Abstract
The selfing syndrome constitutes a suite of floral and reproductive trait changes that have evolved repeatedly across many evolutionary lineages in response to the shift to selfing. Convergent evolution of the selfing syndrome suggests that these changes are adaptive, yet our understanding of the detailed molecular genetic basis of the selfing syndrome remains limited. Here, we investigate the role of cis-regulatory changes during the recent evolution of the selfing syndrome in Capsella rubella, which split from the outcrosser Capsella grandiflora less than 200 ka. We assess allele-specific expression (ASE) in leaves and flower buds at a total of 18,452 genes in three interspecific F1 C. grandiflora x C. rubella hybrids. Using a hierarchical Bayesian approach that accounts for technical variation using genomic reads, we find evidence for extensive cis-regulatory changes. On average, 44% of the assayed genes show evidence of ASE; however, only 6% show strong allelic expression biases. Flower buds, but not leaves, show an enrichment of cis-regulatory changes in genomic regions responsible for floral and reproductive trait divergence between C. rubella and C. grandiflora. We further detected an excess of heterozygous transposable element (TE) insertions near genes with ASE, and TE insertions targeted by uniquely mapping 24-nt small RNAs were associated with reduced expression of nearby genes. Our results suggest that cis-regulatory changes have been important during the recent adaptive floral evolution in Capsella and that differences in TE dynamics between selfing and outcrossing species could be important for rapid regulatory divergence in association with mating system shifts.
Keywords: gene expression, adaptation, self-fertilization, allele-specific expression, mating system evolution, cis-regulatory evolution
Introduction
The transition from outcrossing to predominant self-fertilization has occurred repeatedly in flowering plants (Stebbins 1950). In association with this shift, marked changes in floral and reproductive traits have occurred independently in many different lineages (Barrett 2002). In general, selfers tend to show reduced allocation of resources to traits involved in pollinator attraction and reward (e.g., smaller petals, less nectar per flower, and less scent), exhibit changes in floral morphology that may improve the efficacy of autonomous self-pollination (e.g., reduced separation between stigma and anthers), and show reduced allocation of resources to male function (reduced ratio of pollen to ovules) (reviewed in Sicard and Lenhard 2011). Together, this combination of floral and reproductive traits is termed “the selfing syndrome” (Ornduff 1969).
Despite the striking pattern of convergent floral evolution in association with the shift to selfing, we currently have a limited understanding of the molecular genetic basis of the selfing syndrome. Quantitative trait loci (QTL) for the selfing syndrome have been identified in a handful of systems (e.g., Capsella; Sicard et al. 2011; Slotte et al. 2012; Leptosiphon; Goodwillie et al. 2006; Mimulus; Fishman et al. 2002; Fishman et al. 2015; Oryza; Grillo et al. 2009; Solanum; Bernacchi and Tanksley 1997). In domesticated tomatoes, cis-regulatory changes at the Style2.1 gene have been implicated in reduced stigma exsertion (Chen et al. 2007), but in most other systems, the molecular basis of the selfing syndrome is not known. A major unresolved question thus concerns the general importance of cis-regulatory changes versus other types of molecular changes for the evolution of the selfing syndrome.
Cis-regulatory changes have long been hypothesized to be important for organismal adaptation (Doebley and Lukens 1998; Carroll 2000; Wray 2007; Carroll 2008; Stern and Orgogozo 2008; but see Hoekstra and Coyne 2007), due to their potentially limited negative pleiotropic effects (Wray 2007). The prospects for identifying cis-regulatory changes on a transcriptome-wide scale have greatly improved due to the advent of massively parallel sequencing (Fraser 2011). In particular, methods for assessing allele-specific expression (ASE) that contrast the relative levels of expression of two alleles in an individual allow for transcriptome-scale assessment of cis-regulatory changes. ASE studies require the presence of transcribed polymorphisms and rigorous bioinformatic approaches but have benefits over mapping approaches (e.g., eQTL mapping) in terms of cost and resolution and can identify individual genes with cis-regulatory changes (Pastinen 2010).
As part of our broad goal to examine molecular genetic changes associated with the selfing syndrome, we examine the influence of cis-regulatory changes on the evolution of the selfing syndrome in Capsella rubella. We further test whether silencing of transposable elements (TEs) through the RNA-directed methylation pathway is important for global cis-regulatory divergence in association with the shift to selfing. The crucifer genus Capsella is a promising system for assessing the role of cis-regulatory changes in association with plant mating system shifts and adaptation, because of the availability of a sequenced genome of C. rubella (Slotte et al. 2013) and because it is possible to generate viable offspring from crosses between Capsella species that differ in their mating system (e.g., Slotte et al. 2012, Rebernig et al. 2015).
In C. rubella, the transition to selfing occurred relatively recently (<200 ka) and was associated with speciation from an outcrossing progenitor similar to present-day Capsella grandiflora (Slotte et al. 2013, Foxe et al. 2009, Guo et al. 2009, St Onge et al. 2011, Brandvain et al. 2013). Despite the recent shift to selfing, C. rubella already exhibits a derived reduction in petal size and a reduced pollen-ovule ratio, as well as a reduction of the degree of flower opening (Sicard et al. 2011, Slotte et al. 2012). Capsella rubella therefore exhibits floral and reproductive characters typical of a selfing syndrome. The selfing syndrome of C. rubella is associated with improved efficacy of autonomous self-pollination (Sicard et al. 2011), and regions with QTL for floral divergence between C. rubella and C. grandiflora exhibit an excess of fixed differences and reduced polymorphism in C. rubella (Slotte et al. 2012). Together, these observations suggest that the rapid evolution of the selfing syndrome in C. rubella was driven by positive selection.
While the molecular genetic basis of the selfing syndrome in C. rubella has not been identified, it has been suggested that cis-regulatory changes could be involved, and a previous study found many flower and pollen development genes to be differentially expressed in flower buds of C. grandiflora and C. rubella (Slotte et al. 2013). However, these results could be confounded by differences in floral organ sizes and pollen number between C. rubella and C. grandiflora, and Slotte et al. (2013) did not directly assess cis-regulatory changes or investigate possible causes of cis-regulatory divergence. There is reason to believe that cis-regulatory changes could be partly caused by differences in TE abundance between selfers and outcrossers, as TE silencing can affect nearby gene expression in plants (Hollister and Gaut 2009; Hollister et al. 2011). As C. rubella harbors fewer TEs close to genes than C. grandiflora (Ågren et al. 2014), this system offers an opportunity to investigate the role of TEs for cis-regulatory evolution and for the evolution of floral and reproductive traits in association with the shift to selfing.
In this study, we directly assessed cis-regulatory divergence by analyzing ASE in F1 hybrids of C. grandiflora and C. rubella and investigated the role of cis-regulatory changes for the selfing syndrome in C. rubella. We conducted deep sequencing of transcriptomes, small RNAs, and genomes of C. grandiflora x C. rubella hybrids to identify genes with cis-regulatory divergence in flower buds and leaves and tested whether cis-regulatory changes in flowers were overrepresented in genomic regions responsible for adaptive phenotypic divergence. We further identified TEs in C. rubella and C. grandiflora and tested whether TE insertions targeted by uniquely mapping 24-nt siRNAs were associated with cis-regulatory divergence. Our results provide insight into the role of cis-regulatory changes in association with the shift to selfing in a wild plant system.
Results
Many Genes Exhibit ASE in Interspecific F1 Hybrids
To quantify ASE between C. grandiflora and C. rubella, we generated deep whole transcriptome RNAseq data from flower buds and leaves of three C. grandiflora x C. rubella F1 hybrids (total 52.1 vs. 41.8 Gb with Q ≥ 30 for flower buds and leaves, respectively). We included three technical replicates for one F1 to examine the reliability of our expression data. For all F1s and their C. rubella parents, we also generated deep (38–68x) whole genome resequencing data to reconstruct parental haplotypes and account for read mapping biases.
F1 RNAseq reads were mapped with high stringency to reconstructed parental haplotypes specific for each F1, that is, reconstructed reference genomes containing whole-genome haplotypes for both the C. grandiflora and the C. rubella parent of each F1 (see Materials and Methods). We conducted stringent filtering of genomic regions where single-nucleotide polymorphisms (SNPs) were deemed unreliable for ASE analyses due to, for example, high repeat content, copy number variation, or a high proportion of heterozygous genotypes in an inbred C. rubella line (for details, see Materials and Methods and supplementary text S1, Supplementary Material online); this mainly resulted in removal of pericentromeric regions (supplementary figs. S2–S5, Supplementary Material online). After filtering, we identified approximately 18,200 genes with approximately 274,000 transcribed heterozygous SNPs that were amenable to ASE analysis in each F1 (table 1). The mean allelic ratio of genomic read counts at these SNPs was 0.5 (supplementary fig. S6, Supplementary Material online), suggesting that our bioinformatic procedures efficiently minimized read mapping biases. Furthermore, technical reliability of our RNAseq data was high, as indicated by a mean Spearman's ρ between replicates of 0.98 (range 0.94–0.99).
Table 1.
Genes Amenable to Analysis of ASE in Flower Bud and Leaf Samples from the Three C. grandiflora x C. rubella F1s, Counts of Genes with Evidence for ASE and the Estimated False Discovery Rate (FDR), and Proportion of Genes with ASE.
| F1 Designation | Sample | Genes Amenable to ASE Analysisa | Analyzed Genesb | Heterozygous SNPs in Analyzed Genes | Genes with ASE PP ≥ 0.95 c | FDR | ASE Proportiond |
|---|---|---|---|---|---|---|---|
| Inter3.1 | Flower buds | 18,299 | 16,857 | 262,120 | 4,728 | 0.0013 | 0.38 |
| Inter4.1 | 18,270 | 17,837 | 272,126 | 5,744 | 0.0022 | 0.42 | |
| Inter5.1 | 18,144 | 17,448 | 262,696 | 5,176 | 0.0020 | 0.40 | |
| Inter3.1 | Leaves | 18,299 | 14,877 | 238,786 | 5,105 | 0.0012 | 0.44 |
| Inter4.1 | 18,270 | 15,784 | 249,181 | 8,129 | 0.0024 | 0.62 | |
| Inter5.1 | 18,144 | 15,478 | 240,653 | 4,795 | 0.0018 | 0.41 |
aTotal number of genes with heterozygous SNPs in coding regions remaining after filtering.
bNumber of genes amenable to ASE analyses with expression data in at least one of the replicates of the sample.
cNumber of genes with evidence for ASE (posterior probability ≥ 0.95).
dDirect estimate of the ASE proportion independent of significance cutoffs.
We assessed ASE using a Bayesian statistical method with a reduced false-positive rate compared with the standard binomial test (Skelly et al. 2011). The method uses genomic read counts to model technical variation in ASE and estimates the global proportion of genes with ASE, independent of specific significance cutoffs, and also yields gene-specific estimates of the ASE ratio and the posterior probability of ASE. The model also allows for and estimates the degree of variability in ASE along the gene, through the inclusion of a dispersion parameter.
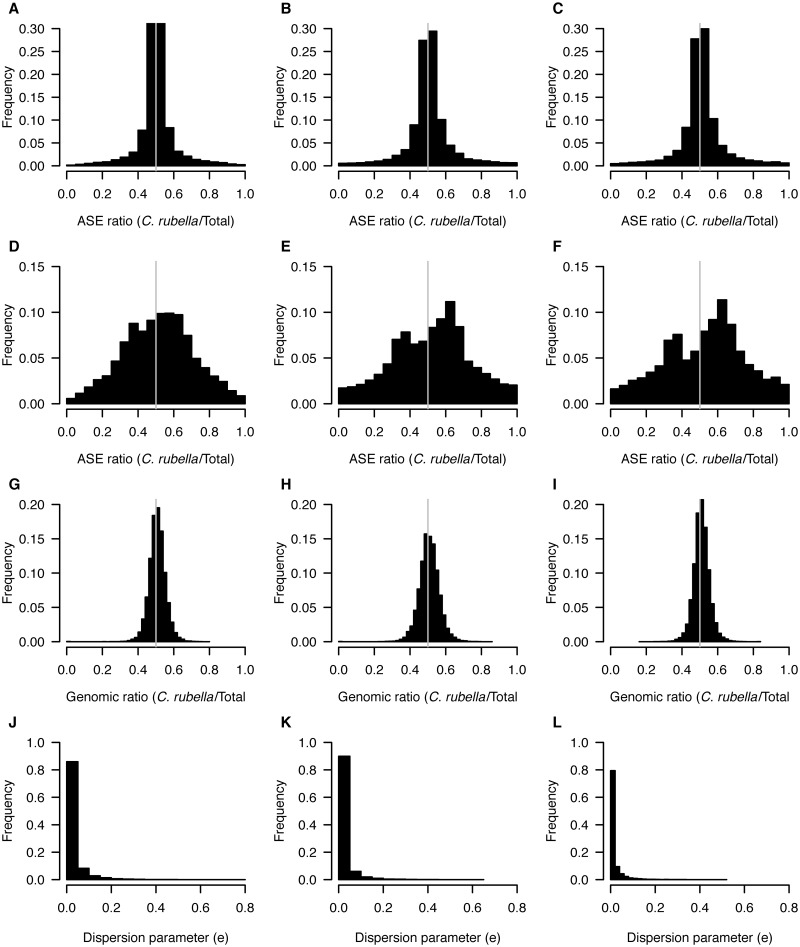
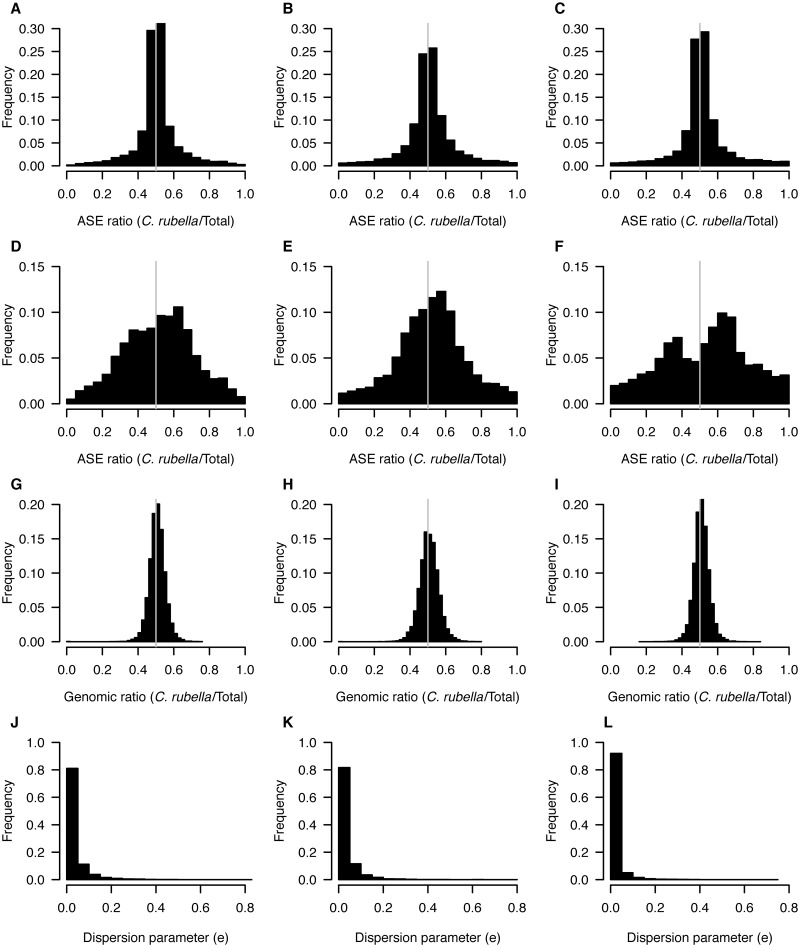
On the basis of this method, we estimate that on average, the proportion of assayed genes with ASE is 44.6% (table 1; supplementary table S8, Supplementary Material online). In general, most allelic expression biases were moderate, and only 5.9% of assayed genes showed ASE ratios greater than 0.8 or less than 0.2 (figs. 1 and 2). There was little variation in ASE ratios along genes, as indicated by the distribution of the dispersion parameter estimates having a mode close to zero and a narrow range (figs. 1 and 2). This suggests that unequal expression of differentially spliced transcripts is not a major contributor to regulatory divergence between C. rubella and C. grandiflora (figs. 1 and 2).
Fig. 1.
ASE in flower buds. Distributions of ASE ratios (C. rubella/total) for all assayed genes (A–C) and for genes with at least 0.95 posterior probability of ASE (D–F). Ratio of C. rubella to total for genomic reads, for genes with significant ASE (G–I), and the distribution of the dispersion parameter that quantifies variability in ASE across genes (J–L). All distributions are shown for each of the three interspecific F1s inter 3.1 (left), inter4.1 (middle), and inter5.1 (right).
Fig. 2.
ASE in leaves. Distributions of ASE ratios (C. rubella/total) for all assayed genes (A–C) and for genes with at least 0.95 posterior probability of ASE (D–F). Ratio of C. rubella to total for genomic reads, for genes with significant ASE (G–I), and the distribution of the dispersion parameter that quantifies variability in ASE across genes (J–L). All distributions are shown for each of the three interspecific F1s inter 3.1 (left), inter4.1 (middle), and inter5.1 (right).
For genes with evidence for ASE (hereafter defined as posterior probability of ASE ≥ 0.95), there was a moderate shift toward higher expression of the C. rubella allele (mean ratio C. rubella/total = 0.56; figs. 1 and 2). This shift was present for all F1s, for both leaves and flowers (figs. 1 and 2). No such shift was apparent for genomic reads, and ratios of genomic read counts for SNPs in genes with ASE were very close to 0.5 (mean ratio C. rubella/total = 0.51; figs. 1 and 2). Furthermore, quantitative polymerase chain reaction (qPCR) with allele-specific probes for five genes validated our ASE results empirically (supplementary table S9, Supplementary Material online). Thus, C. rubella alleles appear to be on average expressed at a higher level than C. grandiflora alleles in our F1s.
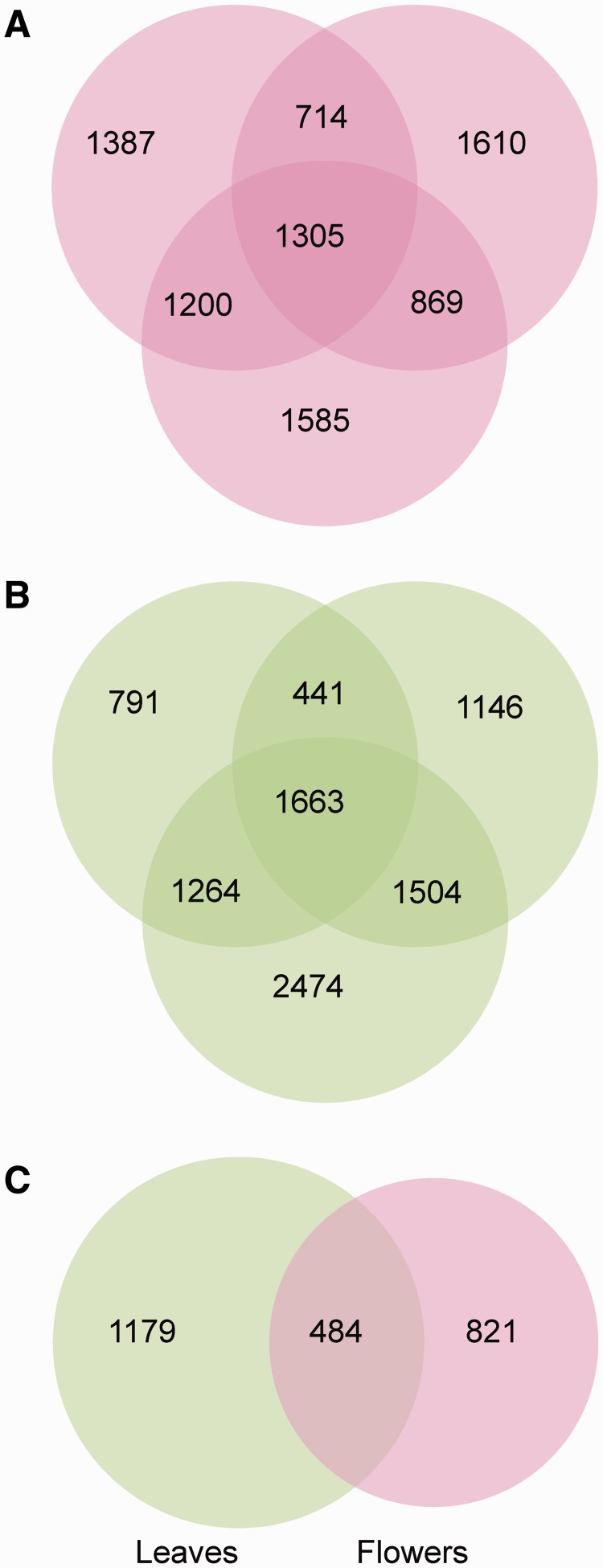
The mean ASE proportion, as well as the absolute number of genes with ASE, was greater for leaves (49%; 6,010 genes) than for flower buds (40%; 5,216 genes), although this difference was largely driven by leaf samples from one of our F1s (table 1). Most instances of ASE were specific to either leaves or flower buds, and on average, only 15% of genes expressed in both leaves and flower buds showed consistent ASE in both organs (fig. 3). Many cases of ASE were also specific to a particular F1, and across all three F1s, there were 1,305 genes that showed consistent ASE in flower buds and 1,663 in leaves (fig. 3).
Fig. 3.
Many cases of ASE are specific to individuals or samples. Venn diagrams showing intersections of genes with ASE in flower buds (A) and leaves (B) of the three F1 individuals, and (C) in all leaf and flower samples, for the set of genes assayed in all F1s.
Enrichment of Cis-Regulatory Changes in Genomic Regions Responsible for Phenotypic Divergence
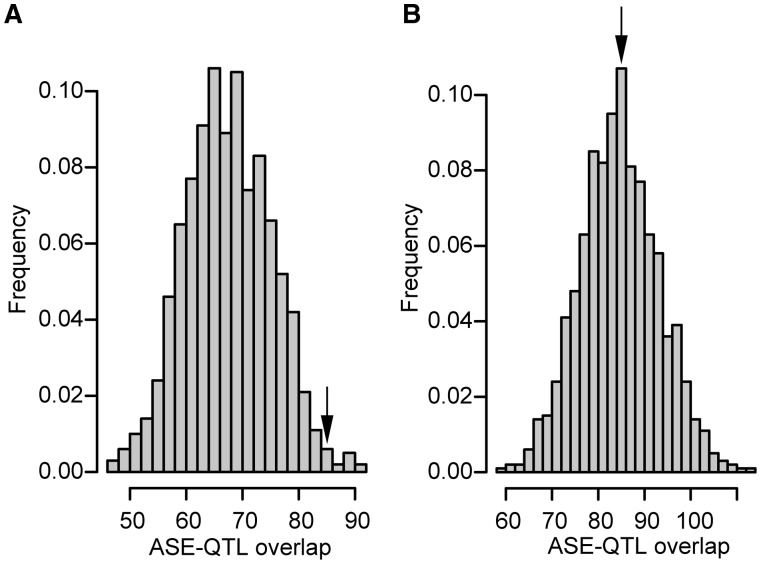
We used permutation tests to check for an excess of genes showing ASE within five previously identified narrow (<2 Mb) QTL regions responsible for floral and reproductive trait divergence (Slotte et al. 2012). These genomic regions harbor major QTL for petal size and flowering time but also encompass part of the confidence intervals for QTL for sepal size, stamen length, and ovule number, as QTL for different floral and reproductive traits are highly overlapping (Slotte et al. 2012). As the selfing syndrome has a shared genetic basis in independent C. rubella accessions (Sicard et al. 2011, Slotte et al. 2012), we reasoned that genes with consistent ASE across all F1s would be most likely to represent candidate cis-regulatory changes underlying QTL. Out of the 1,305 genes with ASE in flower buds of all F1s, 85 were found in narrow QTL regions, and this overlap was significantly greater than expected by chance (permutation test, P = 0.03; fig. 4; see Materials and Methods for details). In contrast, for leaves, there was no significant excess of genes showing ASE in narrow QTL (permutation test, P = 1; fig. 4). Thus, the association between QTL and ASE in flower buds is unlikely to be an artifact of locally elevated heterozygosity facilitating both ASE and QTL detection, which should affect analyses of both leaf and flower samples.
Fig. 4.
Enrichment of genes with ASE in narrow QTL regions. There is an excess of genes with ASE in narrow QTL regions for flower buds (A) but not for leaves (B). Histograms show the distribution of numbers of genes with ASE that fall within narrow QTL regions, based on 1,000 random permutations of the observed number of genes with ASE among all genes where we could assess ASE. Arrows indicate the observed number of genes with ASE that are located in narrow QTL regions.
List Enrichment Analyses Reveal Floral Candidate Genes with ASE
We conducted list enrichment analyses to characterize the functions of genes showing ASE relative to all genes amenable to analysis of ASE (i.e., harboring heterozygous transcribed SNPs and expressed at detectable levels). There was an enrichment of Gene Ontology (GO) terms involved in defense and stress responses for genes with ASE in flower buds and in leaves (supplementary table S10, Supplementary Material online). GO terms related to hormonal responses, including brassinosteroid and auxin biosynthetic processes, were specifically enriched among genes with ASE in flower buds (supplementary table S10, Supplementary Material online). Genes with nearby heterozygous TE insertions were also enriched for a number of GO terms related to reproduction and defense (supplementary tables S11 and S12, Supplementary Material online), suggesting that heterozygous TE insertions could be important for patterns of GO term enrichment for genes with ASE
We further identified 19 genes involved in floral and reproductive development in Arabidopsis thaliana, which are located in QTL regions (see above) and show ASE in flower buds (table 2). These genes are of special interest as candidate genes for detailed studies of the genetic basis of the selfing syndrome in C. rubella.
Table 2.
Selfing Syndrome Candidate Genes Identified Based on ASE, QTL information, and Arabidopsis Annotation.
| C. rubella Ortholog | Arabidopsis Ortholog | Arabidopsis Annotation | GO Biological Process Terms Related to Floral and Reproductive Development |
|---|---|---|---|
| Carubv10012851ma,b | AT3G24340 | CHR40 | Regulation of flower development |
| Carubv10016094ma,b | AT3G24650 | ATABI3, ABI3, SIS10 | Embryo development, cotyledon development |
| Carubv10007602ma,b | AT4G21600 | ENDO5 | Brassinosteroid biosynthetic process |
| Carubv10000655mb,c | AT5G08130 | BIM1 | Brassinosteroid-mediated signaling pathway, primary shoot apical meristem specification |
| Carubv10006681mb,c | AT4G28720 | YUC8 | Brassinosteroid-mediated signaling pathway |
| Carubv10021883ma,d | AT1G68480 | JAG | Sepal formation, flower development, abaxial cell fate specification, anther development, carpel development, stamen development, petal formation, specification of floral organ Identity |
| Carubv10021345ma,d | AT1G68640 | PAN, TGA8 | Petal formation, sepal formation, regulation of flower development |
| Carubv10013321ma,d | AT3G22420 | ATWNK2, WNK2, ZIK3 | Photoperiodism, flowering |
| Carubv10016406ma,d | AT3G23270 | — | Pollen tube growth |
| Carubv10014951ma,d | AT3G23440 | EDA6, MEE37 | Megagametogenesis |
| Carubv10014152ma,d | AT3G23630 | ATIPT7, IPT7 | Pollen tube growth, reciprocal meiotic recombination |
| Carubv10010238ma,d | AT3G62210 | EDA32 | Polar nucleus fusion |
| Carubv10004312ma,d | AT4G16760 | ATACX1, ACX1 | Pollen development |
| Carubv10005585ma,d | AT4G17030 | AT-EXPR, EXPR, ATEXLB1, ATEXPR1, EXLB1 | Sexual reproduction |
| Carubv10007441ma,d | AT4G20370 | TSF | Regulation of flower development, photoperiodism, flowering, Positive regulation of flower development |
| Carubv10004229ma,d | AT4G20910 | CRM2, HEN1 | Specification of floral organ identity, floral organ formation, petal formation, regulation of flower development, sepal formation, meristem initiation, meristem development, ovule development |
| Carubv10015623ma,d | AT4G21380 | ARK3, RK3 | Recognition of pollen |
| Carubv10007227ma,d | AT4G21530 | APC4 | Ovule development |
| Carubv10007633ma,d | AT4G21590 | ENDO3 | Petal development, stamen development, pollen tube growth, ovule development |
aLocated within narrow QTL regions.
bASE in all three F1s.
cLocated within QTL regions but not narrow QTL regions.
dASE in the F1 with data for three replicates, but not in all three F1s.
Intergenic Divergence Is Elevated Near Genes with ASE
To investigate the role of polymorphisms in regulatory regions for ASE, we assessed levels of heterozygosity in intergenic regions 1 kb upstream of genes and in previously identified conserved noncoding regions (Williamson et al. 2014) within 5 kb and 10 kb of genes. Genes with ASE were not significantly more likely to be associated with conserved noncoding regions with heterozygous SNPs than genes without ASE. However, levels of intergenic heterozygosity 1 kb upstream of genes were slightly but significantly higher for genes with ASE than for those without ASE (median heterozygosity of 0.016 vs. 0.014, respectively in leaves [Wilcoxon rank sum test, W = 295,692,325, P value = 2.26*10−115], median heterozygosity of 0.017 versus 0.014, respectively, in flowers [Wilcoxon rank sum test, W = 297,625,040, P value = 6.16*10−142], supplementary table S13, Supplementary Material online), suggesting that polymorphisms in regulatory regions upstream of genes might contribute to cis-regulatory divergence.
Enrichment of TEs Near Genes with ASE
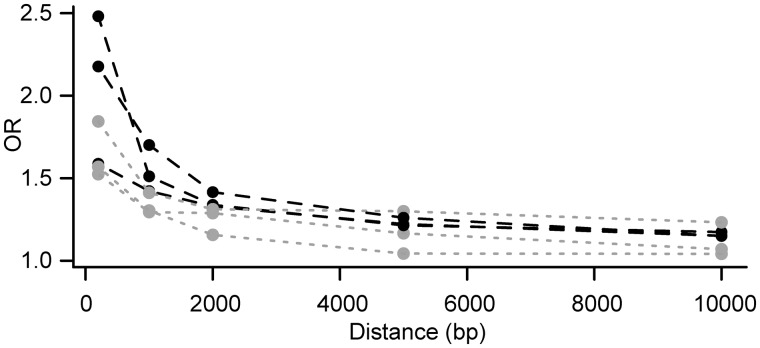
To test whether differences in TE content might contribute to cis-regulatory divergence between C. rubella and C. grandiflora, we examined whether heterozygous TE insertions near genes were associated with ASE. We identified TE insertions specific to the C. grandiflora or C. rubella parents of our F1s using genomic read data, as in Ågren et al. (2014) (table 3; see Materials and Methods). Overall, we found that C. rubella harbored fewer TE insertions close to genes than C. grandiflora (on average, 482 vs. 1,154 insertions within 1 kb of genes in C. rubella and C. grandiflora, respectively). Among heterozygous TE insertions, Gypsy insertions were the most frequent (table 3); they were also the most frequent genome-wide (table 3). There was a significant association between heterozygous TE insertions within 1 kb of genes and ASE, for both leaves and flower buds, and the strength of the association was greater for TE insertions closer to genes (table 4; fig. 5). This was true for individual F1s, as well as for all F1s collectively (table 4; fig. 5; supplementary table S14, Supplementary Material online).
Table 3.
Mean Number of TE Insertions in Three Interspecific F1s.
| TE Superfamily | Mean Copy Number | Heterozygous Insertions | Insertions Specific to the C. rubella Parental Genome | Insertions Specific to the C. grandiflora Parental Genome |
|---|---|---|---|---|
| CACTA | 84 | 40 | 10 | 30 |
| Copia | 710 | 483 | 144 | 339 |
| Gypsy | 1,124 | 602 | 153 | 449 |
| Harbinger | 176 | 109 | 26 | 83 |
| hAT | 83 | 55 | 16 | 40 |
| Helitron | 236 | 127 | 30 | 97 |
| LINE | 229 | 165 | 38 | 128 |
| MuDR | 203 | 109 | 28 | 81 |
| SINE | 113 | 92 | 9 | 83 |
| Total | 2,958 | 1,782 | 454 | 1,330 |
Note.—The overall number and heterozygous insertions with parent of origin information are presented.
Table 4.
Enrichment of Heterozygous TEs Near Genes with ASE.
| Sample | Window Size (bp) | +ASE, +TE | +ASE, −TE | −ASE, +TE | −ASE, −TE | P |
|---|---|---|---|---|---|---|
| Flower buds | 200 | 113 | 5,103 | 136 | 12,029 | 4.32*10−19 |
| 1,000 | 218 | 4,998 | 339 | 11,826 | 5.07*10−16 | |
| 2,000 | 307 | 4,909 | 540 | 11,624 | 6.53*10−12 | |
| 5,000 | 566 | 4,650 | 1,108 | 11,057 | 8.22*10−10 | |
| 10,000 | 958 | 4,258 | 2,006 | 10,159 | 2.32*10−7 | |
| Leaves | 200 | 108 | 5,902 | 115 | 9,255 | 8.52*10−7 |
| 1,000 | 216 | 5,793 | 277 | 9,093 | 1.49*10−4 | |
| 2,000 | 317 | 5,693 | 435 | 8,935 | 2.25*10−3 | |
| 5,000 | 595 | 5,415 | 877 | 8,493 | NS | |
| 10,000 | 1,027 | 4,983 | 1,576 | 7,795 | NS |
Note.—Mean counts over all three F1s and Fisher exact test P values. The four categories of counts correspond to numbers of genes with ASE (posterior probability of ASE ≥ 0.95) and TE insertions within a specific window size near the gene (+ASE,+TE), with ASE but without TEs (+ASE, −TE), without ASE but with TE insertions (−ASE,+TE), and with neither ASE nor TEs (−ASE,−TE). NS, not significant.
Fig. 5.
Enrichment of TEs near genes with ASE. Odds ratios (ORs) of the association between genes with ASE and TEs, with TE insertions scored in four different window sizes (within a distance of 0 bp, 1 kb, 2 kb, 5 kb, and 10 kb of each gene). Odds ratios for flower buds are shown for all three F1s studies, with values for flower buds in black and leaves in gray.
TEs Targeted by Uniquely Mapping 24-nt Small RNAs Are Associated with Reduced Expression of Nearby Genes
To test whether siRNA-based silencing of TEs might be responsible for the association between TE insertions and ASE in Capsella, we analyzed data for flower buds from one of our F1s, for which we had matching small RNA data (see Materials and Methods). We selected only those 24-nt siRNA reads that mapped uniquely, without mismatch, to one site within each of our F1s, because uniquely mapping siRNAs have been shown to have a more marked association with gene expression in Arabidopsis (Hollister and Gaut 2009). For each gene, we then assessed the ASE ratio of the allele on the same chromosome as a TE insertion (i.e., ASE ratios were polarized such that relative ASE was equal to the ratio of the expression of the allele with a TE insertion on the same chromosome over the total expression of both alleles) and then further examined the influence of nearby siRNAs.
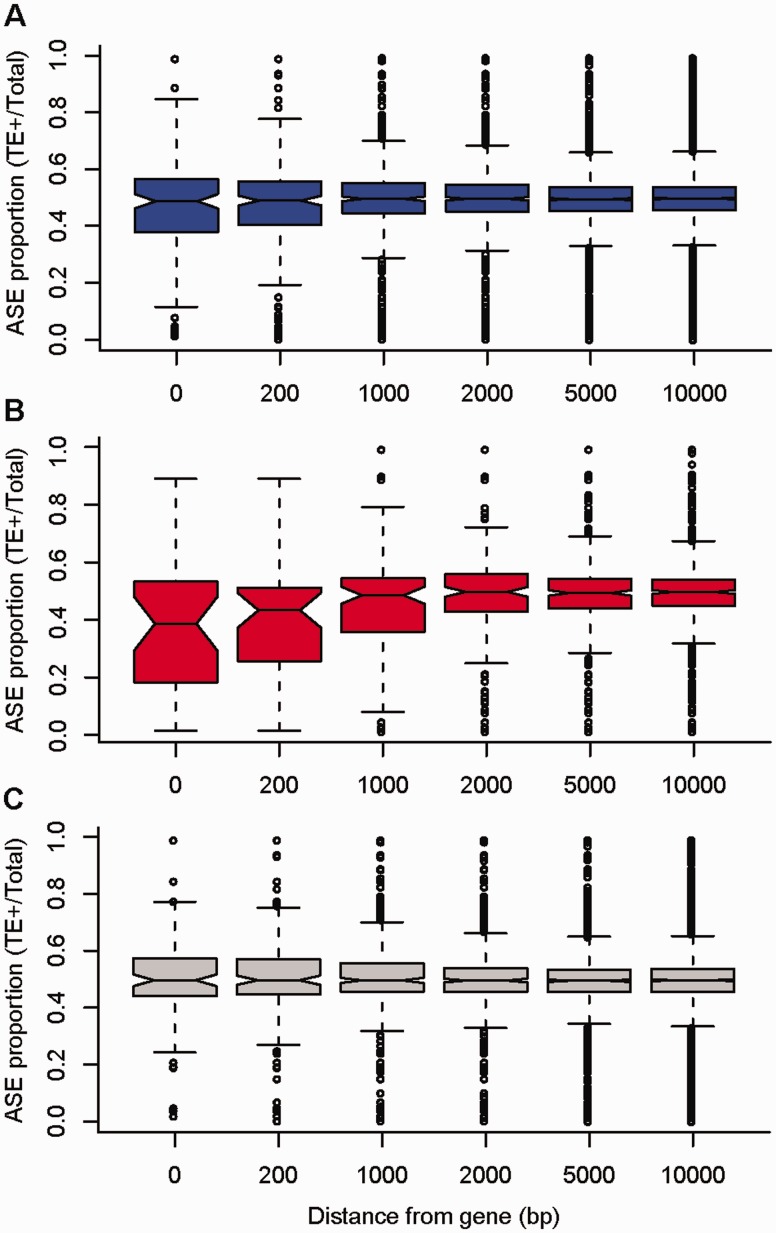
Overall, the mean relative ASE was reduced for genes with nearby TE insertions (fig. 6) with a more pronounced effect for TE insertions within 1 kb (within the gene: Wilcoxon rank sum test, W = 1,392,103, P value = 8.76*10−3; within 200 bp: Wilcoxon rank sum test, W = 1,903,047, P value = 7.17*10−3; within 1 kb: Wilcoxon rank sum test, W = 3,687,972, P value = 8.19*10−3). The magnitude of the effect on ASE was more pronounced for genes near TE insertions targeted by uniquely mapping 24-nt siRNAs (fig. 6; for genes with a TE insertion within the gene: Wilcoxon rank sum test, W = 423,369, P value = 1.36*10−4; within 200 bp: W = 540,926, P value = 1.82*10−5; within 1 kb: W = 983,938, P value = 3.13*10−3). In contrast, no significant effect on ASE was apparent for genes near TE insertions that were not targeted by uniquely mapping 24-nt siRNAs (fig. 6). Thus, uniquely mapping siRNAs targeting TE insertions appear to be responsible for the association we observe between ASE and TE insertions. Globally, Gypsy and hAT insertions made up a greater proportion of the TE insertions that were targeted by siRNA, compared with those that were not (Chi-squared test, χ = 35.9468, P = 1.796*10−5, supplementary fig. S7, Supplementary Material online). However, for heterozygous TE insertions within 1 kb of genes, there were no significant differences in the composition of TEs that were versus were not targeted by uniquely mapping siRNAs.
Fig. 6.
The effect of TE insertions on relative allelic expression. Boxplots show the relative allelic expression (expression of the allele on same haplotype as TE insertion relative to expression of both alleles) for genes near heterozygous TE insertions, scored in a range of window sizes ranging from 0 bp (within the gene) to 10 kb from the gene. (A) The relative allelic expression is reduced for genes with nearby TE insertions. (B) The degree of reduction of relative allelic expression is stronger for genes near TE insertions targeted by uniquely mapping siRNA. (C) There is no reduction of relative allelic expression for genes near TE insertions that are not targeted by uniquely mapping siRNA.
Discussion
In this study, we have quantified ASE to understand the role of cis-regulatory changes in association with a recent plant mating system shift. Our results indicate that many genes, on average over 40%, harbor cis-regulatory differences between C. rubella and C. grandiflora. The proportion of genes with ASE may seem high given the recent divergence (∼100 ka) between C. rubella and C. grandiflora (Brandvain et al. 2013, Slotte et al. 2013). However, the majority of genes with ASE showed relatively mild allelic expression biases, and while our estimates are higher than those in a recent microarray-based study of interspecific Arabidopsis hybrids (<10%) (He et al. 2012a), our results are consistent with recent analyses of RNAseq data from intraspecific F1 hybrids of Arabidopsis accessions (∼30%) (Cubillos et al. 2014). Somewhat higher levels of ASE were found in a recent study of maize and teosinte (∼70% of genes showed ASE in at least one tissue and F1 individual) (Lemmon et al. 2014), and using RNAseq data and the same hierarchical Bayesian analysis that we employed, Skelly et al. (2011) estimated that a substantially higher proportion, >70% of assayed genes, showed ASE among two strains of Saccharomyces cerevisiae. Thus, our estimates of the proportion of genes with ASE fall within the range commonly observed for recently diverged accessions or lines based on RNAseq data.
Two lines of evidence suggest that cis-regulatory changes have contributed to floral and reproductive adaptation to selfing in C. rubella. First, we find an excess of genes with ASE in flower buds within previously identified narrow QTL regions for floral and reproductive traits that harbor a signature of selection (Slotte et al. 2012). This suggests either that multiple cis-regulatory changes were involved in the evolution of the selfing syndrome in C. rubella or that these regions harbor an excess of cis-regulatory changes for other reasons, for instance, due to hitchhiking of cis-regulatory variants with causal variants for the selfing syndrome. Distinguishing between these hypotheses will require identification of causal genetic changes for the selfing syndrome in C. rubella. In contrast, no such excess is present for genes with ASE in leaves, suggesting that this observation is not simply a product of higher levels of divergence among C. rubella and C. grandiflora in certain genomic regions facilitating both QTL delimitation and ASE analysis. Second, we find that genes involved in hormonal responses, including brassinosteroid biosynthesis, are overrepresented among genes with ASE in flower buds but not in leaves. Based on a study of differential expression and functional information from A. thaliana, regulatory changes in this pathway were previously suggested to be important for the selfing syndrome in C. rubella (Slotte et al. 2013). While we do not identify ASE at the same genes as in Slotte et al. (2013), our work nonetheless provides support for cis-regulatory changes at other genes in the brassinosteroid pathway contributing to the selfing syndrome of C. rubella. Future studies should conduct fine-scale mapping and functional validation to fully explore this hypothesis. To facilitate this work, we have identified a set of candidate genes with ASE that are located in genomic regions harboring QTL for floral and reproductive trait divergence between C. rubella and C. grandiflora. Of particular interest in this list is the gene JAGGED (JAG), which is involved in determining petal growth and shape by promoting cell proliferation in A. thaliana (Sauret-Güeto et al. 2013, Schiessl et al. 2014). As C. rubella has reduced petal size due to a shortened period of proliferative growth (Sicard et al. 2011), and the C. rubella allele is expressed at a lower level than the C. grandiflora allele, this gene is a very promising candidate gene for the selfing syndrome.
Our work also provides general insights into the nature of cis-regulatory divergence. Indeed, many instances of ASE were specific to a particular individual or tissue, an observation also supported by recent studies (e.g., Lemmon et al. 2014, He et al. 2012a). This suggests that there is substantial variation in ASE depending on genotype and developmental stage, consistent with the reasoning that cis-regulatory changes can have very specific effects, but expression noise is probably also a contributing factor. It is also difficult to completely rule out the possibility that some cases of subtle ASE may not represent biologically meaningful cis-regulatory variation. However, in our analyses, we took several steps to model and account for technical variation to reduce the incidence of false positives. We also cannot fully rule out imprinting effects as potential causes of ASE, because generating reciprocal F1 hybrids was not possible due to seed abortion in C. rubella x C. grandiflora crosses. However, we do not expect these effects to make a major contribution to the patterns we observed; in Arabidopsis, imprinting effects are only prevalent in endosperm tissue, and are rare in more advanced stage tissues such as those analyzed here (Scott et al. 1998, Wolff et al. 2011, Cubillos et al. 2014), which suggests that imprinting is not likely to be responsible for the patterns we observe.
One somewhat unexpected finding was the global shift in expression levels toward higher relative expression of the C. rubella allele in the F1 hybrids. No marked bias was present for the same SNPs and genes in our genomic data, suggesting that if systematic bioinformatic biases are the cause, the effect is specific to transcriptomic reads. This seems unlikely to completely explain the shift in expression that we observe, as we made considerable effort to avoid reference mapping bias, including high stringency mapping of transcriptomic reads to reconstructed parental haplotypes specific to each F1. Similar global shifts toward higher expression of the alleles from one parent have also been observed in F1s of maize and teosinte (Lemmon et al. 2014) and Drosophila (McManus et al. 2010). An even stronger bias toward higher expression of the Arabidopsis lyrata allele was recently observed in F1s of A. thaliana and A. lyrata (He et al. 2012a) and was attributed to interspecific differences in gene silencing. Our results mirror those seen in some allopolyploids, where homeologs from one parental species can be expressed at a markedly higher level than those from the other parental species (e.g., Chang et al. 2010; Flagel and Wendel 2010; Schnable et al. 2011; Yoo et al. 2013).
To investigate potential mechanisms for cis-regulatory divergence, we first examined heterozygosity in regulatory regions and conserved noncoding regions close to genes. While genes with ASE in general showed slightly elevated levels of heterozygosity upstream of genes, there was no enrichment of conserved noncoding regions with heterozygous SNPs close to genes with ASE. It thus seems likely that divergence in regulatory regions in the proximity of genes, but not specifically in conserved noncoding regions, has contributed to global cis-regulatory divergence between C. rubella and C. grandiflora.
To examine biological explanations for the shift toward a higher relative expression of C. rubella alleles, we examined the relationship between TE insertions and ASE. As C. rubella harbors a lower number of TE insertions near genes than C. grandiflora, we reasoned that TE silencing might contribute to the global shift in expression toward higher relative expression of the C. rubella allele, with C. grandiflora alleles being preferentially silenced due to targeted methylation of nearby TEs, through transcriptional gene silencing mediated by 24-nt siRNAs. Our results are consistent with this hypothesis. Not only is there is an association between genes with TEs and heterozygous TE insertions in our F1s, there is also reduced expression of alleles that reside on the same haplotype as a nearby TE insertion, and the reduction is particularly strong for TEs that are targeted by uniquely mapping siRNAs. In contrast, no effect on ASE is apparent for TEs that are not targeted by uniquely mapping siRNAs. Moreover, the relatively limited spatial scale over which siRNA-targeted TE insertions are associated with reduced expression of nearby genes (<1 kb) is consistent with previous results from Arabidopsis (Hollister and Gaut 2009, Hollister et al. 2011, Wang et al. 2013). Our findings therefore suggest that silencing of TE insertions close to genes is important for global cis-regulatory divergence between C. rubella and C. grandiflora.
Why then do C. rubella and C. grandiflora differ with respect to silenced TEs near genes? In Arabidopsis, methylated TE insertions near genes appear to be predominantly deleterious and exhibit a signature of purifying selection (Hollister and Gaut 2009). The reduced prevalence of TE insertions near genes in C. rubella could be caused by rapid purging of recessive deleterious alleles due to increased homozygosity as a result of self-fertilization (Arunkumar et al. 2014). However, we prefer the alternative interpretation that deleterious alleles that were rare in the outcrossing ancestor were preferentially lost in C. rubella, mainly as a consequence of the reduced effective population size associated with the shift to selfing. This is in line with analyses of polymorphism and divergence at nonsynonymous sites, for which C. rubella exhibits patterns consistent with a general relaxation of purifying selection (Slotte et al. 2013).
If TE dynamics are generally important for cis-regulatory divergence in association with plant mating system shifts, we might expect different effects on cis-regulatory divergence depending not only on the genome-wide distribution of TEs but also on the efficacy of silencing mechanisms in the host (Hollister and Gaut 2009, Hollister et al. 2011, Ågren and Wright 2015). For instance, He et al. (2012a) found a shift toward higher relative expression of alleles from the outcrosser A. lyrata, which harbors a higher TE content, a fact which they attributed to differences in silencing efficacy between A. thaliana and A. lyrata; indeed, TEs also showed upregulation of the A. lyrata allele (He et al. 2012b), and A. lyrata TEs were targeted by a lower fraction of uniquely mapping siRNAs (Hollister et al. 2011). In contrast, we found no evidence for a difference in silencing efficacy between C. rubella and C. grandiflora, which harbor similar fractions of uniquely mapping siRNAs (12% vs. 10% uniquely mapping/total 24-nt RNA reads for C. rubella and C. grandiflora, respectively). Thus, in the absence of strong divergence in silencing efficacy, differences in the spatial distribution of TEs, such as those we observe between C. rubella and C. grandiflora, might be more important for cis-regulatory divergence. More studies of ASE in F1s of selfers of different ages and their outcrossing relatives are needed to assess the general contribution of differences in silencing efficacy versus genomic distribution of TE insertions for cis-regulatory divergence in association with mating system shifts.
Conclusions
We have shown that many genes exhibit cis-regulatory changes between C. rubella and C. grandiflora and that there is an enrichment of genes with floral ASE in genomic regions responsible for phenotypic divergence. In combination with analyses of the function of genes with floral ASE, this suggests that cis-regulatory changes have contributed to the evolution of the selfing syndrome in C. rubella. We further observe a general shift toward higher relative expression of the C. rubella allele, an observation that can in part be explained by elevated TE content close to genes in C. grandiflora and reduced expression of C. grandiflora alleles due to silencing of nearby TEs. These results support the idea that TE dynamics and silencing are of general importance for cis-regulatory divergence in association with plant mating system shifts.
Materials and Methods
Plant Material
We generated three interspecific C. grandiflora x C. rubella F1s by crossing two accessions of the selfer C. rubella as pollen donor with three accessions of the outcrosser C. grandiflora as seed parent (supplementary table S16, Supplementary Material online). No viable seeds were obtained from reciprocal crosses. Seeds from F1s and their C. rubella parental lines were surface sterilized and germinated on 0.5 x Murashige-Skoog medium. We transferred 1-week-old seedlings to soil in pots that were placed in randomized order in a growth chamber (16 h light: 8 h dark; 20 °C: 14 °C). After 4 weeks, but prior to bolting, we sampled young leaves for RNA sequencing. Mixed-stage flower buds were sampled 3 weeks later, when all F1s were flowering. To assess data reliability, we collected three separate samples of leaves and flower buds from one F1 individual and three biological replicates of one C. rubella parental line. For genomic DNA extraction, we sampled leaves from all three F1 individuals and from their C. rubella parents. For small RNA sequencing, we germinated six F2 offspring from one of our F1 individuals and sampled flower buds as described above.
Sample Preparation and Sequencing
We extracted total RNA for whole transcriptome sequencing with the RNEasy Plant Mini Kit (Qiagen, Hilden, Germany). For small RNA sequencing, we extracted total RNA using the mirVana kit (Life Technologies). For whole-genome sequencing, we used a modified CTAB DNA extraction (Doyle and Doyle 1987) to obtain predominantly nuclear DNA. RNA sequencing libraries were prepared using the TruSeq RNA v2 protocol (Illumina, San Diego, CA). DNA sequencing libraries were prepared using the TruSeq DNA v2 protocol. Small RNA libraries were prepared from 1 µg of total RNA using the TruSeq SmallRNA SamplePrep from Illumina according to the manufacturer's protocol (no. 15004197 rev E; Illumina, San Diego, CA). Sequencing was performed on an Illumina HiSeq 2000 instrument (Illumina, San Diego, CA) to gain 100-bp paired end reads, except for small RNA samples for which single end 50 bp reads were obtained. Sequencing was done at the Uppsala SNP & SEQ Technology Platform, Uppsala University, except for accession C. rubella Cr39.1 where genomic DNA sequencing was done at the Max Planck Institute of Developmental Biology, Tübingen. In total, we obtained 93.9 Gb (Q ≥ 30) of RNAseq data, with an average of 9.3 Gb per sample. In addition, we obtained 45.6 Gb (Q ≥ 30) of DNAseq data, corresponding to a mean expected coverage per individual of 52x, and 106,110,000 high-quality (Q ≥ 30) 50 bp small RNA reads. All sequence data have been submitted to the European Bioinformatics Institute (www.ebi.ac.uk, last accessed August 11, 2015), with study accession number: PRJEB9020.
Sequence Quality and Trimming
We merged read pairs from fragment spanning less than 185 nt (this also removes potential adapter sequences) in SeqPrep (https://github.com/jstjohn/SeqPrep, last accessed August 11, 2015) and trimmed reads based on sequence quality (phred cutoff of 30) in CutAdapt 1.3 (Martin 2011). For DNA and RNAseq reads, we removed all read pairs where either of the reads was shorter than 50 nt. We then analyzed each sample individually using fastQC v. 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, last accessed August 11, 2015) to identify potential errors that could have occurred in the process of amplifying DNA and RNA. We assessed RNA integrity by analyzing the overall depth of coverage over annotated coding genes, using geneBody_coverage.py that is part of the RSeQC package v. 2.3.3 (Wang et al. 2012). For DNA reads, we analyzed the genome coverage using bedtools v.2.17.0 (Quinlan and Hall 2010) and removed all potential PCR duplicates using Picard v.1.92 (http://broadinstitute.github.io/picard/, last accessed August 11, 2015). Small RNA reads were trimmed using custom scripts and CutAdapt 1.3 and filtered to retain only reads of 24 nt length.
Read Mapping and Variant Calling
We mapped both genomic reads and RNAseq reads to the v1.0 reference C. rubella assembly (Slotte et al. 2013) (http://www.phytozome.net/capsella). For RNAseq reads, we used STAR v.2.3.0.1 (Dobin et al. 2013) with default parameters. For genomic reads, we modified the default STAR settings to avoid splitting up reads, and for mapping 24-nt small RNA, we used STAR with settings modified to require perfect matches to the parental haplotypes of the F1s as well as to a TE library based on multiple Brassicaceae species and previously used in Slotte et al. (2013).
Variant calling was done in GATK v. 2.5-2 (McKenna et al. 2010) according to GATK best practices (DePristo et al. 2011, Van der Auwera et al. 2013). Briefly, after duplicate marking, local realignment around indels was undertaken, and base quality scores were recalibrated, using a set of 1,538,085 SNPs identified in C. grandiflora (Williamson et al. 2014) as known variants. Only SNPs considered high quality by GATK were kept for further analysis. Variant discovery was done jointly on all samples using the UnifiedGenotyper, and for each F1, genotypes were phased by transmission, by reference to the genotype of its highly inbred C. rubella parental accession.
We validated our procedure for calling variants in genomic data by comparing our calls for the inbred line C. rubella 1GR1 at 176,670 sites sequenced in a different individual from the same line by Sanger sequencing (Slotte et al. 2010). Overall, we found 29 calls that differed among the two sets, resulting in an error rate of 0.00016, considerably lower than the level of divergence among C. rubella and C. grandiflora (0.02; Brandvain et al. 2013).
Reconstruction of Parental Haplotypes of Interspecific F1s
We reconstructed genome-wide parental haplotype sequences for each interspecific F1 and used these as a reference sequence for mapping genomic and transcriptomic reads for ASE analyses. This was done to reduce effects of read mapping biases on our analyses of ASE by increasing the number of mapped reads and reducing mismapping that can result when masking heterozygous SNPs in F1s (Degner et al. 2009).
To reconstruct parental genomes for each F1, we first conducted genomic read mapping, variant calling, and phasing by reference to the inbred C. rubella parent as described in the section “Read Mapping and Variant Calling” above. The resulting phased vcf files were used in conjunction with the C. rubella reference genome sequence to create a new reference for each F1, containing both of its parental genome-wide haplotypes. Read mapping of both genomic and RNA reads from each F1 was then redone to its specific parental haplotype reference genome, and read counts at all reliable SNPs (see Filtering) were obtained using Samtools mpileup and a custom software written in javascript by Johan Reimegård. The resulting files with allele counts for genomic and transcriptomic data were used in all downstream analyses of allelic expression biases (see Analysis of ASE below).
Filtering
We used two approaches to filter the genome assembly to identify regions where we have high confidence in our SNP calls. Genomic regions with evidence for large-scale copy number variation were identified using Control-FREEC (Boeva et al. 2011), and repeats and selfish genetic elements were identified using RepeatMasker 4.0.1 (http://www.repeatmasker.org, last accessed August 11, 2015). Additionally, we identified genomic regions with unusually high proportions of heterozygous genotype calls in a laboratory-inbred C. rubella line, which is expected to be highly homozygous. Regions with evidence for high proportions of repeats, copy number variation, or high proportion of heterozygous calls in the inbred line mainly corresponded to centromeric and pericentromeric regions, and these were removed from consideration in further analyses of ASE (supplementary figs. S2–S5, Supplementary Material online).
Analysis of ASE
Analyses of ASE were done using a hierarchical Bayesian method developed by Skelly et al. (2011). The method requires read counts at heterozygous coding SNPs for both genomic and transcriptomic data. Genomic read counts are used to fit the parameters of a beta-binomial distribution, to obtain an empirical estimate of the distribution of variation in allelic ratios due to technical variation (as there is no true ASE for genomic data on read counts for heterozygous SNPs). This distribution is then used in analyses of RNAseq data where genes are assigned posterior probabilities of exhibiting ASE.
We conducted ASE analyses using the method of Skelly et al. (2011) for each of our three F1 individuals. Prior to analyses, we filtered the genomic data to only retain read counts for heterozygous SNPs in coding regions that did not overlap with neighboring genes, and following Skelly et al. (2011), we also removed SNPs that were the most strongly biased in the genomic data (specifically, in the 1% tails of a beta-binomial distribution fit to all heterozygous SNPs in each sample), as such highly biased SNPs may result in false inference of variable ASE if retained. The resulting data set showed very little evidence for read mapping bias affecting allelic ratios: The mean ratio of C. rubella alleles to total was 0.507 (supplementary fig. S6, Supplementary Material online).
All analyses were run in triplicate and Markov chain Monte Carlo convergence was checked by comparing parameter estimates across independent runs from different starting points and by assessing the degree of mixing of chains. For all analyses of RNA counts, we used median estimates of the parameters of the beta-binomial distribution from analyses of genomic data for all three F1s (supplementary table S8, Supplementary Material online). Runs were completed on a high-performance computing cluster at Uppsala University (UPPMAX) using the pqR implementation of R (http://www.pqr-project.org), for 200,000 generations or a maximum runtime of 10 days. We discarded the first 10% of each run as burn-in prior to obtaining parameter estimates.
ASE Validation by qPCR
We validated ASE results by performing qPCR with TaqMan Reverse Transcription Reagents (LifeTechnologies, Carlsbad, CA) using oligo(dT)16 s to convert mRNA into cDNA using the manufacturer’s protocol and performed qPCR with the Custom TaqMan Gene Expression Assay (LifeTechnologies, Carlsbad, CA) with the colors FAM and VIC using manufacturer’s protocol. The qPCR for both alleles was multiplexed in one well to directly compare the two alleles using a Bio-Rad CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA). To exclude color bias, we used reciprocal probes with VIC and FAM colorant (supplementary table S15, Supplementary Material online). The expression difference between the C. rubella and C. grandiflora allele was quantified using the difference in relative expression between the two alleles, as well as the quantification cycle (Cq value). A lower Cq value correlates with a higher amount of starting material in the sample. If the direction of allelic imbalance inferred by qPCR was the same as for ASE inferred by the method by Skelly et al. (2011), we considered that the qPCR supported the ASE results. For further details see supplementary text S1, Supplementary Material online.
Enrichment of Genes with ASE in Genomic Regions Responsible for Phenotypic Divergence
We tested whether there was an excess of genes with evidence for ASE (posterior probability of ASE ≥ 0.95 in all three F1 hybrids) in previously identified genomic regions harboring QTL for phenotypic divergence between C. rubella and C. grandiflora (Slotte et al. 2012). For this purpose, we concentrated on narrow QTL regions, defined as in a previous study (Slotte et al. 2012) (i.e., QTL regions with 1.5 logarithm of odds [LOD] confidence intervals <2 Mb). The five QTL regions that met our criteria for inclusion as narrow QTL were non-overlapping and corresponded to previously identified QTL for floral and reproductive traits (on scaffolds 2 and 7 for petal width, on scaffold 7 for petal length and on scaffolds 1 and 3 for flowering time). As QTL for floral and reproductive traits are generally highly overlapping, these genomic regions also encompass part of the confidence intervals for other QTL, including a major QTL for petal length on scaffold 2, and QTL for sepal length, stamen length, and ovule number on scaffold 7). Significance was based on a permutation test (1,000 permutations) in R 3.1.2.
List Enrichment Tests of GO Terms
We tested for enrichment of GO biological process terms using Fisher exact tests in the R package TopGO (Alexa et al. 2006). GO terms were downloaded from TAIR (http://www.arabidopsis.org) on September 3, 2013, for all A. thaliana genes that have orthologs in the C. rubella v1.0 annotation, and we only considered GO terms with at least two annotated members in the background set.
We tested for enrichment of GO biological process terms among genes with ASE in all of our F1s. Separate tests were conducted for leaf and flower bud samples, and background sets consisted of all genes where we could assess ASE in either leaves or flower buds.
We used the same approach to test for enrichment of GO biological process terms among genes within 1 kb and 2 kb of heterozygous TE insertions in F1 Inter4.1, for which we had matching small RNA data. For this purpose, separate tests were done for all heterozygous TE insertions, heterozygous TE insertions targeted by uniquely mapping siRNAs, and heterozygous TE insertions not targeted by siRNAs. For these tests, the background sets consisted of all annotated C. rubella genes.
Intergenic Heterozygosity in Regulatory and Conserved Noncoding Regions
We quantified intergenic heterozygosity 1 kb upstream of genes using VCFTools (Danecek et al. 2011) and compared levels of polymorphism among genes with and without ASE using a Wilcoxon rank sum test. We further assessed whether there was an enrichment of conserved noncoding elements (identified in Williamson et al. [2014]) with heterozygous SNPs within 5 kb of genes with ASE, using Fisher exact tests. Separate tests were conducted for each F1.
Identification of TE Insertions and Association with ASE
We used PoPoolationTE (Kofler et al. 2012) to identify TEs in our F1s. While intended for pooled datasets, this method can also be used on genomic reads from single individuals (Ågren et al. 2014). For this purpose, we used a library of TE sequences based on several Brassicaceae species (Slotte et al. 2013). We used the default pipeline for PoPoolationTE, modified to require a minimum of 5 reads to call a TE insertion, and the procedure in Ågren et al. (2014) to determine heterozygosity or homozygosity of TE insertions. Parental origins of TE insertions were inferred by combining information from runs on F1s and their C. rubella parents. We used chi-square tests to assess tested whether the composition of heterozygous TE insertions targeted by uniquely mapping siRNAs differed from those not targeted by siRNAs.
We tested whether heterozygous TE insertions within a range of different window sizes close to genes (200 bp, 1 kb, 2 kb, 5 kb, and 10 kb) were associated with ASE by performing Fisher exact tests. We tested whether the expression of the allele on the same chromosome as a nearby (within 1 kb) TE insertion was reduced compared with ASE at against genes without nearby TE insertions using a Wilcoxon rank sum test. Similar tests were conducted to test for an effect on relative ASE of TE insertions with uniquely mapping siRNAs.
Supplementary Material
Supplementary text S1, figures S2–S7, and tables S8–S16 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Daniel Skelly, Duke University, for helpful advice on ASE analyses, Emily Josephs, University of Toronto, and Adrian Platts, McGill University for information on conserved noncoding regions in Capsella, and Michael Nowak, Stockholm University, for valuable comments on the manuscript. Sequencing was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b2012122. This work was supported by grants from the Swedish Research Council, the Erik Philip-Sörensen foundation, the Nilsson-Ehle foundation, the Magnus Bergvall foundation, and the Royal Swedish Academy of Sciences to T.S. D.K. acknowledges funding from the Human Frontier Science Program (LT000783) and the German Research Foundation Priority Program 1529—“Adaptomics” (WE 2897).
References
- Ågren JA, Wang W, Koenig D, Neuffer B, Weigel D, Wright SI. 2014. Mating system shifts and transposable element evolution in the plant genus Capsella. BMC Genomics 15:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ågren JA, Wright SI. 2015. Selfish genetic elements and plant genome size evolution. Trends Plant Sci. 20:195–196. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607. [DOI] [PubMed] [Google Scholar]
- Arunkumar R, Ness RW, Wright SI, Barrett SCH. 2014. The evolution of selfing is accompanied by reduced efficacy of selection and purging of deleterious mutations. Genetics 199(3):817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SC. 2002. The evolution of plant sexual diversity. Nat Rev Genet. 3:274–284 [DOI] [PubMed] [Google Scholar]
- Bernacchi D, Tanksley SD. 1997. An interspecific backcross of Lycopersicon esculentum x L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147:861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Zinovyev A, Bleakley K, Vert J-P, Janoueix-Lerosey I, Delattre O, Barillot E. 2011. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27:268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9:e1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2000. Endless forms: the evolution of gene regulation and morphological diversity. Cell 101:577–580. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Chang PL, Dilkes BP, McMahon M, Comai L, Nuzhdin SV. 2010. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11:R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-Y, Cong B, Wing R, Vrebalov J, Tanksley SD. 2007. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318:643–645 [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Stegle O, Grondin C, Canut M, Tisné S, Gy I, Loudet O. 2014. Extensive cis-regulatory variation robust to environmental perturbation in Arabidopsis. Plant Cell 26:4298–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Lukens L. 1998. Transcriptional regulators and the evolution of plant form. Plant Cell 10:1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19: 11–15. [Google Scholar]
- Fishman L, Beardsley PM, Stathos A, Williams CF, Hill JP. 2015. The genetic architecture of traits associated with the evolution of self-pollination in Mimulus. New Phytol. 205:907–917 [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Willis JH. 2002. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution 56:2138–2155. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186:184–193 [DOI] [PubMed] [Google Scholar]
- Foxe JP, Slotte T, Stahl EA, Neuffer B, Hurka H, Wright SI. 2009. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci U S A. 106:5241–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB. 2011. Genome-wide approaches to the study of adaptive gene expression evolution: systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. Bioessays 33:469–477. [DOI] [PubMed] [Google Scholar]
- Goodwillie C, Ritland C, Ritland K. 2006. The genetic basis of floral traits associated with mating system evolution in Leptosiphon (Polemoniaceae): an analysis of quantitative trait loci. Evolution 60:491–504 [PubMed] [Google Scholar]
- Grillo MA, Changbao L, Fowlkes AM, Briggeman TM, Zhou A., Schemske DW, Sang T. 2009. Genetic architecture for the adaptive origin of wild rice, Oryza nivara. Evolution 63:870–883 [DOI] [PubMed] [Google Scholar]
- Guo Y-L, Bechsgaard JS, Slotte T, Neuffer B, Lascoux M, Weigel D, Schierup MH. 2009. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci U S A. 106:5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Zhang X, Hu J, Turck F, Dong X, Goebel U, Borevitz J, de Meaux J. 2012a. Genome-wide analysis of cis-regulatory divergence between species in the Arabidopsis genus. Mol Biol Evol. 29:3385–3395. [DOI] [PubMed] [Google Scholar]
- He F, Zhang X, Hu JY, Turck F, Dong X, Goebel U, Borevitz JO, de Meaux J. 2012b. Widespread interspecific divergence in cis-regulation of transposable elements in the Arabidopsis genus. Mol Biol Evol. 29:1081–1091. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. 2009. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo Y-L, Ott F, Weigel D, Gaut BS. 2011. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 108:2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Betancourt AJ, Schlötterer C. 2012. Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet. 8:e1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon ZH, Bukowski R, Sun Q, Doebley JF. 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet. 10:e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, O'Duff M, Eipper-Mains J, Graveley BR, Wittkopp PJ. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20: 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornduff R. 1969. Reproductive biology in relation to systematics. Taxon 18(2):121–133 [Google Scholar]
- Pastinen T. 2010. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 11:533–538. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebernig CA, Lafon-Placette C, Hatorangan MR, Slotte T, Köhler C. 2015. Non-reciprocal interspecies hybridization barriers in the Capsella genus are established in the endosperm. PLoS Genet. 11:e1005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E. 2013. JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol. 11:e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Muiño JM, Sablowski R. 2014. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc Natl Acad Sci U S A. 111:2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M. 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 108:4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG. 1998. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125:3329–3341. [DOI] [PubMed] [Google Scholar]
- Sicard A, Lenhard M. 2011. The selfing syndrome: a model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann Bot. 107(9):1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Stacey N, Hermann K, Dessoly J, Neuffer B, Bäurle I, Lenhard M. 2011. Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell 23:3156–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly DA, Johansson M, Madeoy J, Wakefield J, Akey JM. 2011. A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res. 21:1728–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. 2010. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 27:1813–1821. [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 45(7):831–835 [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Stern D, Andolfatto P, Wright SI. 2012. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution 66:1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge KR, Källman T, Slotte T, Lascoux M, Palmé AE. 2011. Contrasting demographic history and population structure in Capsella rubella and Capsella grandiflora, two closely related species with different mating systems. Mol Ecol. 20:3306–3320. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and Evolution in Plants. New York: Columbia Univ. Press. [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62(9):2155–2177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- Wang X, Weigel D, Smith LM. 2013. Transposon variants and their effects on gene expression in Arabidopsis. PLoS Genet. 9:e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RJ, Josephs EB, Platts AE, Hazzouri KM, Haudry A, Blanchette M, Wright SI. 2014. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. PLoS Genet. 10:e1004622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C. 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7:e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216 [DOI] [PubMed] [Google Scholar]
- Yoo MJ, Szadkowski E., Wendel JF. 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110:171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.