasmid whereas pgroE contained the same plasmid but lacked the gene that encodes the fluorescent protein. To quantify the cost of expressing the fluorescent protein, we competed pΔgroE cells bearing the marker-plasmid and pΔgroE cells transformed with the same plasmid but lacking the fluorescent protein. Cells with the fluorescent marker grew 12.5% worse than the same cells without the marker (i.e., the cost of the marker resulted in 12.5% [C = 0.125] fewer colonies than the same cells without the marker). We took this measure into account when calculating the fitness of pgroE cells competing against pΔgroE that contained the pAmCyan plasmid. Specifically, we first calculated the CFUs of pgroE and pΔgroE as:
CFUs = (number of colonies × dilution factor)/(the plated volume in ml).
Then, we calculated the relative fitness of pgroE as:
where F and i refer to the counts of CFUs at the end and start of the competition assays, respectively. Competition assays were performed in 12 replicates.
Supplementary Material
Supplementary tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank lab members for discussion on the manuscript. They also thank J. Blazquez and I. Matic for the Escherichia coli hypermutable strains provided. This study was supported by a grant from Science Foundation Ireland (12/IP/1673) and a grant from the Spanish Ministerio de Economía y Competitividad (BFY2009-12022) to M.A.F. C.T. was supported by an EMBO long-term fellowship (EMBO ALTF 730-2011) and a grant Juan de la Cierva from the Ministerio de Economía y Competitividad (JCA-2012-14056). J.A. acknowledges support through the Forschungskredit program of the University of Zurich, grant FK-14-076. A.W. acknowledges support through the Swiss National Science Foundation grant 31002A_1461317, as well as through the University Priority Research Program in Evolutionary Biology at the University of Zurich.
References
- Amitai G, Gupta RD, Tawfik DS. 2007. Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J. 1:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baty F, Delignette-Muller ML. 2004. Estimating the bacterial lag time: which model, which precision? Int J Food Microbiol. 91:261–277. [DOI] [PubMed] [Google Scholar]
- Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 5:1675–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. 2013. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 49:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Dasgupta CN, Slade D, Le Blastier S, Selva M, Matic I. 2007. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics 176:1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. 2010. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2:602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogumil D, Dagan T. 2012. Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry 51:9941–9953. [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Ami GOH, Web Presence Working Group. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KK, Lee BS, Masuda T, Ito T, Ikeda K, Hirayama A, Deng L, Dong J, Shimizu K, Soga T, et al. 2014. Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat Commun. 5:3233. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, et al. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57:1959–1972. [DOI] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 1151:165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragosits M, Mattanovich D. 2013. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact. 12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M. 1971. Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:59. [DOI] [PubMed] [Google Scholar]
- Eigen M. 2002. Error catastrophe and antiviral strategy. Proc Natl Acad Sci U S A. 99:13374–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Thompson JW, Dubois LG, Moseley MA, Wernegreen JJ. 2013. Proteomic analysis of an unculturable bacterial endosymbiont (Blochmannia) reveals high abundance of chaperonins and biosynthetic enzymes. J Proteome Res. 12:704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares MA, Barrio E, Sabater-Munoz B, Moya A. 2002. The evolution of the heat-shock protein GroEL from Buchnera, the primary endosymbiont of aphids, is governed by positive selection. Mol Biol Evol. 19:1162–1170. [DOI] [PubMed] [Google Scholar]
- Fares MA, Moya A, Barrio E. 2004. GroEL and the maintenance of bacterial endosymbiosis. Trends Genet. 20:413–416. [DOI] [PubMed] [Google Scholar]
- Fares MA, Moya A, Barrio E. 2005. Adaptive evolution in GroEL from distantly related endosymbiotic bacteria of insects. J Evol Biol. 18:651–660. [DOI] [PubMed] [Google Scholar]
- Fares MA, Ruiz-Gonzalez MX, Moya A, Elena SF, Barrio E. 2002. Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature 417:398. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. 2010. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29:1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan CG, O’Mahony J, Hill C. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect Immun. 69:3924–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 16:574–581. [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Ferrada E, Wagner A. 2011. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474:92–95. [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Wagner A. 2012. Environmental change exposes beneficial epistatic interactions in a catalytic RNA. Proc Biol Sci. 279:3418–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RL, Gates J, Scott MP, Carroll SB. 1999. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283:532–534. [DOI] [PubMed] [Google Scholar]
- Lachowiec J, Lemus T, Borenstein E, Queitsch C. 2015. Hsp90 promotes kinase evolution. Mol Biol Evol. 32:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE. 1991. Quantifying fitness and gene stability in microorganisms. Biotechnology 15:173–192. [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Roth JR, Fredriksson A, Nystrom T, Berg OG, Andersson DI. 2005. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 37:1376–1379. [DOI] [PubMed] [Google Scholar]
- Malouin F, Chamberland S, Brochu N, Parr TR., Jr 1991. Influence of growth media on Escherichia coli cell composition and ceftazidime susceptibility. Antimicrob Agents Chemother. 35:477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. 2009. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5:e1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93:2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res. 106:2–9. [DOI] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. 2001. SSAHA: a fast search method for large DNA databases. Genome Res. 11:1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol. 64:1694–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C, Aoyagi A, Duval B, Baca A, Silva FJ, et al. 2014. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 6:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: rapid annotation transfer tool. Nucleic Acids Res. 39:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. [DOI] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. 2013. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science 342:1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Arnold FH. 2009. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 10:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396:336–342. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll S, Feldhaar H, Gross R. 2009. Transcriptional profiling of the endosymbiont Blochmannia floridanus during different developmental stages of its holometabolous ant host. Environ Microbiol. 11:877–888. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS. 2012. Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun. 3:1257. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Serrano L, Tawfik DS. 2008. How protein stability and new functions trade off. PLoS Comput Biol. 4:e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–673. [DOI] [PubMed] [Google Scholar]
- True JR, Carroll SB. 2002. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 18:53–80. [DOI] [PubMed] [Google Scholar]
- Turrientes MC, Baquero F, Levin BR, et al. 2013. Normal mutation rate variants arise in a Mutator (Mut S) Escherichia coli population. PLoS One 8:e72963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham RC, Kamerbeek J, Palacios C, et al. 2003. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci U S A. 100:581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Minasov G, Shoichet BK. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 320:85–95. [DOI] [PubMed] [Google Scholar]
- Warringer J, Blomberg A. 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20:53–67. [DOI] [PubMed] [Google Scholar]
- Wielgoss S, Barrick JE, Tenaillon O, et al. 2013. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc Natl Acad Sci U S A. 110:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox JL, Dunbar HE, Wolfinger RD, Moran NA. 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 48:1491–1500. [DOI] [PubMed] [Google Scholar]
- Williams TA, Fares MA. 2010. The effect of chaperonin buffering on protein evolution. Genome Biol Evol. 2:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser MJ, Ribeck N, Lenski RE. 2013. Long-term dynamics of adaptation in asexual populations. Science 342:1364–1367. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. 1999. Hox genes in digit development and evolution. Cell Tissue Res. 296:19–25 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.