Abstract
Polymorphic inversions are a type of structural variants that are difficult to analyze owing to their balanced nature and the location of breakpoints within complex repeated regions. So far, only a handful of inversions have been studied in detail in humans and current knowledge about their possible functional effects is still limited. However, inversions have been related to phenotypic changes and adaptation in multiple species. In this review, we summarize the evidences of the functional impact of inversions in the human genome. First, given that inversions have been shown to inhibit recombination in heterokaryotes, chromosomes displaying different orientation are expected to evolve independently and this may lead to distinct gene-expression patterns. Second, inversions have a role as disease-causing mutations both by directly affecting gene structure or regulation in different ways, and by predisposing to other secondary arrangements in the offspring of inversion carriers. Finally, several inversions show signals of being selected during human evolution. These findings illustrate the potential of inversions to have phenotypic consequences also in humans and emphasize the importance of their inclusion in genome-wide association studies.
Keywords: inversions, human genome, phenotypic effects, disease, gene expression, evolution
Introduction
Chromosomal inversions were first discovered in Drosophila almost a century ago and they were the first type of genetic variants to be studied [1–3]. An inversion involves a change in orientation of a segment of DNA within a chromosome, but this balanced nature together with the fact that many of them are mediated by repeats complicate their analysis. Therefore, for a long time inversions have been largely overlooked, especially in humans. Recent genomic information, however, has revealed that structural variation is more common than previously thought, increasing the interest in this type of variants [4–7].
Inversions are often generated by non-allelic homologous recombination (NAHR) between inverted repeats, but they can also be originated by double-strand break repair mechanisms, like non-homologous end joining, or replication-based mechanisms mediated by microhomology, like fork stalling and template switching [8–11] (see also the accompanying review by Escaramís et al. [12] for a detailed explanation of each mechanism). Thus, in most cases no significant DNA gain or loss is produced. However, inversions have been demonstrated to affect phenotype in multiple species: from size or developmental time in Drosophila [13], to flowering time in plants [14], or adaptation to freshwater in sticklebacks [15], although the mechanisms by which they are able to do so are far from clear. Accordingly, depending on their phenotypic consequences, evolutionary forces such as natural selection or random drift will determine the fate of inversions (fixation or elimination), shape their geographical distributions and determine their frequencies in populations [16].
Inversions are considered to have both direct and indirect effects at the molecular level [3, 13, 16]. Indirect effects involve the effective suppression of recombination within the inverted sequence in heterozygotes. This can be caused either by an inhibition of crossovers in this region or by selection against the unbalanced gametes generated by single crossovers that take place within oppositely oriented segments. As a consequence, standard (Std) and inverted (Inv) chromosomes evolve separately and diverge, accumulating changes that remain associated with the arrangement in which they first appeared. Over time, this leads to the generation of distinct haplotypes in linkage disequilibrium (LD) with each arrangement that maintain together combinations of alleles that may be favorable under certain environmental conditions. This in turn could provide an advantage to one of the arrangements and cause its increase in frequency in the population [13, 17–19]. Alternatively, inversion breakpoints can directly disrupt coding sequences or alter gene expression of adjacent genes by separating regulatory elements from the corresponding coding sequences, providing new regulatory sequences, or moving the genes to different regulatory domains [20, 21]. Finally, for some inversions an additional direct consequence has been observed: the predisposition to other rearrangements, mainly copy number alterations and translocations [22].
In this review, we will focus on the current knowledge about the functional consequences of human polymorphic inversions, ranging from their role in disease to their effects on gene expression, as well as any possible signals of natural selection.
Human polymorphic inversions
Even though the existence of inversions during primate evolution and speciation has been long known [23, 24], we still lack a complete picture of how many polymorphic inversions exist in the human genome and their association with changes in gene expression, adaptation or disease. Human inversions have been typically studied by cytogenetic techniques such as G-banded karyotypes [25] that only allowed the identification of variants of several megabases. Other traditional molecular approaches that have been applied to the targeted detection of inversions are fluorescence in situ hybridization [26, 27], southern blot hybridization [28] or pulsed-field gel electrophoresis [29]. This resulted in the initial identification of a handful of human inversions, most of which were relatively large [30, 31].
In the past years, sequencing techniques have provided valuable applications to the global discovery of inversions (and other structural variants). First, whole-genome sequence comparison allowed us to identify inverted segments between independently assembled individuals [6, 32, 33]. Second, paired-end sequencing and mapping (PEM) supposed a major breakthrough in the field, and most predicted inversions so far have been detected using this strategy [4, 5, 7, 11, 34–36]. The method consists in sequencing the two ends of DNA segments of known size from a given individual and mapping the resulting reads to a reference genome (see the accompanying review by Escaramís et al. [12]). In the case of inversions, abnormal orientation mappings of the read pairs are indicative of the presence of a breakpoint. Both approaches should work well on simple, non-repetitive genomes, but they have important limitations when applied to complex genomes such as that of humans. Because they rely on a reference genome, miss-assembled regions in the reference are a source of spurious structural variation in the testing genome [33]. In addition, PEM usually fails to detect inversions flanked by long inverted repeats of high identity, such as repetitive elements and segmental duplications (SDs), owing to problems in identifying unique placements for the sequence reads close to the breakpoints [37, 38], and it also generates a high (and yet unknown) rate of false positives [39].
Given the limitations and the relatively low-throughput of the PEM strategy, computational algorithms have been developed that benefit from the interplay between structural and nucleotide variation to predict inversions in multiple individuals and/or estimate inversion genotypes [40–45]. These methods rely on two main assumptions: (i) inversions lie in regions of strong LD and generate specific LD patterns across the breakpoints, and (ii) SNP haplotypes are indicative of the inversion status. It is still unclear how accurate these predictions are, but a high rate of false positives (i.e. regions of high LD owing to low recombination or recent selective sweeps) and false negatives (i.e. small, rare, recent or recurrent inversions) is expected. Therefore, it is always necessary to perform targeted validations and population-wide genotyping using independent methods. PCR-based techniques are among the most widely used experimental methods to validate and genotype inversions, including regular or long-range PCR [5, 11, 24], haplotype-fusion PCR [46, 47] and inverse PCR [48, 49]. Other promising methodologies that could be applied to study inversions include optical mapping, which generates high-resolution restriction maps from single DNA molecules [50], and BioNano technology, which allows the construction of genomic maps by imaging linearized DNA molecules with fluorescent marks introduced at specific sites [51], although in both cases samples have to be processed one by one and the ability of these techniques to detect different types of inversions has to be assessed yet. In addition, we are currently developing a new method to genotype simultaneously multiple inversions with known breakpoints based on probe hybridization, which could be very useful to analyze a large number of individuals [52].
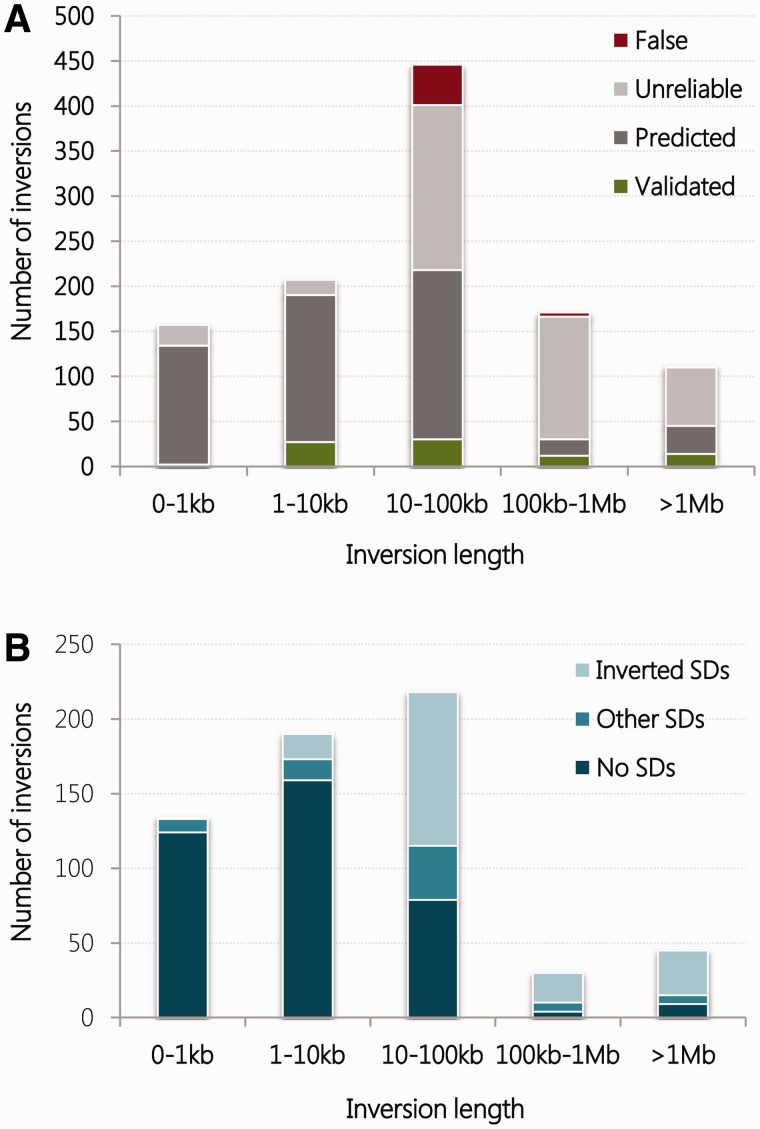
Over the past decade, the Database of Genomic Variants (http://dgv.tcag.ca) [53] has provided a publicly accessible, comprehensive catalogue of all types of structural variation in the human genome, including inversions. More recently, the new InvFEST database (http://invfestdb.uab.cat/) [39] represents an effort to generate a curated, non-redundant catalogue of human polymorphic inversions, which provides refined breakpoint locations, associations with genes and SDs, plus data on experimental validation, population frequency, known functional effects and evolutionary history (for easier access, InvFEST names will be provided for all polymorphic inversions mentioned in the review). In its current version, it contains 1092 candidate inversions, of which only 18% are reported by multiple studies and 85 have been validated experimentally (Figure 1). Interestingly, for those inversions with no experimental data (either validated or false), InvFEST provides information on their reliability according to different bioinformatic internal scores based on the PEM support of each prediction and the mapping to simple repetitive sequences (see [39] for further details). This categorization suggests that more than half of the predictions might be artifacts, supporting the suspicion that large-scale detection methods (which account for 98% of InvFEST inversions) have a high false-positive discovery rate. Still, an analysis of the inversions in InvFEST database reveals some features of human polymorphic inversions. First, although the size distribution of inversions is similar to that reported 5 years ago considering the set of inversions included in the Database of Genomic Variants [30], most inversions >10 kb could be false positives (Figure 2A). Second, as suggested by Pang et al. [11], when only validated and predicted inversions are considered, their size distribution according to the association with SDs (Figure 2B) shows that the longer the inversion, the more probable it is flanked by SDs. Nevertheless, the uncertainty about the final set of human polymorphic inversions complicates making reliable conclusions about the observed patterns.
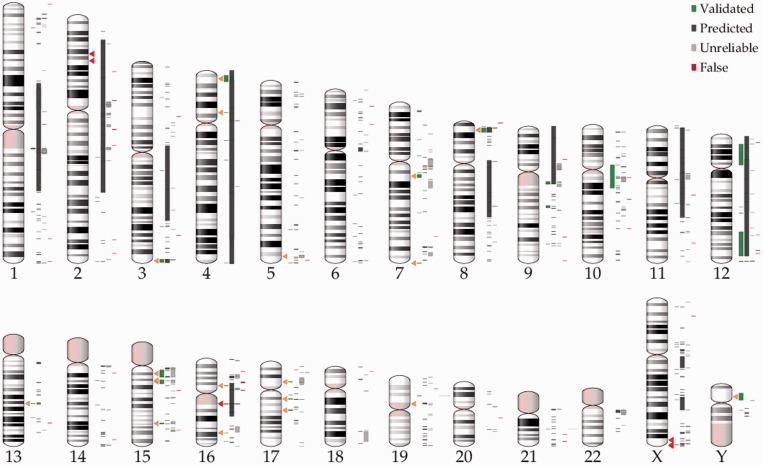
Figure 1.
Distribution of inversion polymorphisms in the human genome. The chromosomal positions of the 1092 inversions reported in InvFEST [39] are indicated to the right of each ideogram (with the status of inversions according to InvFEST represented in the same order as shown in the legend; see main text for details). Triangles mark the position of individual inversions discussed in the main text, with 5 pathogenic inversions in dark and 21 polymorphic inversions in light color. Please note that while triangle marks indicate the center of the inversion, some big inversions span several megabases, specially the pathogenic inversion in chromosome 16, which inverts almost the whole chromosome. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Figure 2.
Size distribution of human polymorphic inversions. (A) The size distribution of inversions in InvFEST [39] shows that the majority of inversions reported to date are <100 kb long, and that while inversions <10 kb long are mostly reliable, more than half of >10 kb inversions might be false positives. (B) If unreliable or false predictions are discarded, the majority of <10 kb inversions have simple breakpoints, while the majority of the remaining inversions are flanked and appear to be mediated by SDs. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Inversions as simple mutations causing disease
Many inversions traditionally detected in human karyotypes do not appear to have any phenotypic effects of clinical significance. This is the case of pericentric inversions (the inverted sequence includes the centromere) in chromosomes 1, 2, 3, 5, 9, 10 and 16, which mainly invert heterochromatic sequences and are frequently observed in cytogenetic analysis [25]. For example, the pericentric inversion in chromosome 9 shows variable frequencies in different human populations ranging from 0.26% in Asians to 3.57% in individuals from African origin [54]. In addition, despite the possible effects on fertility owing to the unbalanced chromosomes resulting from a crossover within the inverted region in heterozygotes, for some of these inversions no increased risk for miscarriage or problems in chromosome segregation during meiosis have been observed [55, 56]. Not all inversions are harmless, however, and several diseases have been found to be occasionally caused by inversions, mostly by direct disruption of one gene [57, 58] or by altering its gene expression [59, 60]. These inversions appear de novo in patients or are inherited mutations restricted to a given family, and thus they do not represent polymorphic variants segregating in human populations. Nevertheless, they have clinical importance and can contribute to the identification of genes underlying certain rare disorders [61, 62]. In this sense, it is possible that the contribution of structural variants to disease is currently underestimated owing to the frequent use of exome sequencing to identify the causal genes, a strategy that fails to detect genes truncated by a breakpoint.
Besides, there are a few inversions that have been demonstrated to produce some diseases in a recurrent manner, such as in hemophilia A and Hunter syndrome (a metabolic disorder called mucopolysaccharidosis type II). In hemophilia A, an inversion is generated by recombination between a 9.5 kb repeat in intron 22 of the F8 gene and one of two other copies located ∼565 kb away on the X chromosome [46, 63, 64]. This gene encoding coagulation factor VIII is disrupted by these inversions in ∼45% of severe hemophilia A patients [65, 66]. Another ∼140 kb inversion mediated by repeats, one of which is located in F8 intron 1, accounts for 4.8% of hemophilia A cases [66]. Apart from disrupting the gene, this second inversion also generates hybrid transcripts including the first F8 exon and exons of the VBP1 gene. The two types of inversions have been detected in familial hemophilia in which the mutation was inherited by the male patients from their unaffected mothers and in isolated cases where the inversion has been generated de novo [65]. In Hunter syndrome, an inversion truncates the X-linked IDS gene, which encodes the iduronate 2-sulfatase enzyme, and occurs by recombination with the homologous sequences of an adjacent partial pseudogene (IDS2) in 13% of patients [67]. Similarly, in some cancers, inversions disrupting coding regions [68] or creating fusion genes that can be directly related to the prognosis of the patient [69, 70], have been found in several unrelated individuals.
Polymorphic inversions associated to phenotypes or diseases
One of the main limitations in the study of the functional effects of human polymorphic inversions is the lack of genotyping information for most of them. However, although the underlying molecular mechanisms remain unclear, phenotypic effects have been associated to the two orientations of a few human inversion polymorphisms. The most striking and well-studied case is that of inversion 17q21.31 (HsInv0573) that inverts ∼970 kb of single-copy sequence containing the gene MAPT among others, and maintains two separated haplotypes: H1 and H2 (inverted with respect to the reference genome) [71]. The two haplotypes have an estimated divergence of 0.476% [72] and include different combinations of duplicated sequences at the breakpoints [73, 74]. H1 haplotype has been associated with several neurodegenerative diseases [75], including progressive supranuclear palsy [76], corticobasal degeneration [77], Alzheimer’s disease [78] and Parkinson’s disease [79–81], which exhibit aggregation of hyperphosphorylated Tau protein (encoded by gene MAPT) in neuronal cell bodies [77]. On the other hand, H2 haplotype has been associated with increased mean rates of recombination in females [71, 82]. Women with high recombination rates tend to have more children [83] and both traits were detected in H2 female carriers in the Icelandic population [71]. Similarly, in the 8p23.1 inversion (HsInv0501), the longest inversion polymorphism (∼4.5 Mb) described in humans, which is mediated by large complex repeats [26], risk alleles for systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), are apparently transmitted only on one of the Std haplotypes [44, 84], so the Inv haplotypes would protect against these diseases.
The recently developed computational methods to predict inversions based in nucleotide variation data [42–45] have opened a new way to investigate potential phenotypic effects of inversions from available genome wide association studies (GWAS). In the first successful application of this strategy, a previously identified inversion in 16p11 (HsInv0786), a 171 kb inverted region flanked by 134 kb inverted SDs [32], was genotyped from SNP data by the inveRsion [42] and PFIDO [44] algorithms, and association analysis revealed that the Inv allele protects against asthma and, more significantly, against the co-occurrence of asthma and obesity [85]. This association between inversion genotypes and the asthma-obesity phenotype was replicated in five independent studies and was not found when independent SNPs were considered, indicating that the observed effect is most likely owing to the whole haplotype [85]. In addition, Ma et al. [86] incorporated the inversions predicted with their principal components analysis-based approach from SNP data [43] in a GWAS analysis to find association of these regions with psoriasis, a chronic, inflammatory skin disease affecting 2–3% of the world population. They found significant associations of this disease with several candidate inversions, of which only two replicated in two different data sets. It turns out that neither of the two candidate inversions overlaps those reported in the InvFEST database [39]. Therefore, these results should be taken cautiously given that the SNP signatures found might not be owing to an inversion, although the regions detected by the association test may still shed light on the genetic architecture of the disease, as the authors point out [86].
Inversions predisposing to other rearrangements
While some inversions apparently do not have phenotypic consequences by themselves, carrying a certain allele may predispose to other rearrangements in the same genomic region, which in turn leads to a disease. In these cases, the parents of affected individuals show an increased frequency of one of the alleles of the inversion compared with the general population. One of the first described examples was that of the Williams-Beuren syndrome. This disease is caused by a 1.5 Mb deletion in chromosome 7 and an inversion involving the same region has been detected with a frequency of 12.4% in the transmitting parents of patients, although it has a frequency of 2.9% in control individuals [29, 87, 88]. Similar results have been observed for deletions causing Angelman syndrome, where the inversion has a frequency of ∼33% in mothers of patients with the class II deletion but only 4.5% in controls [89], and Sotos [90] or 17q21.31 microdeletion [91] syndromes, among others (Table 1). Most of these diseases are thought to be caused by haploinsufficiency of one of the deleted genes, since affected individuals usually carry one normal chromosome (the exceptions being Emery-Dreifuss muscular dystrophy, which is inherited in an X-linked recessive manner, and Angelman syndrome, which is caused by the loss of maternal expression of the imprinted UBE3A gene). In addition, while the causal gene is known in some cases (e.g. EMD for Emery-Dreifuss muscular dystrophy [95] and NSD1 for Sotos syndrome [96]), in others, like Williams-Beuren or 17q21.31 microdeletion syndromes, it has not yet been identified because the deletions are large and eliminate several genes [29, 91].
Table 1.
Human polymorphic inversions predisposing to other rearrangements
| Cytogenetic band | Inversion (InvFEST) | Inversion length | Secondary arrangement | Disorder | References |
|---|---|---|---|---|---|
| 3q29 | HsInv0264 | 1.9 Mb | Deletion | 3q29 microdeletion syndrome | [27] |
| 4p16 | HsInv0472 | 6 Mb | t(4;8)(p16;p23) | Wolf-Hirschhorn syndrome | [26] |
| 5q35 | HsInv0687 | 1.9 Mb | Deletion | Sotos syndromea | [90] |
| 7q11.23 | HsInv0301 | 1.5 Mb | Deletion | Williams-Beuren syndrome (WBS)a | [29, 88] |
| 8p23.1 | HsInv0501 | 4.5 Mb | Deletions/Duplications | Mental retardation and other problems | [92] |
| t(4;8)(p16;p23) | Wolf-Hirschhorn syndrome | [26] | |||
| 15q11-q13 | HsInv0549 | 4 Mb | Deletion | Angelman syndromea | [89] |
| 15q13.3 | HsInv1049 | 1.8 Mb | Deletion | 15q13 microdeletion syndrome | [93] |
| 15q24 | HsInv0544 | 1.2 Mb | Deletion | 15q24 microdeletion syndrome | [27] |
| 17q12 | HsInv1048 | 1.5 Mb | Deletion | RCAD syndrome | [27] |
| 17q21.31 | HsInv0573 | 970 kb | Deletion | 17q21.31 microdeletion syndromea | [91] |
| Xq28 | HsInv0389 | 48 kb | Deletion | Emery-Dreifuss muscular distrophy (EMD) | [28] |
| Yp11.2 | HsInv0415 | 4 Mb | PRKX/PRKY translocation | Sex reversal/male infertility | [94] |
aDisorders in which a higher frequency of the inversion has been found in the transmitting parents of the affected individuals compared with the general population.
Apart from deletions, inversions could also promote more complex intrachromosomal arrangements and translocations (Table 1). This is what happens in region 8p23.1, where the 4.5 Mb inversion could predispose to the formation of a dicentric chromosome carrying a single copy of the inverted region but duplications or deletions of a substantial fraction of chromosome 8 that have been associated with developmental delay and mental retardation, among other problems [92]. In mothers that are doubly heterozygous for the 8p23.1 inversion and another common inversion in chromosome 4 mediated by similar complex repeats, this same inverted region is involved in the generation of the recurrent unbalanced t(4;8)(p16;p23) translocation, which is one of the causes of Wolf-Hirschhorn syndrome [26]. Also, sex reversal caused by a translocation between homologous genes PRKX and PRKY (located on chromosomes X and Y, respectively) seems to occur preferentially on Y chromosomes carrying a ∼4 Mb inversion [46, 94].
The main mechanism by which one of the inversion alleles is thought to preferentially predispose to deletion rearrangements is through specific repetitive sequences in direct orientation. This can be achieved either by the switch of the relative orientation of pre-existing repetitive sequences by the inversion, or by accumulation of structural changes that increase the direct repeat content exclusively in one haplotype. The repeats could then recombine to generate structural variants that were not possible from the original arrangement. This is the situation found in inversion 17q21.31, where both H1 and H2 haplotypes have separately experienced independent duplications at the inverted repeats that mediated the inversion, which resulted in diverse complex structures ranging between 518 kb and 1 Mb of duplicated sequence associated to different sub-haplotypes [73, 74]. One of these sub-haplotypes, H2D, which is found at a higher frequency in European populations, predisposes to the 17q21.31 microdeletion syndrome [91] caused by the deletion of the inverted region by NAHR between 155 kb directly oriented repeats found only in this particular H2 haplotype [74]. Accordingly, the 17q21.31 microdeletion syndrome has been reported almost exclusively in individuals of European descent [97]. Furthermore, in the Emery-Dreifuss muscular dystrophy, the EMD gene deletion may be facilitated by recombination between two Alu elements located respectively in a Std and an Inv chromosome [28]. However, other factors may also play a role in the production of secondary rearrangements. For example, heterozygosity for a polymorphic inversion has been proposed to increase susceptibility to unequal recombination because it may lead to abnormal meiotic pairing [29, 98]. This is the case of the unstable dicentric chromosomes that are generated by recombination between the two arrangements within the 8p23.1 inverted region [92].
Despite the above evidences, we have to take into account that the predisposition to other rearrangements is weak given the low incidence of these diseases and the relatively high frequency of some inversions. Thus, the probability for an inversion carrier to have an affected child, although slightly higher than for a non-carrier, is still extremely low [88, 92]. Besides, not all patients for the diseases carry a deletion, and for those with other types of mutations, the presence of the inversion in their parents might be totally irrelevant. Even in some cases like the Williams-Beuren syndrome, recent studies have not found significant evidence of the association between inversion and deletion [99], suggesting that the increased susceptibility may not be completely clear or that it may be restricted to certain haplotypes. Similarly, an increase in the direct repeat length not always predisposes to deletion. For example, a 130 kb 15q13.3 inversion, which generates ∼188 kb of directly oriented SDs with 99.4% identity at deletion breakpoints (compared with 58 kb in the Std haplotypes), shows the same frequency among microdeletion cases than in the general population [100]. Therefore, further studies are necessary to quantify better the role of inversions in other rearrangements.
Effects of inversions on genes and gene expression
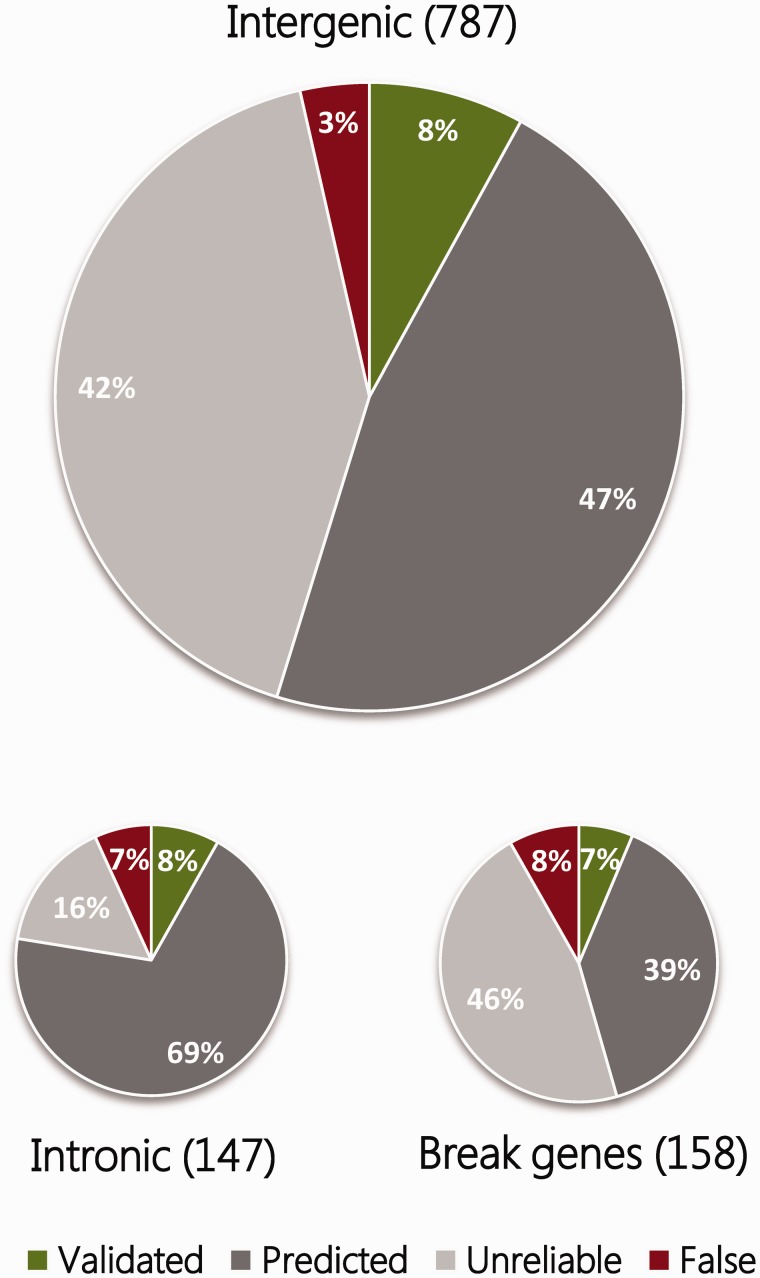
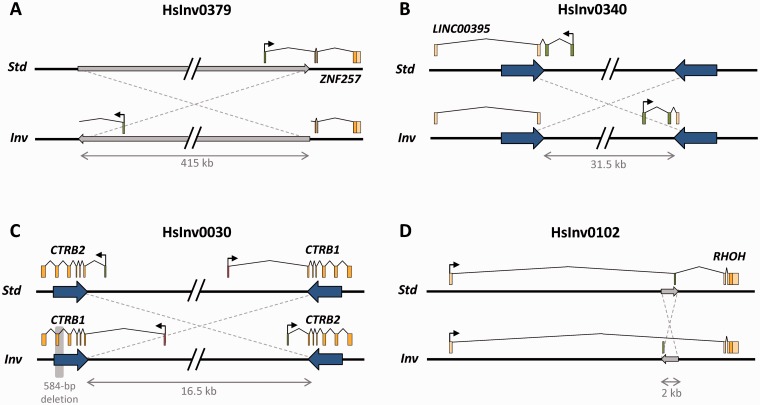
As mentioned in the introduction, inversions can directly affect genes and their regulation at different levels, with consequences that range from drastic to more subtle, and not all have been studied at the same detail. While it is relatively simple to determine if an inversion disrupts a gene (assuming that the exact position of the breakpoints is known), predicting the effects on gene regulation is much more difficult. Considering the most reliable inversions in the InvFEST database (i.e. those with a validated or predicted status), the majority of them are either intergenic or intronic, but ∼12% might break genes or at least some alternative gene transcripts (Figure 3). In several cases, two gene copies located within highly similar SDs at inversion breakpoints are exchanged in the inverted arrangement and no significant expression changes are expected [49]. For example, in HsInv0030 the first exon and promoter of the two chymotrypsinogen precursor genes CTRB1 and CTRB2 are exchanged in one of the inversion alleles [11, 36] (Figure 4). In addition, hybrid transcripts including exons from both genes have been detected [11], confirming the effect of the inversion in the transcript sequence. Other validated polymorphic inversions disrupt genes completely and move part of them several kb away (Figure 4), as happens for long non-coding RNA LINC00395 in HsInv0340 [49], gene ZNF257 in HsInv0379 [39], gene VIPR2 in HsInv0626 [101] and the putative pseudogene CCDC144B in HsInv1051 [49]. Inversions can also affect gene structure with unknown consequences to gene expression, like in HsInv0102 that inverts an alternative non-coding exon of gene RHOH (S. Villatoro and M. Cáceres, unpublished results) (Figure 4). For these cases, detailed analyses are needed to determine how the inversion affects the expression of both the broken genes and more distant genes. Another possibility is that an inversion relocates a gene to a region close to heterochromatin, thus suppressing its expression, as it occurs in position-effect variegation in Drosophila [102]. Although this phenomenon has not been reported in humans, topological domains associated with coordinately regulated gene clusters have been described in mammalian chromatin [103, 104] and the disruption of such domains by inversions might influence gene regulation.
Figure 3.
Classification of inversions according to their overlap with gene regions. The 1092 inversions reported in InvFEST [39] are classified into three main categories: (i) Intergenic, which do not disrupt any genic sequence, even though they might invert complete genes; (ii) Intronic, which are completely included within the intron of a gene; and (iii) Break genes, which are inversions that disrupt genes, either at their ends or inverting any internal exons. Colors indicate the status of inversions as shown in the legend, and illustrate that more than half of inversions that disrupt genic sequences might be false positives. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
Figure 4.
Examples of genes affected by polymorphic inversion breakpoints. (A) Disruption of the protein-coding sequence of gene ZNF257 by one inversion breakpoint of HsInv0379 located in the first intron of the gene [39]. (B) Disruption of a long non-coding RNA gene of unknown function by HsInv0340 [49]. (C) Exchange of CRTB1 and CRTB2 first exons by inversion HsInv0030 generated between inverted repeats overlapping the two genes [11, 36]. (D) Change of orientation of an alternatively spliced exon of gene RHOH by inversion HsInv0102, which no longer can be included in the transcript in the inverted orientation (S. Villatoro and M. Cáceres, unpublished results). Exons are depicted as light boxes with different shades indicating coding and non-coding parts of the transcripts and an arrow showing the direction of transcription. Exons affected by the inversion are represented as dark boxes. The minimum size of the inverted region is indicated below and in those inversions without inverted repeats at the breakpoints (large arrows); it is represented as a narrow gray arrow. In HsInv0030, a polymorphic deletion that removes one exon of CTRB1 in some inverted chromosomes is shaded in gray. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
To date, associations between inversion alleles and gene expression have been performed only for a few of the most studied human polymorphic inversions for which it was possible to predict the genotypes in many individuals [44, 85, 105]. All studies have focused on total expression levels with only a few tissues analyzed in each case, so differences relative to tissues and times of expression remain largely unexplored. Lymphoblastoid cell lines (LCLs) are the most used tissue because they are a common source for DNA and RNA, and several established collections like HapMap samples have available expression data. Also, the analysis has been usually limited to genes located within and around the inversion.
For inversions 8p23.1, 17q21.31 and 16p11, several genes located within the inverted region have been detected as differentially expressed between Std and Inv chromosomes [44, 85, 105]. These expression differences could be caused either by inversion breakpoints separating coding regions from regulatory elements, or by functional SNPs fixed in one of the divergent haplotypes maintained by the inversion, with the last option being generally favored [44, 85, 105]. Of the observed differences, some persist across tissues and populations, indicating a strong association with inversion alleles. This is the case for gene PPP1R3B in inversion 8p23.1, which is downregulated in Inv chromosomes in three different populations both in blood and LCLs [44], or for gene MAPT in inversion 17p21.31, which shows a higher expression in H1 chromosomes in frontal cortex and cerebellum [105, 106]. Some other genes show consistent differences across diverse data sets of the same origin. For example, BLK expression exhibits a positive correlation with the number of 8p23.1 Inv alleles in European populations that is not found in Asians or Africans [44]. Finally, the expression levels of several genes have been associated with the inversion only in a given data set, like genes XKR6, CTSB, NEIL2 and MSRA in LCLs for inversion 8p23.1 [44, 107], PLEKHM1 and CRHR1 in cerebellum for inversion 17q21.31 [105] or genes CCDC101 (blood) and IL27 (LCLs) for inversion 16p11 [85], which illustrates the complexity of determining the effects of inversions in gene expression. In inversion 16p11, other genes outside the inverted region but adjacent to the inversion breakpoints appear to be differentially expressed between arrangements as well (genes TUFM and SPNS1 show a higher expression in Inv alleles in two different tissues), suggesting that the inversion effects extend beyond the inverted region [85]. In addition, most of these expression differences follow an additive model in which the heterozygote shows an intermediate expression compared with both homozygotes, although there are also genes with higher expression in heterozygotes, like IL27 in inversion 16p11 [85].
On the other hand, expression differences have been detected in multiple-copy genes located within the SDs at the inversion breakpoints (e.g. LRRC37A and A4 in 17q21.31, or EIF3C and EIF3CL in 16p11) or in gene families with copies located both within and outside the inverted region (e.g. genes SULTA1, SULTA2 and SULTA4 in 16p11) [85, 105]. However, these differences should be taken with caution because it is difficult to quantify accurately gene expression from each individual copy either by microarrays or RNA-Seq without specific probes or specific mappings. This is exemplified by LRRC37A, whose expression shows highly significant differences between the 17q21.31 inversion alleles in several brain tissues, but the probes used are able to bind more than one target gene in the family [105]. In inversion 17q21.31 the situation is further complicated by the existence of H1 and H2 chromosomes with different numbers of partial copies of genes KANSL1 and NSF at the inversion breakpoints [73, 74], which might result in expression differences owing to the extra copies rather than to the inversion itself. In fact, new KANSL1 transcripts that may have an impact on female fertility are produced from both H1 and H2 haplotypes with partial KANSL1 duplications [73].
Remarkably, a few of the detected gene expression differences have been linked to phenotype, providing a mechanism by which the inversion could have functional consequences. BLK expression might mediate the increased risk for SLE and RA associated with one of the 8p23.1 inversion Std haplotypes [44], and overproduction of Tau protein encoded by MAPT (increased in inversion 17q21.31 H1 haplotypes) in neuronal cell bodies has been linked to several neurodegenerative disorders [77]. In inversion 16p11, expression changes in genes involved in energy balance and immunity like TUFM and IL27 might explain the protective effect of the Inv allele against co-occurrence of asthma and obesity [85]. However, in none of these cases a genome-wide differential expression analysis has been performed to identify possible downstream expression changes due to those generated within and around the inverted region. Such analysis, together with a more complete screening of human tissues, could provide a better understanding of the functional consequences of inversions at the organism level.
Inversions and natural selection
An alternative way to identify the functional impact of inversions is through the detection of the action of natural selection. Although inversions may have played an important role in primate speciation [108, 109], their role in shaping human populations is still uncertain. Basically, we lack tests to study the selective advantage or disadvantage of inversions, even at a small scale, and thus for just a handful of inversions their selective effects have been reported in terms of the frequency and geographic distribution of the alleles.
The 17q21.31 inversion is the most intensively studied inversion also in this sense. The inversion has been reported to be under positive selection in Europeans [71], where the Inv haplotype (H2) reaches a relatively high frequency (∼10–35%) [74]. Specifically, as mentioned before, women carrying either one or two copies of the inversion tend to have more children [71], but at the same time, H2D, the most common H2 haplotype in European populations, has been associated to disease-causing microdeletions [91]. Therefore, the haplotype associated to the detrimental duplication architecture predisposing to intellectual disability is also the one conferring advantage to female carriers, and is protective against neurodegenerative diseases [75–81]. These results, together with the global distribution of the inversion 17q21.31 haplotypes, led some authors to hypothesize that the high frequency of H2 in Europeans would be the result of founder effects instead of selection [72, 110]. However, the finding that H2 seems to be the ancestral haplotype [72], together with the analysis of nucleotide variation in the region and the dating of the different characterized haplotypes has raised the alternative hypothesis that the original H1 haplotypes first appeared and increased in frequency in Africa, and the complex duplicated haplotypes were independently generated later on, being maintained predominantly in populations that migrated out of Africa [74]. Advances in population genetics inference might help better explain the nucleotide variation patterns found in this region and differentiate between selection and demographic processes in the expansion of the H2D haplotype in Europe [31, 74].
For other inversions like 8p23.1, no clear signals of positive selection have been found in spite of multiple loci within the inverted region being putative targets of natural selection, including loci associated with autoimmune and cardiovascular disease [44]. In this case, the Inv allele is the ancestral but no SNPs are in perfect LD with the inversion haplotype, which suggests some level of gene flow between Std and Inv haplotypes, or that the inversion has appeared recurrently throughout evolution [27, 107]. Even though these observations hinder the study of the inversion, its worldwide clinal distribution is consistent with neutral demographic models of the human expansion out of Africa [44].
Finally, inversions HsInv0030 and 16p11 share some characteristics with the two inversions above [11, 85]. In HsInv0030, the Inv orientation is the ancestral one and is the major allele [11]. Furthermore, ∼6% of inverted alleles are associated to a 585 bp deletion that eliminates exon 6 of CTRB2. The authors found evidence of population differentiation in the haplotype frequencies, and together with the possible functional effect of the inversion and deletion alleles on the genes, they hypothesized that the observed differences could be the result of adaptation to different diets across populations [11]. Estimated inversion 16p11 allele frequencies also show remarkable worldwide population stratification, with the derived allele frequency (Std orientation) ranging from 90% in East Africa to 51% in Northern Europe, which has led to suggest the existence of selective sweeps for the inverted allele after human migration out of Africa [85]. Nevertheless, for none of the two inversions formal tests of selection have been performed.
Concluding remarks
As we have seen, different types of evidence indicate that inversions could have important consequences in the human genome. However, for most human inversions this has not been investigated yet and more work needs to be done to determine their effects in humans. In this sense, it is important to note that a high proportion of the inversions mediated by inverted repeats have apparently occurred multiple times in the human lineage [27, 49, 98], and the number of recurrent inversions could increase considerably as more individuals and more populations are studied. As a result, in contrast to copy number variants (CNVs), in which most of the disease associations found are mimicked by SNPs and have been already indirectly explored [111], recurrent inversions that are not tagged by SNPs have likely been missed from current GWAS studies and their potential effects remain hidden. Therefore, high-throughput methods are needed to directly genotype inversions in a high number of individuals and should contribute to unveil their real functional and evolutionary impact in the near future.
Key points.
While there are multiple examples of particular inversions leading to disease by disruption of coding regions or by affecting gene expression, only a few recurrent inversions have been described that repeatedly cause disease by these mechanisms.
Some polymorphic inversions segregating in human populations with no apparent phenotype predispose to pathogenic microdeletions in the offspring of carriers.
Diverse studies demonstrate that several inversions are linked to haplotypes that carry alleles with different expression between inverted and non-inverted chromosomes.
Although they are difficult to analyze, phenotypic effects have been associated to certain inversions that might be evolving under positive selection.
Acknowledgements
We thank the members of the Comparative and Functional Genomics group for helpful discussions and comments on the manuscript.
Biographies
Marta Puig is a postdoctoral researcher at the Institut de Biotecnologia i de Biomedicina of the Universitat Autònoma de Barcelona (Spain) working in the functional effects of inversions in humans.
Sònia Casillas is a postdoctoral researcher at the Institut de Biotecnologia i de Biomedicina and the Departament de Genètica i de Microbiologia of the Universitat Autònoma de Barcelona (Spain) with a long trajectory in bioinformatics and molecular evolution.
Sergi Villatoro is a PhD student at the Institut de Biotecnologia i de Biomedicina of the Universitat Autònoma de Barcelona (Spain) studying human genetic diseases related to structural variants.
Mario Cáceres is an ICREA Research Professor and leads the Comparative and Functional Genomics group at the Institut de Biotecnologia i de Biomedicina of the Universitat Autònoma de Barcelona (Spain) focused on the study of human inversions.
Funding
This work was supported by the European Research Council (ERC) under the European Union Seventh Research Framework Programme (FP7) [ERC Starting Grant 243212 (INVFEST) to M.C.] and the Ministerio de Economía y Competitividad (Spain) [BFU2013-42649-P to M.C.].
References
- 1.Sturtevant AH. Genetic factors affecting the strength of linkage in Drosophila. Proc Natl Acad Sci USA 1917;3:555–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobzhansky TG. Genetics of the Evolutionary Process. New York: Columbia University Press, 1970. [Google Scholar]
- 3.Krimbas CB, Powell JR. Drosophila Inversion Polymorphism. Boca Raton: CRC Press, 1992. [Google Scholar]
- 4.Tuzun E, Sharp AJ, Bailey JA, et al. Fine-scale structural variation of the human genome. Nat Genet 2005;37:727–32. [DOI] [PubMed] [Google Scholar]
- 5.Korbel JO, Urban AE, Affourtit JP, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science 2007;318:420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol 2007;5:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature 2008;453:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Khajavi M, Connolly AM, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 2009;41:849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam HYK, Mu XJ, Stütz AM, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol 2010;28:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd JM, Graves T, Newman TL, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 2010;143:837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang AWC, Migita O, Macdonald JR, et al. Mechanisms of formation of structural variation in a fully sequenced human genome. Hum Mutat 2013;34:345–54. [DOI] [PubMed] [Google Scholar]
- 12.Escaramís G, Docampo E, Rabionet R. A decade of structural variants: description, history, and methods to detect structural variation. Brief Funct Genomics 2015;14:305–14. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst 2008;39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol 2010;8:e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones FC, Grabherr MG, Chan YF, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 2012;484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biol 2010;8:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics 2006;173:419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joron M, Frezal L, Jones RT, et al. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 2011;477:203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson MJ, Jiggins CD. Supergenes and their role in evolution. Heredity 2014;113:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sperlich D, Pfriem P. Chromosomal polymorphisms in natural and experimental populations. In: Ashburner M, Carson HL, Thompson JN. (eds). The Genetics and Biology of Drosophila, Vol. 3e New York: Academic Press, 1986, 257–309. [Google Scholar]
- 21.Kleinjan D-J, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet 1998;7:1611–8. [DOI] [PubMed] [Google Scholar]
- 22.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet 2006;7:407–42. [DOI] [PubMed] [Google Scholar]
- 23.Yunis JJ, Prakash O. The origin of man: a chromosomal pictorial legacy. Science 1982;215:1525–30. [DOI] [PubMed] [Google Scholar]
- 24.Feuk L, MacDonald JR, Tang T, et al. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet 2005;1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas NS, Bryant V, Maloney V, et al. Investigation of the origins of human autosomal inversions. Hum Genet 2008;123:607–16. [DOI] [PubMed] [Google Scholar]
- 26.Giglio S, Calvari V, Gregato G, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 2002;71:276–85.12058347 [Google Scholar]
- 27.Antonacci F, Kidd JM, Marques-Bonet T, et al. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Genet 2009;18:2555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small K, Iber J, Warren ST. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat Genet 1997;16:96–9. [DOI] [PubMed] [Google Scholar]
- 29.Osborne LR, Li M, Pober B, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 2001;29:321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feuk L. Inversion variants in the human genome: role in disease and genome architecture. Genome Med 2010;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alves JM, Lopes AM, Chikhi L, et al. On the structural plasticity of the human genome: chromosomal inversions revisited. Curr Genomics 2012;13:623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin J, Han C, Gordon LA, et al. The sequence and analysis of duplication-rich human chromosome 16. Nature 2004;432:988–94. [DOI] [PubMed] [Google Scholar]
- 33.Khaja R, Zhang J, MacDonald JR, et al. Genome assembly comparison identifies structural variants in the human genome. Nat Genet 2006;38:1413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wang W, Li R, et al. The diploid genome sequence of an Asian individual. Nature 2008;456:60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn S-M, Kim T-H, Lee S, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res 2009;19:1622–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKernan KJ, Peckham HE, Costa GL, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res 2009;19:1527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onishi-Seebacher M, Korbel JO. Challenges in studying genomic structural variant formation mechanisms: the short-read dilemma and beyond. BioEssays 2011;33:840–50. [DOI] [PubMed] [Google Scholar]
- 38.Lucas Lledó JI, Cáceres M. On the power and the systematic biases of the detection of chromosomal inversions by paired-end genome sequencing. PLoS One 2013;8:e61292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Fundichely A, Casillas S, Egea R, et al. InvFEST, a database integrating information of polymorphic inversions in the human genome. Nucleic Acids Res 2014;42:D1027–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bansal V, Bashir A, Bafna V. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res 2007;17:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sindi SS, Raphael BJ. Identification and frequency estimation of inversion polymorphisms from haplotype data. J Comput Biol 2010;17:517–31. [DOI] [PubMed] [Google Scholar]
- 42.Cáceres A, Sindi SS, Raphael BJ, et al. Identification of polymorphic inversions from genotypes. BMC Bioinformatics 2012;13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J, Amos CI. Investigation of inversion polymorphisms in the human genome using principal components analysis. PLoS One 2012;7:e40224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salm MPA, Horswell SD, Hutchison CE, et al. The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genome Res 2012;22:1144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cáceres A, González JR. Following the footprints of polymorphic inversions on SNP data: from detection to association tests. Nucleic Acids Res 2015;43:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner DJ, Shendure J, Porreca G, et al. Assaying chromosomal inversions by single-molecule haplotyping. Nat Methods 2006;3:439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turner DJ, Tyler-Smith C, Hurles ME. Long-range, high-throughput haplotype determination via haplotype-fusion PCR and ligation haplotyping. Nucleic Acids Res 2008;36:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossetti LC, Radic CP, Larripa IB, et al. Genotyping the hemophilia inversion hotspot by use of inverse PCR. Clin Chem 2005;51:1154–8. [DOI] [PubMed] [Google Scholar]
- 49.Aguado C, Gayà-Vidal M, Villatoro S, et al. Validation and genotyping of multiple human polymorphic inversions mediated by inverted repeats reveals a high degree of recurrence. PLoS Genet 2014;10:e1004208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teague B, Waterman MS, Goldstein S, et al. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci USA 2010;107:10848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam ET, Hastie A, Lin C, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol 2012;30:771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cáceres M, Villatoro S, Aguado C. Inverse Multiplex Ligation-dependent Probe Amplification (iMLPA), an in vitro method of genotyping multiple inversions. Spain: European Patent Office, WO2015011200 (A1), 2015. [Google Scholar]
- 53.MacDonald JR, Ziman R, Yuen RKC, et al. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014;42:D986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu LY, Benn PA, Tannenbaum HL, et al. Chromosomal polymorphisms of 1, 9, 16, and Y in 4 major ethnic groups: a large prenatal study. Am J Med Genet 1987;26:95–101. [DOI] [PubMed] [Google Scholar]
- 55.Ferfouri F, Clement P, Gomes DM, et al. Is classic pericentric inversion of chromosome 2 inv(2)(p11q13) associated with an increased risk of unbalanced chromosomes? Fertil Steril 2009;92:1497.e1–4. [DOI] [PubMed] [Google Scholar]
- 56.Ait-Allah A, Ming P. The clinical importance of pericentric inversion of chromosome 9 in prenatal diagnosis. J Matern Investig 1997;7:126–8. [Google Scholar]
- 57.Jones ML, Murden SL, Brooks C, et al. Disruption of AP3B1 by a chromosome 5 inversion: a new disease mechanism in Hermansky-Pudlak syndrome type 2. BMC Med Genet 2013;14:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Utami KH, Hillmer AM, Aksoy I, et al. Detection of chromosomal breakpoints in patients with developmental delay and speech disorders. PLoS One 2014;9:e90852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anger GJ, Crocker S, McKenzie K, et al. X-linked deafness-2 (DFNX2) phenotype associated with a paracentric inversion upstream of POU3F4. Am J Audiol 2014;23:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brown KK, Reiss JA, Crow K, et al. Deletion of an enhancer near DLX5 and DLX6 in a family with hearing loss, craniofacial defects, and an inv(7)(q21.3q35). Hum Genet 2010;127:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blake J, Riddell A, Theiss S, et al. Sequencing of a patient with balanced chromosome abnormalities and neurodevelopmental disease identifies disruption of multiple high risk loci by structural variation. PLoS One 2014;9:e90894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lettice LA, Daniels S, Sweeney E, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat 2011;32:1492–9. [DOI] [PubMed] [Google Scholar]
- 63.Lakich D, Kazazian HH, Antonarakis SE, et al. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 1993;5:236–41. [DOI] [PubMed] [Google Scholar]
- 64.Bagnall RD, Giannelli F, Green PM. Polymorphism and hemophilia A causing inversions in distal Xq28: a complex picture. J Thromb Haemost 2005;3:2598–9. [DOI] [PubMed] [Google Scholar]
- 65.Antonarakis SE, Rossiter JP, Young M, et al. Factor VIII gene inversions in severe hemophilia A: results of an international consortium study. Blood 1995;86:2206–12. [PubMed] [Google Scholar]
- 66.Bagnall RD, Waseem N, Green PM, et al. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood 2002;99:168–74. [DOI] [PubMed] [Google Scholar]
- 67.Bondeson ML, Dahl N, Malmgren H, et al. Inversion of the IDS gene resulting from recombination with IDS-related sequences is a common cause of the Hunter syndrome. Hum Mol Genet 1995;4:615–21. [DOI] [PubMed] [Google Scholar]
- 68.Rhees J, Arnold M, Boland CR. Inversion of exons 1-7 of the MSH2 gene is a frequent cause of unexplained Lynch syndrome in one local population. Fam Cancer 2014;13:219–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–6. [DOI] [PubMed] [Google Scholar]
- 70.Gruber TA, Larson Gedman A, Zhang J, et al. An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 2012;22:683–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet 2005;37:129–37. [DOI] [PubMed] [Google Scholar]
- 72.Zody MC, Jiang Z, Fung H-C, et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 2008;40:1076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boettger LM, Handsaker RE, Zody MC, et al. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 2012;44:881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinberg KM, Antonacci F, Sudmant PH, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet 2012;44:872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pittman AM, Fung H-C, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Hum Mol Genet 2006;15:R188–95. [DOI] [PubMed] [Google Scholar]
- 76.Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 1999;8:711–5. [DOI] [PubMed] [Google Scholar]
- 77.Webb A, Miller B, Bonasera S, et al. Role of the tau gene region chromosome inversion in progressive supranuclear palsy, corticobasal degeneration, and related disorders. Arch Neurol 2008;65:1473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Myers AJ, Kaleem M, Marlowe L, et al. The H1c haplotype at the MAPT locus is associated with Alzheimer’s disease. Hum Mol Genet 2005;14:2399–404. [DOI] [PubMed] [Google Scholar]
- 79.Skipper L, Wilkes K, Toft M, et al. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am J Hum Genet 2004;75:669–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tobin JE, Latourelle JC, Lew MF, et al. Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology 2008;71:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Setó-Salvia N, Clarimón J, Pagonabarraga J, et al. Dementia risk in Parkinson disease: disentangling the role of MAPT haplotypes. Arch Neurol 2011;68:359–64. [DOI] [PubMed] [Google Scholar]
- 82.Fledel-Alon A, Leffler EM, Guan Y, et al. Variation in human recombination rates and its genetic determinants. PLoS One 2011;6:e20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kong A, Barnard J, Gudbjartsson DF, et al. Recombination rate and reproductive success in humans. Nat Genet 2004;36:1203–6. [DOI] [PubMed] [Google Scholar]
- 84.Namjou B, Ni Y, Harley ITW, et al. The Effect of Inversion at 8p23 on BLK Association with Lupus in Caucasian Population. PLoS One 2014;9:e115614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.González JR, Cáceres A, Esko T, et al. A common 16p11.2 inversion underlies the joint susceptibility to asthma and obesity. Am J Hum Genet 2014;94:361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma J, Xiong M, You M, et al. Genome-wide association tests of inversions with application to psoriasis. Hum Genet 2014;133:967–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bayés M, Magano LF, Rivera N, et al. Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 2003;73:131–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hobart HH, Morris CA, Mervis CB, et al. Inversion of the Williams syndrome region is a common polymorphism found more frequently in parents of children with Williams syndrome. Am J Med Genet C Semin Med Genet 2010;154C:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gimelli G, Pujana MA, Patricelli MG, et al. Genomic inversions of human chromosome 15q11-q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet 2003;12:849–58. [DOI] [PubMed] [Google Scholar]
- 90.Visser R, Shimokawa O, Harada N, et al. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am J Hum Genet 2005;76:52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koolen DA, Vissers LELM, Pfundt R, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet 2006;38:999–1001. [DOI] [PubMed] [Google Scholar]
- 92.Hollox EJ, Barber JC, Brookes AJ, et al. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res 2008;18:1686–97. [DOI] [PubMed] [Google Scholar]
- 93.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008;40:322–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jobling MA, Williams G, Schiebel K, et al. A selective difference between human Y-chromosomal DNA haplotypes. Curr Biol 1998;8:1391–4. [DOI] [PubMed] [Google Scholar]
- 95.Bione S, Maestrini E, Rivella S, et al. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 1994;8:323–7. [DOI] [PubMed] [Google Scholar]
- 96.Kurotaki N, Imaizumi K, Harada N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 2002;30:365–6. [DOI] [PubMed] [Google Scholar]
- 97.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet 2011;43:838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao PN, Li W, Vissers LELM, et al. Recurrent inversion events at 17q21.31 microdeletion locus are linked to the MAPT H2 haplotype. Cytogenet Genome Res 2010;129:275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frohnauer J, Caliebe A, Gesk S, et al. No significantly increased frequency of the inversion polymorphism at the WBS-critical region 7q11.23 in German parents of patients with Williams-Beuren syndrome as compared to a population control. Mol Cytogenet 2010;3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Antonacci F, Dennis MY, Huddleston J, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat Genet 2014;46:1293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beri S, Bonaglia MC, Giorda R. Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. Eur J Hum Genet 2013;21:757–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vogel MJ, Pagie L, Talhout W, et al. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenetics Chromatin 2009;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nora EP, Lajoie BR, Schulz EG, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012;485:381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Le Dily F, Baù D, Pohl A, et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev 2014;28:2151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Jong S, Chepelev I, Janson E, et al. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics 2012;13:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet 2007;39:1494–9. [DOI] [PubMed] [Google Scholar]
- 107.Bosch N, Morell M, Ponsa I, et al. Nucleotide, cytogenetic and expression impact of the human chromosome 8p23.1 inversion polymorphism. PLoS One 2009;4:e8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marquès-Bonet T, Cáceres M, Bertranpetit J, et al. Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet 2004;20:524–9. [DOI] [PubMed] [Google Scholar]
- 109.Marquès-Bonet T, Sànchez-Ruiz J, Armengol L, et al. On the association between chromosomal rearrangements and genic evolution in humans and chimpanzees. Genome Biol 2007;8:R230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Donnelly MP, Paschou P, Grigorenko E, et al. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am J Hum Genet 2010;86:161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Craddock N, Hurles ME, Cardin N, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 2010;464:713–20. [DOI] [PMC free article] [PubMed] [Google Scholar]