Abstract
There is increasing evidence that dosage compensation is not a ubiquitous feature following sex chromosome evolution, especially not in organisms where females are the heterogametic sex, like in birds. Even when it occurs, compensation can be incomplete and limited to dosage-sensitive genes. However, previous work has mainly studied transcriptional regulation of sex-linked genes, which may not reflect expression at the protein level. Here, we used liquid chromatography–tandem mass spectrometry to detect and quantify expressed levels of more than 2,400 proteins in ten different tissues of male and female chicken embryos. For comparison, transcriptome sequencing was performed in the same individuals, five of each sex. The proteomic analysis revealed that dosage compensation was incomplete, with a mean male-to-female (M:F) expression ratio of Z-linked genes of 1.32 across tissues, similar to that at the RNA level (1.29). The mean Z chromosome-to-autosome expression ratio was close to 1 in males and lower than 1 in females, consistent with partly reduced Z chromosome expression in females. Although our results exclude a general mechanism for chromosome-wide dosage compensation at translation, 30% of all proteins encoded from Z-linked genes showed a significant change in the M:F ratio compared with the corresponding ratio at the RNA level. This resulted in a pattern where some genes showed balanced expression between sexes and some close to 2-fold higher expression in males. This suggests that proteomic analyses will be necessary to reveal a more complete picture of gene regulation and sex chromosome evolution.
Keywords: sex chromosome evolution, dosage compensation, chicken, proteomics, mass spectrometry
Introduction
One important scenario for adaptive regulation of gene expression is when there are changes or differences in gene dose (Veitia et al. 2008, 2013). Maintaining ancestral protein levels to sustain stoichiometric relationships in networks and pathways might in such cases be critical for proper function of protein–protein interactions (Zhang et al. 2010). A classical question in genetics to which this applies is the potential need for sex-linked dosage compensation upon sex chromosome evolution and the concomitant degeneration of nonrecombining genes (Ohno 1967; Disteche 2012). A common model of sex chromosome evolution posits that selection for linkage disequilibrium between a sex-determining locus and sexually antagonistic genes on the same chromosome promotes cessation of recombination in intervening regions, for example, by fixation of inversion mutations (Charlesworth et al. 2005; Ellegren 2011b). The nonrecombining chromosome—the Y chromosome for male heterogamety or the W chromosome for female heterogamety—will then inevitably suffer from a number of degenerative forces associated with lack of recombination. As a consequence, it is likely to degenerate and eventually lose a large fraction of its original gene content (Charlesworth B and Charlesworth D 2000; Bachtrog 2013). This leads in turn to an imbalance in sex-linked gene dose, with the dose halved in the heterogametic sex compared with the ancestral situation still present in the homogametic sex, with potentially harmful effects.
Sex-linked dosage compensation is a well-known phenomenon that occurs in many male heterogametic systems through a variety of molecular mechanisms (Disteche 2012; Mank 2013). In eutherian mammals, female X chromosome inactivation balances the number of active X chromosomes between females and males (Lyon 1974), but does not in itself restore ancestral expression levels of sex-linked genes. If anything, it increases the problem by halving the effective gene dose in both sexes, relative to that of autosomes; recall that most protein–protein interactions including sex-linked genes are likely to involve autosomal interaction partners. It has subsequently been suggested that the expression of autosomal and sex-linked genes is equalized in humans and other mammals by upregulation of gene expression in the single active X chromosome of both sexes (Nguyen and Disteche 2006; Deng et al. 2011; Kharchenko et al. 2011; Yildirim et al. 2012), although there are opposing views on such a potential mechanism (Xiong et al. 2010; Lin et al. 2012; Chen and Zhang 2015) with an on-going debate related to way of data analysis (Deng et al. 2011). Nevertheless, depending on their dosage-sensitivity, genes may very well vary as to whether they are compensated or not, and to what extent (Julien et al. 2012; Pessia et al. 2012).
Dosage compensation was previously considered ubiquitous and the default expectation in organisms with differentiated sex chromosomes (Ohno 1967; Mank 2013; Gartler 2014). However, this view had to be revised when the first large-scale transcriptome studies in a female heterogametic system (males ZZ, females ZW) revealed that chromosome-wide dosage compensation of chicken sex chromosomes was absent or, better phrased, incomplete (Ellegren et al 2007; Itoh et al 2007). This was later confirmed for other bird species (Itoh et al. 2010; Naurin et al. 2011; Wolf and Bryk 2011; Adolfsson and Ellegren 2013; Uebbing et al. 2013) as well as for other organisms with female heterogamety including butterflies and other insects, trematods, and snakes (Vicoso and Bachtrog 2011; Harrison et al. 2012; Vicoso et al. 2013; but see Smith et al. 2014). In birds, mRNA levels of expressed Z-linked genes are on average ≈1.5 times higher in males than in females, with considerable variation among genes (Mank and Ellegren 2009). This variation could be taken to suggest that compensatory regulation has evolved on a gene-by-gene basis according to the dosage-sensitivity of individual genes (Mank and Ellegren 2009; Naurin et al. 2010; Julien et al. 2012).
As for most other genome-wide aspects of gene regulation, sex-linked dosage compensation has up until now only been studied on a large scale by examining the output of gene expression at the RNA level, not at the functionally more relevant protein level. In general, whether it is appropriate to use mRNA concentrations as proxies for the abundance of the corresponding proteins has been an open question (Abreu et al. 2009; Vogel and Marcotte 2012). Emerging evidence now suggests that this is not necessarily the case (de Sousa Abreu et al. 2009; Ghazalpour et al. 2011; Juschke et al. 2013). Regarding dosage compensation it cannot be excluded that regulation of sex-linked gene expression occurs after mRNA synthesis, resulting in balanced expression at the protein level between sex-linked and autosomal genes, and between sex-linked genes in males and females. This may include regulation in connection with processing and degradation of mRNA, at translation and/or through posttranslational degradation; we will for simplicity collectively refer to these posttranscriptional stages as “translation.”
Testing the possibility of dosage compensation at the translation stage has been hindered by a lack of quantitative, high-throughput proteomic methods for measuring the abundance of large sets of proteins from biological samples. This is bound to change by the introduction and development of quantitative mass spectrometry methodologies, such as liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Vogel and Marcotte 2008), which can simultaneously quantify thousands of annotated proteins (Lu et al. 2007; Wilhelm et al. 2014). Such data are still limited to a handful of model species (Laurent et al. 2010; Ghazalpour et al. 2011; Johansson et al. 2013; Wilhelm et al. 2014) and there is so far only a single report using large-scale proteomic data for addressing dosage compensation, in humans (Chen and Zhang 2015). However, provided a well-annotated transcriptome or proteome is available, it should in principle have wide applicability.
Here we use LC-MS/MS to measure protein abundance in multiple tissues of male and female chicken, and perform deep transcriptome sequencing in the same individuals, with the aim of studying the relationship between sex-linked gene dose and gene expression at the functionally relevant protein level. The main conclusion of this work is that avian dosage compensation is generally incomplete also at the protein level, although for individual genes the relative levels of male and female expression differ between RNA and protein.
Results
We collected samples from ten different tissues (brain, bursa, heart, kidney, liver, lung, muscle, ovary, spleen, and testis) of five female and five male chicken, and analyzed all 90 samples with RNA-seq and tandem mass spectrometry for transcriptomic and proteomic quantification, respectively. Based on Ensembl annotations, the transcriptome analysis detected and quantified RNA expression from a total of 14,575 genes in at least one tissue (11,380–12,787 per tissue). The proteome analysis, based on Uniprot annotations, identified and quantified 2,420 unique proteins (764–1,291 per tissue) by the iBAQ label-free approach. In total, 2,388 genes were common to the RNA and protein sets, of which 108 were located on the chicken Z chromosome (18–51 per tissue). RNA and protein expression data from these genes constitute the primary data set for analysis of regulation of Z-linked genes at the transcription and translation levels, respectively.
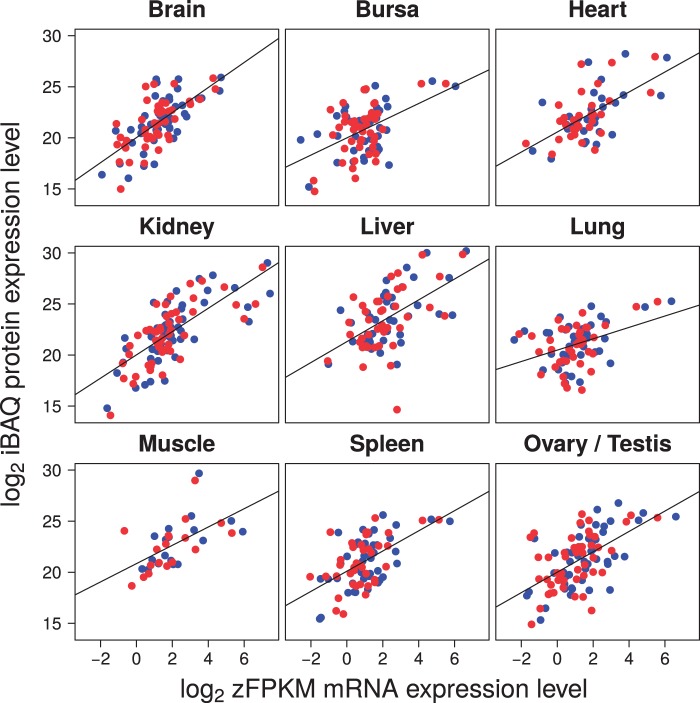
RNA-seq corroborated previous work in chicken, demonstrating largely male-biased expression and thus incomplete dosage compensation of Z-linked genes (median male-to-female [M:F] ratio per tissue ranging from 1.17 to 1.44, with a mean of 1.29; table 1 and supplementary fig. S1, Supplementary Material online). Proteomic data revealed a similar pattern, with a median M:F ratio per tissue of 1.19–1.49 (mean 1.32; table 1 and supplementary fig. S1, Supplementary Material online). The ratios were not significantly different between RNA and protein levels, neither in tests within each tissue nor in a paired test of the mean values of each tissue (V = 12.5, P = 0.48, Wilcoxon signed rank test). As a comparison, the median M:F ratio of autosomal genes was 0.98 and 1.00 for RNA and proteins, respectively. Overall, this directly indicates that full dosage compensation of avian sex-linked genes was not achieved by compensatory regulation at translation. Moreover, we found that levels of RNA and protein expression from Z-linked genes were well correlated (fig. 1), both in males (Spearman’s ρ = 0.272–0.713 for the different tissues, P < 0.05 in all but one tissue) and in females (ρ = 0.303–0.705, P < 0.05 in all but one tissue) (supplementary table S1, Supplementary Material online).
Table 1.
Median M:F Expression Ratios of Z-Linked Genes and Autosomal Genes per Tissue.
| Tissue | Z-Linked |
Autosomal |
||||
|---|---|---|---|---|---|---|
| RNA | Protein | n | RNA | Protein | n | |
| Brain | 1.35 | 1.24 | 20 | 0.98 | 1.01 | 645 |
| Bursa | 1.22 | 1.21 | 19 | 0.98 | 0.99 | 576 |
| Heart | 1.44 | 1.49 | 16 | 1.01 | 0.93 | 471 |
| Kidney | 1.27 | 1.44 | 26 | 0.97 | 1.01 | 654 |
| Liver | 1.28 | 1.30 | 26 | 0.99 | 0.98 | 508 |
| Lung | 1.17 | 1.28 | 16 | 0.97 | 1.03 | 528 |
| Muscle | 1.26 | 1.40 | 13 | 0.98 | 1.01 | 382 |
| Spleen | 1.34 | 1.19 | 18 | 1.00 | 1.00 | 570 |
| Mean | 1.29 | 1.32 | 0.98 | 1.00 | ||
Fig. 1.
Correlation between protein and mRNA expression for Z-linked genes in females (red) and males (blue).
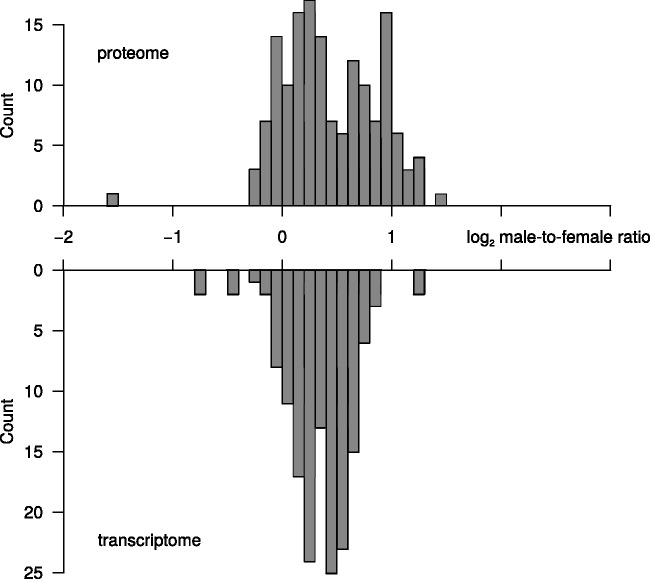
The M:F ratio of expression levels for Z-linked genes showed considerable variation at both RNA and protein levels (fig. 2). Ratios at the RNA level were approximately normally distributed. This included some genes with equal expression levels in the two sexes, as expected at fully compensated gene dose, whereas others approached 2-fold higher expression in males than in females, corresponding to the difference in gene dose. Despite a similar mean value, the distribution of M:F ratios at the protein level was more heterogeneous (fig. 2 and supplementary fig. S2, Supplementary Material online).
Fig. 2.
Distribution of log2 M:F expression ratios of Z-linked genes for proteins (upper part, proteome) and RNA (lower part, transcriptome).
We next compared expression levels of Z-linked and autosomal genes. In contrast to the analyses of M:F ratios, which always implied comparing expression of the same set of genes between two groups (males and females), by necessity this implied comparing expression in completely different sets of genes between two groups (Z chromosome and autosomes). Coupled with the much smaller sample size of Z-linked than autosomal genes, this meant that estimates of the Z-to-A (Z:A) ratio were associated with large variances (supplementary fig. S3, Supplementary Material online) and should be treated with some caution. However, the prediction under incomplete dosage compensation is that the Z:A ratio in males should be higher than that in females. Moreover, although in males the Z:A ratio should be close to 1, in females it should be lower than 1.
At the RNA level, the Z:A ratio in males (ZZ:AA; mean for nine tissues = 0.836 ± 0.192 SD) was higher than that in females (Z:AA; 0.682 ± 0.146). There was considerable variation in median Z:A ratios among tissues (table 2 and supplementary fig. S3, Supplementary Material online), a likely consequence of the high variances, but the trend of a higher ratio in males than in females per tissue was seen more often than expected by chance (in eight of eight tissues; sign test, P = 0.004, one-tailed). The same overall pattern was seen at the protein level, with mean ratios of 1.047 ± 0.624 in males and 0.837 ± 0.448 in females (P = 0.004), again with significant variation among tissues. These findings corroborate the notion that translation generally does not seem to act as an alternative means for dosage compensation of sex-linked genes. As a consequence, sex-linked expression was on average lower than the expression of autosomal genes in females, but not in males.
Table 2.
The Ratio between Mean Expression Level of Z-Linked and Autosomal Genes in Females (Z:AA) and Males (ZZ:AA).
| Tissue | Female (Z:AA) |
Male (ZZ:AA) |
nZ | nA | ||
|---|---|---|---|---|---|---|
| RNA | Protein | RNA | Protein | |||
| Brain | 0.60 | 0.58 | 0.76 | 0.75 | 45 | 1,217 |
| Bursa | 0.52 | 0.48 | 0.62 | 0.61 | 43 | 1,101 |
| Heart | 0.66 | 0.68 | 0.88 | 0.79 | 31 | 974 |
| Kidney | 0.74 | 0.83 | 0.91 | 1.13 | 51 | 1,108 |
| Liver | 1.01 | 1.61 | 1.23 | 1.94 | 40 | 849 |
| Lung | 0.60 | 0.52 | 0.69 | 0.69 | 39 | 974 |
| Muscle | 0.75 | 1.56 | 0.85 | 2.15 | 18 | 716 |
| Spleen | 0.58 | 0.44 | 0.68 | 0.56 | 40 | 1,078 |
| Ovary | 0.67 | 0.83 | 49 | 1,205 | ||
| Testis | 0.91 | 0.80 | 50 | 1,178 | ||
| Mean | 0.68 (0.62) | 0.84 (0.62) | 0.84 (0.78) | 1.05 (0.76) | ||
Note.—Mean values in parentheses are with liver and muscle excluded.
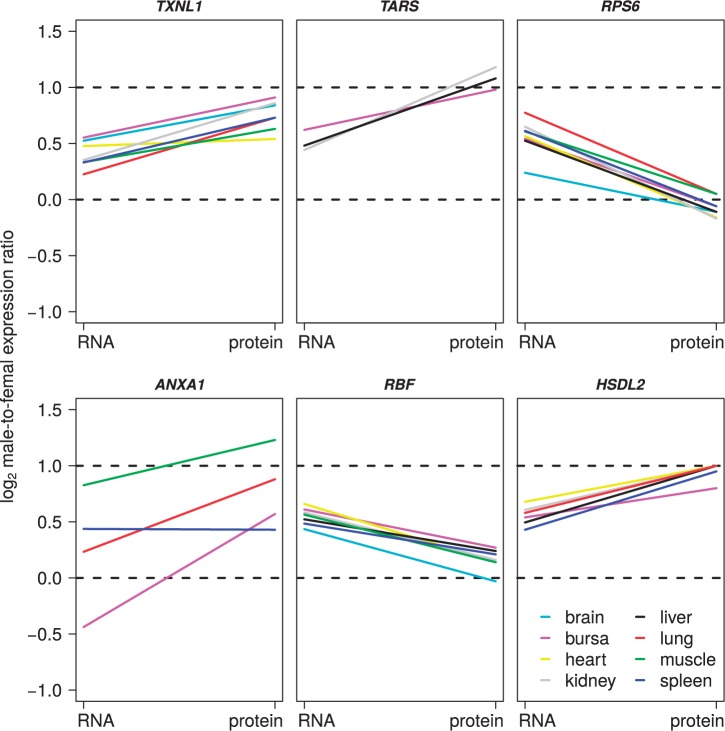
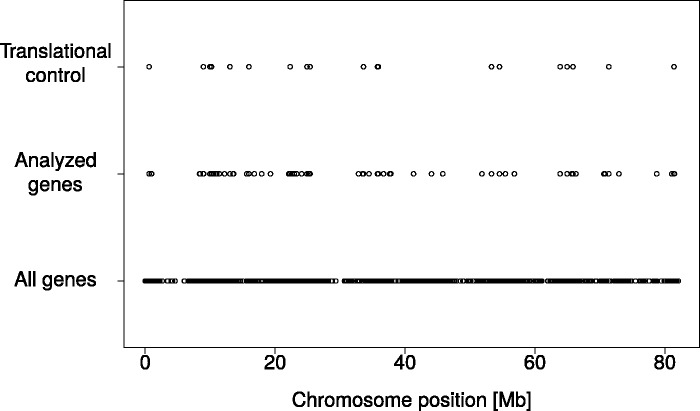
Although the results presented above do not support a general mechanism for chromosome-wide complete dosage compensation at the translation level, we asked whether there are individual genes for which translational regulation significantly alters the sex-differences in expression seen at the RNA level. Specifically, we screened for Z-linked genes where |(log2 M:FRNA) − (log2 M:Fprotein)| > 0.5; the screening was thus not set to enrich for genes that reached a certain M:F ratio at the protein level, but simply queried for a pronounced change in M:F ratio between RNA and protein levels (fig. 3 and supplementary fig. S5, Supplementary Material online). Five genes had (log2 M:FRNA) − (log2 M:Fprotein) > 0.5 in at least one tissue and generally showed close to balanced sex-specific levels of protein expression (supplementary table S2, Supplementary Material online). Another 14 genes had (log2 M:FRNA) − (log2 M:Fprotein) < −0.5 (supplementary table S2, Supplementary Material online). In contrast to the former category of genes, this group had close to 2-fold higher protein expression in males than in females. Thus, in total, 19 of 63 (30%) genes showed a pronounced change in the M:F ratio between the RNA and protein levels. These genes were evenly distributed along the Z chromosome and showed no evidence of clustering (fig. 4).
Fig. 3.
Examples of Z-linked genes showing a consistent difference in the M:F expression ratio (log2) between RNA and protein levels. Each line represents data from one tissue. Note that genes may differ in the number of tissues in which they were expressed and detected. Supplementary figure S5, Supplementary Material online, shows all 19 genes with a consistent difference in the M:F expression ratio (log2) between RNA and protein levels.
Fig. 4.
Distribution of genes along the chicken Z-chromosome. The figure shows the location of all ENSEMBL annotated genes (bottom), genes analyzed in this study for which both proteomic and RNA-seq data were available (middle) and genes that showed a consistent difference in the M:F expression ratio between RNA and protein levels.
It was clear that these changes were consistent among tissues for the same genes (fig. 3 and supplementary fig. S5, Supplementary Material online), with one gene, RPS6, showing (log2 M:FRNA) − (log2 M:Fprotein) > 0.5 in seven of eight tissues in which it was expressed, in all cases resulting in almost balanced expression between sexes. Intriguingly, 8 of the 14 genes that had (log2 M:FRNA) − (log2 M:Fprotein) < −0.5 showed translation regulation in liver (five with liver-specific expression), compared with 0–3 genes with translation regulation in the other tissues.
We asked whether additional regulation at translation level was more prevalent for Z-linked genes than for autosomal genes. Indeed, the proportion of genes with |(log2 M:FRNA) − (log2 M:Fprotein)| > 0.5 was significantly higher among Z-linked genes than among autosomal genes (250 of 1,344, 19%; χ2 = 5.23, P = 0.022, two-tailed). Moreover, the correlation between RNA and protein levels was less strong for Z-linked genes than for autosomal genes (supplementary tables S1 and S3, Supplementary Material online; mean ρ for Z-linked genes = 0.520, for autosomal genes = 0.642; ρ was lower for Z-linked genes than for autosomal genes in 14 of 18 sex-tissue combinations, P = 0.031 binomial test, two-tailed).
Discussion
The use of transcriptome data for assessing how regulation of gene expression affects biological function generally assumes that RNA levels adequately reflect protein levels, that is, that gene regulation mainly occurs at transcription, not translation. A body of recent literature testifies that this can be a gross oversimplification and that the correlation between gene expression at transcript and protein levels in some cases is only modest (Lu et al. 2007; de Sousa Abreu et al. 2009; Ghazalpour et al. 2011). In principle, this means that conclusions on protein abundance, and thereby their biological activity, from transcriptome studies may be misleading.
We focused on chicken as previous work in this main avian model (as well as in other female heterogametic species) had indicated that sex chromosome dosage compensation is incomplete. These observations were based on RNA data from microarray experiments (Ellegren et al. 2007; Itoh et al. 2007) or RNA-seq (e.g., Wolf and Bryk 2011) and it cannot be excluded that sex-specific regulation of protein expression modulates or alters the patterns seen at the RNA level. To test this, we made an extensive proteomic analysis of chicken by identifying and quantifying 2,420 different proteins in multiple tissues using tandem mass spectrometry. The two main conclusions from this work were that 1) overall, dosage compensation in chicken is incomplete also at the protein level, and 2) there are consistent changes in various tissues in the M:F expression ratio between RNA and protein levels for individual genes.
Complete dosage compensation implies hypertranscription or hypertranslation of the entire X- or Z-chromosome in the heterogametic sex; epigenetically mediated hypertranscription of the X-chromosome is seen in Drosophila (Larschan et al. 2011; Conrad and Akhtar 2012) and Caenorhabditis elegans (Ercan et al. 2007). Our results exclude a chromosome-wide mechanism for dosage compensation at translation in the avian model and confirm conclusions from studies at the RNA level that avian dosage compensation is incomplete. Indeed, such a mechanism is not easily perceived unless mRNA encoded from sex chromosomes would carry a mark that affects ribosomal translation or the stability of mRNA during the transport from the nucleus. With incompleteness of dosage compensation thus confirmed at the protein level, the question that naturally follows is why complete compensation has not evolved. At least two models have been put forward to explain this situation (Mank 2013). First, due to feedback regulation and other network interactions, a change in gene dose does not necessarily give rise to a change of the corresponding magnitude at the RNA/protein level (Malone et al. 2012). Second, due to their special inheritance, sex chromosomes are hotspots for sexually antagonistic loci (Rice 1984), at which sex-biased gene expression might actually be selected for Ellegren and Parsch (2007). Accumulation of male-beneficial genes on the Z chromosome (Ellegren 2011a) may thus introduce trade-offs between masculinization and dosage compensation (Naurin et al. 2010; Mank et al. 2011; Wright et al. 2012). Both these models are consistent with observations at both RNA (e.g., Mank and Ellegren 2009; Itoh et al. 2010) and protein (this study) levels, demonstrating considerable variation among Z-linked genes in the M:F expression ratio.
Our other major conclusion has bearing on the mentioned variation among genes, namely the observation of gene-specific changes in the M:F expression ratio from RNA to the protein level in 30% of the Z-linked genes. Specifically, for individual genes we found evidence for regulation at translation leading either to a reinforced excess of male expression (14 genes; supplementary table S2, Supplementary Material online) or to equalized expression between males and females (from a stage of moderate male excess at the RNA level; 5 genes; supplementary table S2, Supplementary Material online). For the latter category of genes, translational regulation could be seen as a complementary means for reaching balanced expression in males and females, that is, dosage compensation. Formally, the present data cannot reveal if this restored ancestral expression levels prior to sex chromosome differentiation, although that would seem reasonable. If so, compensation may have been in the form of increased expression from the single Z chromosome in females or the result of lowered expression from the Z chromosomes in males. That regulation of protein expression to equalize sex-specific expression levels would occur for some but not all genes is in line with the idea that avian dosage compensation is mediated on a gene-by-gene basis, depending on the dosage sensitivity and “masculinity” of individual genes (Mank and Ellegren 2009; Naurin et al. 2010; Mank 2013; Uebbing et al. 2013). We note that the frequency of genes with significant regulation at the translation level was higher for Z-linked than for autosomal genes, indicating that translation constitutes an important means to handle differences in gene dose of sex-linked genes. Two of the five genes with close to sex-equal expression after translation regulation are members of large mitochondrial membrane complexes; the RBF protein is a subunit of ATP synthase and NDUFS4 is part of respiratory chain complex 1. A third, RPS6 (ribosomal protein S6) is part of the ribosomal complex. As imbalanced expression of protein complex members can be detrimental (Birchler and Veitia 2012), the impetus for reaching sex-equal expression of these genes may be particularly strong.
The pattern seen in the group of genes for which the sex difference in expression was reinforced at the protein level is perhaps less intuitively understood. A common denominator for these 14 genes was a M:F expression ratio in the range 1–1.5 at the RNA level and in the range 1.5–2 at the protein level. This would suggest that the difference in gene dose between males and females was to some extent overly/inadequately compensated at transcription. Subsequent regulation at translation to render sex-differences in protein levels more similar to the sex-difference in gene dose could potentially be related to male-beneficial functions of the corresponding genes. However, it is not intuitive from the genes’ functional annotations why this would be the case—most are involved with general metabolic functions that should be important for both sexes. The number of genes is too small to make a gene ontology analysis meaningful although we note that 5 of the 14 proteins localize to mitochondria (DMGDH, OXCT1), peroxisomes (HSD17B4), or both (AMACR, HSDL2). Neither this group of genes or the group of five genes reaching sex-equal expression after translational regulation showed any evidence of clustering along the Z chromosomes.
Seen in a wider context, this study illustrates a general development in the fields of molecular evolution and evolutionary genomics, with an increased focus on the proteome (Diz et al. 2012). The concept of “evolution at two levels” originally introduced by King and Wilson (1975) refers to that phenotypic evolution may either be due to changes in gene sequences or changes in the way expression of gene sequences is regulated. In a functional context, this translates into changes in protein sequences and changes in the levels (and the temporal and spatial distribution) of encoded proteins. The relative role of these genetic mechanisms for evolutionary change is an issue of long-standing debate in evolutionary biology (Carroll 2005; Hoekstra and Coyne 2007). Although gene expression is by now appreciated as important to phenotypic evolution, the vast majority of work has been on RNA, not proteins (but see, e.g., Laurent et al. 2010; Artieri and Fraser 2013). With proteomic methods becoming both more widely accessible and applicable to many different organisms, we foresee that quantitative studies such as this will come to provide a more comprehensive portray of how regulation of gene expression contributes to evolution.
Materials and Methods
Sample Collection
Chicken eggs (White Leghorn) were purchased from Ova Production AB (Vittinge, Sweden). Eggs were incubated at 37.5 °C and 60% relative humidity with automatic turning every 6 h until sampled. Five embryos of each sex were collected after incubation of the eggs for 18 days and a number of tissues were sampled and flash frozen in liquid nitrogen. Spleen, testicles, ovarium, bursa Fabricii (hereafter referred to as just bursa), and brain were collected whole, whereas pieces of tissue were taken from heart, liver, kidney, lung, and breast muscle (muscle). Samples were stored at −80 °C until analysis.
RNA-seq and Transcriptome Analysis
A minimum of 4 mg and a maximum of 25 mg of each sampled tissue was put in RNase-free, round-bottom Eppendorf tubes together with RNA Stabilization Reagent (Qiagen Inc., Valencia, CA) and 5-mm steel bullets, baked at 190 °C for 4 h followed by tissue rupture by shaking at 25 Hz for 30 s using the Retsch MM300 mixer mill (Retsch Inc., Haan, Germany). Total RNA was extracted from a partition of the homogenate using the RNeasy Mini Kit (Qiagen Inc.); the yield of total RNA ranged between 3 and 58 µg for individual tissue samples. First flow through from the RNA extraction was used for subsequent protein mass spectrometry analysis.
Sequencing libraries (each for every individual/tissue combination) were prepared using the TruSeq stranded mRNA sample preparation kit (Illumina, San Diego, CA; cat# RS-122-2101, RS-122-2102). Libraries were sequenced on an Illumina HiSeq instrument at the Uppsala University SNP & SEQ technology platform for 100 cycles and produced more than 48 million read pairs per sample (48.9–112.6 million). Three samples were subsequently found to be mislabeled and were excluded from downstream analyses (one female liver, one female spleen, and one male liver).
RNA-seq reads were mapped onto the chicken genome (Galgal4, ENSEMBL release 77) using TopHat v. 2.0.12 (Kim et al. 2013) and summarized per gene (ENSEMBL annotations, release 77) using Cufflinks v. 2.2.1 (Trapnell et al. 2012) with multiread correction option (-u). All samples had a mapping rate of 76.9–88.0%, except for one female spleen sample that had 64.5%. The number of mapped reads ranged from 39.4 to 92.1 million, with a mean value of 50.0 million. We extracted FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) normalized gene expression values and normalized further using the method of Hart et al. (2013), a between-sample normalization method of log2 FPKM values similar to a z-transformation (zFPKM). We applied an expression cutoff of 0.125 zFPKM as suggested in that study.
M:F ratios were calculated for genes with data available for all five males and all five females; inclusion of genes with partially missing data did not affect the results other than adding noise. Gonads were excluded from estimates of M:F expression ratios as testis and ovary are different tissues. Z chromosome-to-autosomal (i.e., female Z:AA and male ZZ:AA) expression ratios were calculated by dividing the median expression of Z-linked genes by the median expression of autosomal genes. All genes with data available for at least one individual were in this case included as it was important to maximize the amount of data when two completely different gene sets (Z-linked and autosomal, respectively) were compared. Confidence intervals of the ratios were determined by 10,000 bias-corrected (using the cumulative normal distribution) bootstrap replicates, see Uebbing et al. (2013). Gonadal tissues were included in Z chromosome-to-autosome comparisons in males (testes) and females (ovary).
Protein Sample Preparation and Mass Spectrometry
During RNA extraction the first flow through of each sample was saved and used for protein sample preparation. Proteins were precipitated at −20 °C for 1 h using ice-cold acetone, sedimented, and dissolved in SDS (sodium dodecyl sulfate) lysis buffer (4% SDS in 100 mM Tris/HCl pH 7.6) at 70 °C for 30 min. Protein concentration was determined by DC (detergent compatible) protein assay (Bio-Rad, Hercules, CA). Enzymatic fragmentation of proteins was performed by in-solution digestion (Andersen et al. 2005). In brief, precipitation of proteins by acetone was repeated prior to dissolving the protein pellet in urea buffer (6 M urea, 2 M thiourea, 10 mM HEPES, pH 8.0). In the next step, protein disulfide bonds were reduced with 10 mM dithiothreitol and alkylated with 55 mM iodoacetamide. Protein digestion was performed by Lys-C (protein-to-enzyme ratio 100:1) (Wako Chemicals GmbH, Neuss, Germany) at room temperature for 3 h followed by trypsin treatment (protein-to-enzyme ratio 100:1) (Promega Corporation, Madison, WI) at room temperature over night. Resulting peptides were purified and concentrated by stop and go extraction (STAGE) tips (Rappsilber et al. 2003).
To perform a quantitative proteome analysis, peptides were modified by stable isotope dimethyl labeling as described in Boersema et al. (2009). In brief, samples from male chicken embryos were light labeled in 4% formaldehyde and 0.6 M sodium cyanoborohydride for 1 h at room temperature. For heavy labeling of female chicken samples, stable isotope substituted formaldehyde (CD2O) was used. Next, labeling reaction was stopped by acidification with 1% ammonia solution and 5% formic acid prior to mixing heavy (female) and light (male) samples. STAGE tips were used for sample clean-up and purification before MS analysis.
Reverse-phase chromatography for peptide separation was performed using an Easy nano flow system (Thermo Fisher Scientific, Waltham, MA) coupled to a Q-Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were separated by precolumn (100 µm ID, 5 µm C18-beads) and analytical columns (75 µm ID, 3 µm C18-beads) (Thermo Fisher Scientific) using a linear gradient from 4% to 48% acetonitril with 0.1% formic acid for 138 min at a flow rate of 250 nl/min, followed by 75% acetonitril for 6 min and 4% acetonitril for 6 min for re-equilibration. After separation, peptides were ionized using a nano electrospray ionization source and transferred into the mass spectrometer. Positive ion MS spectra (m/z = 400–1,750) were acquired using an automatic gain control (AGC) target of 3E6 at a resolution of 70,000. The ten most intense peaks were isolated for higher-energy collisional dissociation (HCD) fragmentation (25% normalized collision energy) and MS/MS spectra were generated with an AGC target of 5E5 at a resolution of 17,500. Mass spectrometer worked in data-dependent mode.
Raw data were processed using MaxQuant (1.5.0.25) (Cox et al. 2009). Database searches were performed using the implemented Andromeda search engine to correlate MS/MS spectra to the Uniprot chicken database (release 2014-08). The following parameters were used for data processing: Maximum of two miss cleavages, mass tolerance of 4.5 ppm for main search, trypsin as digesting enzyme, carbamidomethylation of cysteins as fixed modification, oxidation of methionine, and acetylation of the protein N-terminus as variable modifications. For dimethyl labeling Lys0 and Nter0 were set for light label, and Lys4 and Nter4 for heavy label. Furthermore, intensity-based absolute quantification (iBAQ) normalization of peptide intensities was used for label free quantification (Nagaraj et al. 2011). For protein identification only, peptides with a minimum of seven amino acids and at least one unique peptide were required. For quantification of proteins, two ratio counts were set as a minimum. Only proteins with at least two peptides and at least one unique peptide were considered as identified and used for further data analysis.
Similar to RNA, M:F protein ratios were calculated for genes without missing data whereas Z:A ratios were calculated for all genes where at least one individual produced data, in order to maximize the number of genes included. This way, M:F ratios were calculated for 13–26 constitutively expressed Z-linked genes per tissue, whereas Z:A ratios were derived from 18–51 Z-linked and 716–1,217 autosomal genes per tissue. Z:A ratios were calculated from iBAQ values. iBAQ is a quantification algorithm designed for label-free abundance measurements (Schwanhausser et al. 2011). Although we used a chemical labeling approach, application of iBAQ measurements was important for getting absolute quantity estimates in the comparisons between protein and RNA expressions, and between protein expression from autosomal and Z-linked genes. M:F expression ratios correlated well between dimethyl labeling ratios and ratios derived from iBAQ (mean Spearman’s ρ = 0.74 over tissues, range: 0.65–0.87), suggesting that iBAQ measurements adequately capture the biological signals.
RNA sequence data have been deposited to the European Nucleotide Archive under the accession number PRJEB8390. The mass spectrometry proteomics data have been deposited to the ProteomeXchange via the PRIDE partner respository with the dataset identifier PXD002403.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Swedish Research Council (grant numbers 2010-5650 and 2013-8271 to H.E., and 2011-4423 to J.B.), the European Research Council (AdG 249976 to H.E), and the Knut and Alice Wallenberg Foundation (to H.E).
References
- Abreu RD, Penalva LO, Marcotte EM, Vogel C. 2009. Global signatures of protein and mRNA expression levels. Mol BioSyst. 5:1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfsson S, Ellegren H. 2013. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol. 30:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AKL, Ong S-E, Lyon CE, Lamond AI, Mann M. 2005. Nucleolar proteome dynamics. Nature 433:77–83. [DOI] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. 2013. Evolution at two levels of gene expression in yeast. Genome Res. 24:963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2012. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 109:14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. 2009. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 4:484–494. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 2000. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 355:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J. 2015. No X-chromosome dosage compensation in human proteomes. Mol Biol Evol. 32:1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Akhtar A. 2012. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 13:123–134. [DOI] [PubMed] [Google Scholar]
- Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, Mann M. 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 4:698–705. [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. 2009. Global signatures of protein and mRNA expression levels. Mol BioSyst. 5:1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XX, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 43:1179–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. 2012. Dosage compensation of the sex chromosomes. Annu Rev Genet. 46:537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz AP, Martinez-Fernandez M, Rolan-Alvarez E. 2012. Proteomics in evolutionary ecology: linking the genotype with the phenotype. Mol Ecol. 21:1060–1080. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2011a. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 21:2082–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2011b. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 12:157–166. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Hultin-Rosenberg L, Brunstrom B, Dencker L, Kultima K, Scholz B. 2007. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 8:689–698. [DOI] [PubMed] [Google Scholar]
- Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. 2007. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 39:403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM. 2014. A brief history of dosage compensation. J Genet. 93:591–595. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. 2011. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 7:e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Mank JE, Wedell N. 2012. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 4:1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Komori HK, LaMere S, Podshivalova K, Salomon DR. 2013. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band M, Clayton D, et al. 2007. Dosage compensation is less effective in birds than in mammals. J Biol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Replogle K, Kim YH, Wade J, Clayton DF, Arnold AP. 2010. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 20:512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson Å, Enroth S, Palmblad M, Deelder AM, Bergquist J, Gyllensten U. 2013. Identification of genetic variants influencing the human plasma proteome. Proc Natl Acad Sci U S A. 110:4673–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10:e1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juschke C, Dohnal I, Pichler P, Harzer H, Swart R, Ammerer G, Mechtler K, Knoblich J. 2013. Transcriptome and proteome quantification of a tumor model provides novel insights into posttranscriptional gene regulation. Genome Biol. 14:r133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi RB, Park PJ. 2011. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 43:1167–1169. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg S. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M-C, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent JM, Vogel C, Kwon T, Craig SA, Boutz DR, Huse HK, Nozue K, Walia H, Whiteley M, Ronald PC, et al. 2010. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10:4209–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. 2012. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 109:11752–11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 25:117–124. [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1974. Evolution of X-chromosome inactivation in mammals. Nature 250:651–653. [DOI] [PubMed] [Google Scholar]
- Malone J, Cho D-Y, Mattiuzzo N, Artieri C, Jiang L, Dale R, Smith H, McDaniel J, Munro S, Salit M, et al. 2012. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 13:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. 2013. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 29:677–683. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. 2009. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102:312–320. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. 2011. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution 65:2133–2144. [DOI] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Bensch S, Hassequist D. 2010. Why does dosage compensation differ between XY and ZW taxa? Trends Genet. 26:15–20. [DOI] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Hasselquist D, Kim YH, Bensch S. 2011. The sex-biased brain: sexual dimorphism in gene expression in two species of songbirds. BMC Genomics 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet. 38:47–53. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1967. Sex chromosomes and sex linked genes. Berlin (Germany): Springer Verlag. [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. 2012. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 109:5346–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75:663–670. [DOI] [PubMed] [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38:735–742. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. [DOI] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD. 2014. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol Evol. 6:526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebbing S, Künstner A, Mäkinen H, Ellegren H. 2013. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol Evol. 5:1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R, Bottani S, Birchler J. 2008. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 24:390–397. [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. 2013. Gene dosage effects: nonlinearities, genetic interactions, and dosage compensation. Trends Genet. 29:385–393. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2011. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 3:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11:e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. 2008. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat Protoc. 3:1444–1451. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. 2014. Mass-spectrometry-based draft of the human proteome. Nature 509:582–587. [DOI] [PubMed] [Google Scholar]
- Wolf J, Bryk J. 2011. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics 12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Moghadam HK, Mank JE. 2012. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics 192:1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong YY, Chen XS, Chen ZD, Wang XZ, Shi SH, Wang XQ, Zhang JZ, He XL. 2010. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 42:1043–1047. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Sadreyev RI, Pinter SF, Lee JT. 2012. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol. 19:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, MacAlpine DM, Oliver B. 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8:e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.