Abstract
Heterologous expression of many proteins in bacteria, yeasts, and plants is often limited by low titers of functional protein. To address this problem, we have created a two-tiered directed evolution strategy in Escherichia coli that enables optimization of protein production while maintaining high biological activity. The first tier involves a genetic selection for intracellular protein stability that is based on the folding quality control mechanism inherent to the twin-arginine translocation (Tat) pathway, while the second is a semi-high-throughput screen for protein function. To demonstrate the utility of this strategy, variants of the endoglucanase Cel5A from the plant pathogenic fungus Fusarium graminearum were isolated whose production was increased by as much as 30-fold over the parental enzyme. This gain in production was attributed to just two amino acid substitutions, and was isolated after two iterations through the two-tiered approach. There was no significant trade-off in activity on soluble or insoluble cellulose substrates. Importantly, by combining the folding filter afforded by the Tat quality control mechanism with a function-based screen, we show enrichment for variants with increased protein abundance in a manner that does not compromise catalytic activity, providing a highly soluble parent for engineering of improved or new function.
Introduction
Obtaining high levels of stable and functional proteins in Escherichia coli is of vital importance for industrial and medical endeavors [1, 2] as well as for structure determination by x-ray crystallography and nuclear magnetic resonance [3]. However, many recombinant proteins, especially those of non-bacterial origin, are produced inefficiently in E. coli as a result of improper folding, aggregation, and inclusion body formation [4]. To address the molecular bottlenecks associated with recombinant protein production in E. coli, a variety of strategies have been explored including optimization of expression temperature, introduction of different promoters and other upstream/downstream genetic elements, codon harmonization, chaperone co-expression, and genetic fusion to solubility-enhancing partner proteins [4–6]. Numerous success stories notwithstanding, these techniques are often inadequate because they have no direct effect on the intrinsic properties of the target protein. As a result, even when soluble expression is achieved, the protein may become inactive or aggregate following the processes of purification, fusion partner removal, and storage.
An alternative strategy for optimizing gene expression is fine-tuning of the protein sequence in a manner that improves intrinsic folding yield, stability, and solubility. Protein stability can be enhanced by site-directed rational mutation [7] using general strategies such as entropic stabilization [8, 9], helix capping [10, 11], and computational modeling of these and other complex contributions (e.g., optimized core packing, increased burial of hydrophobic surface area) simultaneously [12]. Rational design has similarly been used to generate proteins with improved solubility in a particular host environment [13]. However, the use of rational design can be extremely challenging because the “rules” for protein stability –i.e., how stability of a protein is encoded in its sequence and how specific amino acid changes influence stability– are poorly understood. Furthermore, rational design is only applicable to proteins of known three-dimensional structure.
Directed evolution, in which protein diversity libraries are interrogated for stability- or solubility-enhanced variants, provides an alternative route to obtaining appreciable levels of recombinant proteins, especially when a protein’s structure is unknown [14–16]. Even in cases where a high-resolution structure is available, directed evolution is often the preferred strategy because of its propensity for revealing mutational combinations that would be impossible to deduce by rational means (e.g., phylogenetic comparisons, sequence statistics, or statistical structural comparisons). The crux of this approach is an assay that permits screening or selecting for expressed library members for improved folding robustness, stability, and solubility. Function-based in vivo screening and selection methods have been used to increase the production of diverse protein targets in E. coli [17–19]; however, the protein-specific nature of these assays makes them intractable to other unrelated targets.
A more general approach involves structure- and function-independent assays that couple the folding and solubility of a protein-of-interest with a screenable or selectable activity [20–24]. The advantage of these assays is that they do not require any prior structural or functional knowledge and thus can be applied to virtually any protein. For example, using a genetic selection that relies on the folding quality control (QC) inherent to the E. coli twin-arginine translocation (Tat) pathway [22], we previously optimized the stability and solubility of single-chain Fv (scFv) antibodies [25]. However, because the isolation of solubility-enhanced variants proceeds without a need to maintain function, it is anticipated that many of the hits recovered using these selections will exhibit decreased biological activity as a result of stability-function tradeoffs [26–28]. Indeed, while solubility-enhanced scFv antibodies could be readily identified by genetic selection in E. coli, the binding activity for many of the selected clones was significantly diminished [25].
To remedy this situation, we developed a two-tiered directed evolution strategy for optimizing intracellular stability of protein, where ‘intracellular stability’ is used here to denote both the chemical solubility related to folding and the absence of aggregation or degradation, while retaining its native function. The first tier, based on our previous Tat QC-based folding reporter [22], enables rapid genetic selection of highly expressed variants which are subsequently input to a second tier involving a semi-high-throughput activity assay for functional screening. The utility of this strategy was demonstrated by optimizing production of the glycoside hydrolase family 5 (GH5) enzyme FG03795 from the phytopathogen Fusarium graminearum (hereafter called FgCel5A). Cellulases such as FgCel5A are of preeminent importance for agriculture, biotechnology and bioenergy uses, especially in the development of renewable liquid biofuels [29]. Unfortunately, a major impediment to the advancement of many cellulolytic enzymes is the fact that their expression in a tractable host such as E. coli is often met with low solubility, improper folding, and weak activity [1]. Using our two-tiered directed evolution strategy, we isolated an FgCel5A variant whose production was increased by 30-fold and whose enzymatic activity was virtually unchanged compared to the parental enzyme. Importantly, the ability to optimize FgCel5A abundance without impairing its activity may provide a highly evolvable starting point from which enhanced cellulolytic function can be engineered in the future. Moreover, given the general nature of this methodology, there is the potential for intracellular stabilization of other unrelated enzymes.
Results
Target identification for directed evolution
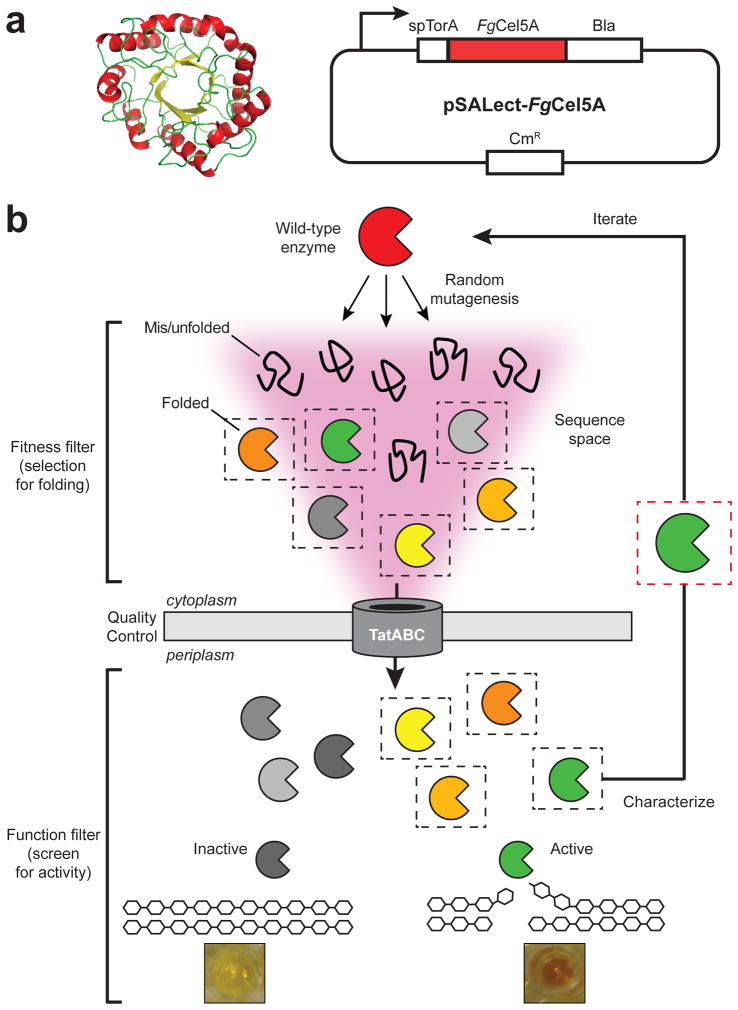
Previous studies suggested that F. graminearum produces one or more highly active cell wall-degrading enzymes (CWDEs) that could be used in enzyme cocktails for breaking down cellulosic materials [30]. Here, we focused on FgCel5A, a GH5 family endo-β-1,4-glucanase. The rationale for choosing FgCel5A was based on the following observations. First, FgCel5A is highly expressed when the fungus is grown on cell walls and in planta during the infection process, indicating a strong natural selection with pathogenicity on grasses. Second, the GH5 family is abundant (at least 5 are expressed) in this fungus and has the widely distributed (β/α)8 barrel structure, which has been proposed as a scaffold for both natural and artificial selection and evolution [31]. However, because the structure of FgCel5A is unknown, we opted for a random directed evolution approach. It should be pointed out that even though the structure for FgCel5A has not been solved, it shares sequence identity with PDB entries 1GZJ (the catalytic domain (CD) of a GH5 from Thermoascus aurantiacus) and 1H1N (the carbohydrate-binding module (CBM) of a GH7 from Trichoderma reesei). Thus, a homology model of the FgCel5A CD was created (Fig. 1a), which could help to estimate the structural context of any identified mutations.
Figure 1. Optimizing protein production by directed evolution.
(a) Homology model of the FgCel5A CD (left) and plasmid pSALect-FgCel5A in which a Tat-dependent export signal (spTorA) directs the FgCel5A-Bla fusion to the periplasm of E. coli (right). (b) Schematic of the two-tiered directed evolution strategy. In the first tier, Tat QC-based genetic selection is applied, linking protein stability in vivo with resistance to β-lactam antibiotics. Since Bla must be in the periplasm to confer resistance to β-lactam antibiotics, only correctly folded fusion proteins that pass the Tat QC are capable of rendering cells drug resistant. Proteins that misfold and/or aggregate are not exported from the cytoplasm, and thus cells expressing these proteins are sensitive to β-lactam antibiotics. This selection allows for rapid isolation of well-folded FgCel5A library members while eliminating those that are poorly folded, thereby focusing the sequence space. In the second tier, an activity screen is imposed to ensure that the proteins that pass Tat QC retain high activity. For FgCel5A, this involves an enzymatic activity assay using the soluble cellulose substrate CMC. Following isolation of variants, the enzymes are characterized for soluble production and activity, and the fittest members are used for further iterations through the two-step selection and screen.
As a preliminary test, a codon-optimized version of FgCel5A without its native signal peptide was expressed from a pET vector in the cytoplasm of E. coli strain BL21(DE3). Consistent with the low solubility that is typically associated with fungal cellulase expression in E. coli [1], FgCel5A accumulated exclusively in the insoluble fraction that was harvested from BL21(DE3) cells with only a faint band detected in the soluble fraction (Supplementary Fig. S1). This extremely poor solubility was observed regardless of whether the affinity tags were placed N- or C-terminally. Given that FgCel5A is naturally secreted and contains six cysteine residues, we suspected that soluble expression could be improved through the use of Origami B(DE3) cells, an isogenic derivative of BL21(DE3) that carries mutations allowing the formation of disulfide bonds in its cytoplasm [32]. Consistent with our hypothesis, the soluble fraction derived from Origami B(DE3) cells showed a small increase in the amount of soluble FgCel5A (Fig. S1). Nonetheless, the majority of FgCel5A still partitioned to the insoluble fraction. Taken together, the limited solubility of FgCel5A, especially in BL21(DE3) cells, makes this enzyme an ideal target for intracellular stability optimization.
Enhanced production of active FgCel5A by two-tiered directed evolution
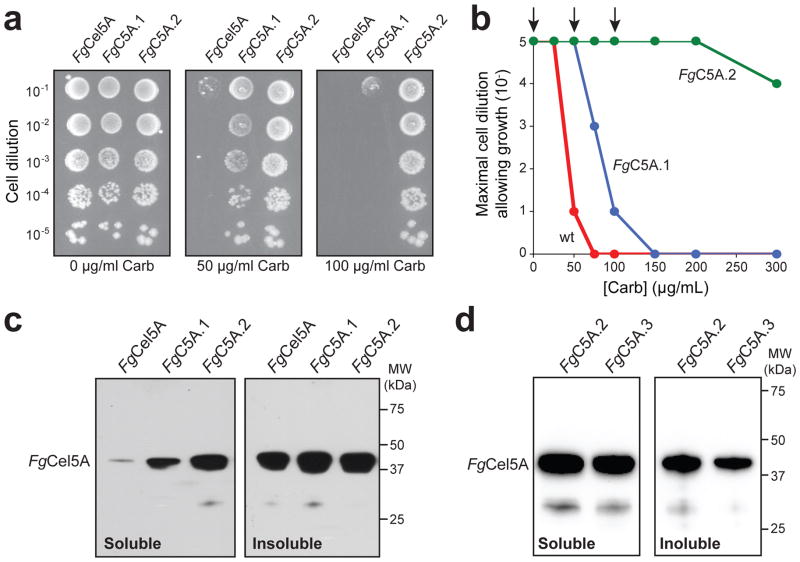
To improve FgCel5A production, we developed a two-tiered directed evolution strategy for selecting protein variants with increased stability in vivo (Fig. 1b). The first tier involved a genetic selection that links intracellular stability, as assessed by the QC mechanism inherent to the Tat pathway, with antibiotic resistance [22]. This selection is based on the observation that Tat QC in E. coli restricts export to properly folded proteins [33, 34] and can thus provide a selective filter for correct folding and robust expression that effectively narrows the sequence space to be searched [22, 24, 25]. Here, the gene encoding codon-optimized FgCel5A lacking its native export signal was cloned between genes encoding the Tat-dependent signal peptide of E. coli trimethylamine N-oxide reductase (spTorA) and TEM-1 β-lactamase (Bla), creating the plasmid pSALect-FgCel5A (Fig. 1a). To showcase the potential of this strategy for improving production, we opted to use E. coli cells with a reducing cytoplasm, namely strain BL21(DE3), because soluble FgCel5A production in these cells was extremely limited (Fig. S1) and thus represented a significant protein engineering challenge. Indeed, when cells expressing FgCel5A from pSALect were plated on media containing different concentrations of carbenicillin (Carb), resistance was only observed at very low levels (25 μg/mL) of Carb (Fig. 2a and b). To improve intracellular stability, a plasmid library was created by subjecting the gene encoding FgCel5A to error-prone PCR mutagenesis. Cells transformed with the library were plated on media containing 50 μg/mL Carb, a concentration of antibiotic to which cells expressing the wild-type (wt) FgCel5A fusion were sensitive (Fig. 2a and b). From the colonies that grew at this selective pressure, 178 hits were isolated for further characterization.
Figure 2. Tat QC-based selection of improved FgCel5A variants.
(a) Representative spot plating of serially diluted E. coli MC4100 cells expressing wt or mutant FgCel5A enzymes from pSALect as indicated. Overnight cultures were normalized to an equivalent number of cells, serially diluted in liquid LB, and plated on LB agar supplemented with varying concentrations of Carb. The spot plates shown are for no selective antibiotic (0 μg/mL Carb) and the concentrations used for the two library selections (50 μg/mL Carb and 100 μg/mL Carb). FgC5A.1 was selected from the first round library while FgC5A.2 was selected from the second round library. (b) Kill curves for wt FgCel5A (red circle), FgC5A.1 (blue circle) and FgC5A.2 (green circle). Plotted data represents the lowest cell dilution that allows for detectable growth at each concentration of Carb. Arrows indicate data that corresponds to the spot plates in (a). (c) Western blot analysis of soluble and insoluble fractions derived from BL21(DE3) cells expressing the wt FgCel5A and isolated mutants from pET28a as indicated. Enzymes were produced in the cytoplasm without the Tat export signal or Bla reporter. Each lane corresponding to the soluble fraction was loaded with 3 μg total protein while the insoluble lanes contained 1 μg total protein. Detection of wt and mutant FgCel5A enzymes was performed using an anti-FLAG antibody. (d) Same as (c) except fractions were derived from BL21(DE3) cells expressing the wt FgCel5A and site-directed FgC5A.3 mutant from pET28a as indicated.
Next, to identify solubility-focused mutants that retained high cellulolytic activity, the selected clones from the first tier were subjected to a semi-high-throughput screen for enzymatic activity. Specifically, a 96-well plate-based screen using carboxymethyl cellulose (CMC) followed by reaction with 3,5-dinitrosalicylic acid (DNS) was used to determine the amount of reducing ends produced [35, 36]. Following this second tier, a total of 12 clones were identified that maintained a similar level of activity compared to the parental enzyme. To ensure that the growth phenotype associated with these clones was not due to spontaneous mutations in the host genome or elsewhere on the plasmid, the FgCel5A genes corresponding to these variants were back-cloned into a fresh plasmid backbone and tested for sensitivity to antibiotic. Clone FgC5A.1 conferred the highest resistance to cells compared to other isolates and much greater resistance than wt FgCel5A (Fig. 2a and b); hence, this clone was chosen for further studies. DNA sequencing of FgC5A.1 revealed two amino acid substitutions: G10D in the CBM and T332I in the CD (Table 1). To further enhance intracellular stability, a second round of directed evolution was performed exactly as described above but with FgC5A.1 as the starting point and 100 μg/mL Carb as the selection pressure. Of the 186 hits that were screened for cellulase activity, clone FgC5A.2 was chosen because it retained high activity while conferring the highest resistance to cells compared to other round-two isolates (Fig. 2a and b). DNA sequencing of FgC5A.2 identified two additional amino acid changes: T166S and D342G, both in the CD (Table 1).
Table 1.
Production and specific activity of FgCel5A and related variants
| Enzyme1 | Mutations | Production (U/L)2 | Specific activity (U/nmol)3 | ||
|---|---|---|---|---|---|
| CBM | CD | 50°C | 37°C | ||
|
|
|||||
| FgCel5A | - | - | 0.15 ± 0.06 | 1.19 ± 0.12 | 0.61 ± 0.06 |
| FgC5A.1 | G10D | T332I | 0.80 ± 0.44 | 1.28 ± 0.07 | 0.66 ± 0.04 |
| FgC5A.2 | G10D | T166S, T332I, D342G | 4.49 ± 1.24 | 1.25 ± 0.06 | 0.63 ± 0.03 |
| FgC5A.3 | - | T332I, D342G | 4.51 ± 0.11 | 1.21 ± 0.09 | 0.65 ± 0.05 |
Each enzyme was expressed from pET28a without a Tat export signal or Bla reporter.
Cellulase production involved hydrolysis of CMC to a desired value and determination of the lysate volumes necessary to obtain such a value according to IUPAC standards. Hydrolysis was carried out using 1% CMC for 13 h at 37°C and sugar reducing ends were determined by reaction with DNS. Error is reported as the standard deviation from at least three biological replicates of the experiment.
Specific activity on CMC was measured using purified enzymes at 50°C and 37°C according to the IUPAC standard protocol for β-1,4-endoglucanase on CMC. Error is reported as the propagated error from three replicates of the experiment and error in the concentration measurement for the purified enzymes.
Evolved FgCel5A mutants retain cellulase activity
We next sought to verify that the Tat QC selection step yielded target enzymes whose production in vivo was enhanced, as opposed to improvements in translocation or folding of the entire fusion. This involved expressing the wt FgCel5A and two engineered variants in the cytoplasm of BL21(DE3) cells without the export signal peptide or Bla reporter. In agreement with the resistance data, soluble accumulation of each enzyme increased significantly with each round of directed evolution (Fig. 2c). To quantify this increase in soluble production, the amount of cellulase per culture volume was determined by assaying cell lysates on CMC. Using the volume of lysate required to hydrolyze a fixed amount of CMC, it was determined that the activity of FgC5A.1 per culture volume increased by >5 fold compared to wt FgCel5A (Table 1). Likewise, the activity of FgC5A.2 on CMC per culture volume increased by nearly 6 fold compared to FgC5A.1 (Table 1).
Next, the wt and two FgCel5A variants were purified and their specific activity was measured in vitro. Each was recovered at greater than 85% purity using Ni2+-affinity chromatography (Fig. S2) and subsequently assayed for endocellulase activity on CMC (CMCase activity) at both 50°C and 37°C, which are the standard reaction temperature for endocellulases [37] and the temperature used for the semi-high-throughput activity screen, respectively. Importantly, all three enzymes exhibited the same specific CMCase activity at both temperatures tested (Table 1), confirming the ability of the secondary screen to yield clones whose activity was not compromised during the selection for intracellular stability. Moreover, because the specific CMCase activities of the three enzymes were comparable, the production of FgC5A.2, as measured by CMCase activity per culture volume, was enhanced by 30-fold over the parental enzyme after just two rounds of directed evolution.
Deconstruction of the mutational pathway to enhanced production
To evaluate the contribution of each mutation to the increased stability of FgC5A.1 and FgC5A.2, each was generated individually in wt FgCel5A by site-directed mutagenesis. Of the four mutations tested – G10D, T166S, T332I, and D342G – all except for T166S were able to increase soluble production of the unfused enzyme compared to wt FgCel5A (Fig. S3a and b). It should be pointed out that the soluble production levels seen for FgC5A.1 required both G10D and T332I mutations (Fig. S3a). Next, we generated four additional mutants by introducing the individual T166S or D342G mutations to either the G10D/T332I double mutant (i.e., clone FgC5A.1) or to the T332I single point mutant. When T166S was added to the T332I mutant or to FgC5A.1, production levels were unchanged (Fig. S3b). In contrast, the addition of D342G to either the T332I mutant or to FgC5A.1 resulted in a dramatic increase in soluble accumulation (Fig. S3b). In fact, the T332I and D342G mutations together were able to completely recapitulate the enhanced solubility observed for FgC5A.2 (Figs. 2d and S3b). In addition to their nearly identical solubility, the activity on CMC per culture volume of both FgC5A.2 and this T332I/D342G double mutant, which we named FgC5A.3, was enhanced by ~30 fold (Table 1). In both cases, this increased production was achieved without compromising specific activity of the enzymes on CMC (Table 1). In light of these findings, all remaining enzyme characterization was performed using the FgC5A.3 clone.
Engineered FgCel5A mutant degrades insoluble substrates
Because activity on CMC does not require CBM binding, it is not representative of catalysis on insoluble substrates that are industrially relevant [29]. Therefore, we compared the activity of wt FgCel5A and FgC5A.3 using the insoluble substrates Avicel and bacterial microcrystalline cellulose (BMCC). As expected, the extent of reaction on Avicel and BMCC were low due to the endoglucanase being tested in the absence of an exoglucanase [38]. Nonetheless, the extent of reaction was similar between the two enzymes (Table 2), suggesting that the mutations that increased soluble production of FgC5A.3 did not negatively impact its ability to degrade insoluble substrates. When a β-glucosidase was added for synergistic degradation of the substrates, the extent of reaction was increased similarly for both enzymes (Table 2), suggesting that the mutations in FgC5A.3 did not interfere with its ability to function synergistically in a minimal cocktail.
Table 2.
Extent of reaction on insoluble substrates
| Enzyme | Avicel1,2 | BMCC1,3 | ||
|---|---|---|---|---|
|
|
||||
| − β-glucosidase | + β-glucosidase4 | − β-glucosidase | + β-glucosidase4 | |
|
|
||||
| FgCel5A | 0.0222 ± 0.0017 | 0.0258 ± 0.0010 | 0.0288 ± 0.0009 | 0.0394 ± 0.0016 |
| FgC5A.3 | 0.0227 ± 0.0015 | 0.0277 ± 0.0014 | 0.0256 ± 0.0039 | 0.0437 ± 0.0029 |
Extents of reaction were measured using purified enzymes at 50°C according to the IUPAC standard protocol for β-1,4-endoglucanase on insoluble substrates. Reducing sugars formed were detected using standard DNS protocol and the extent of reaction was calculated based on the theoretical maximum of reducing ends based on substrate loading. Error represents the standard deviation from a minimum of three replicate experiments.
340 nmol enzyme per gram Avicel substrate.
2,500 nmol enzyme per gram BMCC substrate.
30 CBU per gram substrate.
Biophysical characterization of engineered FgCel5A mutant
Soluble production of a protein is a complex trait that can result from a combination of numerous factors such as thermodynamic stability, proteolytic sensitivity, and protein-protein interactions. To determine which, if any, of these factors contributed to the enhanced production of FgC5A.3, we biophysically characterized this mutant. First, we measured the thermodynamic stability, which for cellulases often involves identifying a thermal inactivation temperature where half of the enzyme’s activity is lost. We measured a thermal inactivation temperature for wt FgCel5A of 68.1 ± 0.3°C, which was 4°C higher than that for FgC5A.3 (63.9 ± 0.1°C) (Fig. S4a). Second, we determined the Gibb’s free energy for these enzymes by measuring their unfolding through intrinsic tryptophan fluorescence as a function of the chemical denaturant guanidine hydrochloride (GdnHCl). The wt enzyme unfolded at a denaturant concentration of 3.87 M GdnHCl with a Gibb’s free energy of 9.64 ± 0.64 kcal/mol, whereas FgC5A.3 denatured at 2.80 M GdnHCl with a free energy of 10.01 ± 0.58 kcal/mol (Fig. S4b). Hence, the two CD mutations in FgC5A.3 appear to slightly decrease the overall thermodynamic stability of the enzyme, perhaps as compensation for the increase in soluble production.
Third, we evaluated protease sensitivity, which was assayed by treatment with Proteinase K (PK). Both wt FgCel5A and FgC5A.3 were degraded to a similar extent by PK, which appeared to attack the less structured CBM and linker portions of both enzymes as evidenced by the appearance of a ~30-kDa degradation product (Fig. S4c). Based on this result, we conclude that resistance to non-specific proteolysis is not the cause for increased production. Fourth, we used size exclusion chromatography to assess the oligomerization state of both enzymes, with higher ordered structures indicative of protein aggregation and possible susceptibility to intracellular proteases. Given that the elution profiles of wt FgCel5A and FgC5A.3 were nearly identical (Fig. S4d), oligomerization state does not appear to explain the production enhancement.
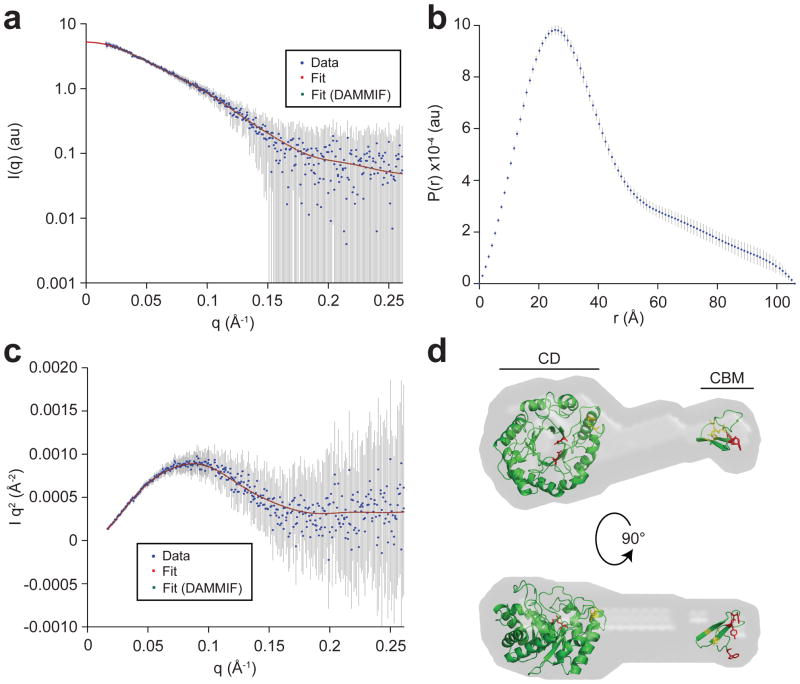
Fifth, small angle X-ray scattering (SAXS) was used to further shed light on the shape, dimension, and conformation of FgC5A.3. The distance distribution function, P(r), was deduced from the scattering intensities of FgC5A.3 (Fig. 3a). The P(r) curve of FgC5A.3 has a bell-shape, which is characteristic of a globular protein (Fig. 3b). Additionally, the Kratky plot shows a prominent small-angle peak with a relatively flat wide-angle data suggesting the protein is well folded (Fig. 3c). Moreover, this data reveals that the FgC5A.3 protein was monodisperse at concentrations below 1.5 mg/mL. Ab initio 3-D envelopes of FgC5A.3 were determined using the program DAMMIF [39]; the best model provided a fit to the experimental data with χ2 = 0.11 with a Dmax of 106 Å (Supplementary Table S1). Of the ten ab initio shapes that were predicted, nine had a nearly identical elongated envelope confirmation containing a globular disc-shaped part and a thin protuberance with a thicker cap (Fig. 3d). The homology-modeled CD (Fig. 1a) fit well in the disc-shaped region of the envelope, while the CBM is predicted to be at the far end of the protuberance. The central portion of the envelope is thought to be the flexible linker that connects the CD and CBM. It should be noted that similar “tadpole”-like envelope shapes and radii of gyration (Rg; Table S1) have been observed for other two-domain endoglucanases that contain a CD and CBM [40, 41], providing evidence that the overall structure of FgC5A.3 was conserved through the directed evolution process.
Figure 3. SAXS analysis and envelope prediction for FgC5A.3 variant.
(a) Scattering intensity (I) as a function of scattering angle (q) for a 1.28 mg/mL sample of FgC5A.3. Scattering intensity is the average of 10 spectra subtracted from the average of 10 blank spectra (sodium acetate buffer only) collected for 2 sec each. Intensity is shown in blue and error is shown as grey bars. The best-fit curve is shown in red and the DAMMIF fit in green. (b) Probability distance distribution as a function of particle diameter predicted by primus (ATSAS) for FgC5A.3. Dmax was set to 106 Å. (c) Kratky plot (I*q2 vs q) for FgC5A.3 to show the quality of the data and fit. (d) DAMMIF predicted envelope for FgC5A.3. The envelope is shown in grey with superimposed CBM and CD homology models depicted in green. Catalytic residues and those involved in cellulose binding are shown in red and predicted disulfide bonds in yellow. Parameters, structural dimensions, and software used to predict the envelope are found in Table S1.
Genetic selection focuses protein library towards active members
To test the extent to which the Tat QC focuses protein libraries towards active members, the original FgCel5A library was selected at several different antibiotic concentrations and isolated colonies were subjected to the plate-based activity screen to determine the number of functional members. Specifically, one library was subjected to “neutral selective pressure” whereby clones were isolated on a concentration of antibiotic to which wt FgCel5A was insensitive (25 μg/mL Carb). This selective pressure was expected to favor the isolation of variants with stability equal to wt FgCel5A. A second library was subjected to “moderate selective pressure” whereby a higher concentration of antibiotic (50 μg/mL Carb) above the point where wt FgCel5A is sensitive was used. Under these conditions, the number of colony forming units (CFUs) was reduced 4-fold compared to those selected using neutral selective pressure. A third library was subjected to “high selective pressure” whereby an even higher concentration of antibiotic (100 μg/mL Carb) was used, which decreased CFUs >10-fold compared to neutral pressure. Finally, “no selective pressure” (0 μg/mL Carb) was performed and represented the scenario where QC-based selection was not applied and isolated cells were selected based simply on plasmid retention.
A comparison of the percentage of active cellulases recovered from each library selection revealed that the neutral selective condition contained the greatest percentage of clones having detectable CMCase activity (defined as signal above background but not necessarily as active as wt FgCel5A) as well as high CMCase activity (defined as signal that was the same or greater than wt FgCel5A) (Table S2). Moderate selective pressure, which was the level used to isolate FgC5A.1, resulted in slightly fewer active and highly active clones coming through the selection step (Table S2). Interestingly, no selection pressure resulted in 14–18% fewer members with detectable activity and 57–125% fewer members with high activity, relative to moderate and neutral selection (Table S2). In contrast to neutral and moderate selection pressure, high selective pressure caused a sharp decline in active and highly active clones (Table S2). This suggests that imposition of a more stringent selection for the purpose of larger gains in intracellular stability comes at the expense of enzymatic activity, reflective of a stability-function tradeoff [26, 27]. Overall, these results suggest that (i) selective pressure for enzyme production is linked with maintenance of enzyme activity; (ii) imposing high selective pressure for folding enhancement comes at the expense of activity; and (iii) moderate gains in fitness through multiple rounds of directed evolution at moderate or even neutral selective pressure may provide the best conditions for optimizing production while maintaining activity.
Discussion
The two-tiered directed evolution approach described here was used to rapidly enhance the soluble production of an endocellulase thirty-fold through just two rounds of mutagenesis/selection/screening. This production enhancement occurred without compromising enzymatic activity on soluble or insoluble cellulose substrates due in part to the inclusion of a secondary activity screen. The increase in production required just two amino acid substitutions in the CD of FgCel5A and was independent of the original genetic-based selection context (i.e., Tat signal peptide and Bla reporter). Exactly how these mutations promote enhanced solubility of FgC5A.3 is currently unknown. One possibility is that mutational changes in the Cel5A structure may allow for productive interactions with cytoplasmic molecular chaperones that lead to increased soluble production. Another possibility is that the acquired mutations may remove kinetic traps in enzyme folding, which is an attractive possibility given how close the mutations are in the primary sequence.
Based on our homology model, the D324G mutation appears to be surface exposed while the T332I mutation is buried. The T332I mutation is located within 10 Å of the active site, but is sufficiently far so as not to compromise enzymatic activity. While activity was spared, the folding stability of the enzyme was decreased which might be the cost associated with remodeling the enzyme’s solubility for an intracellular environment. It should be pointed out that solubility can be increased without compromising stability. For example, the Tat QC filter was recently used to isolate human VH variants with increased solubility in vivo that corresponded with greater protein stability in vitro [42]. However, the activity of the best VH clones was not measured, so it is unknown whether this greater thermodynamic stability came at the expense of antigen-binding activity. Clearly, when soluble expression is increased while maintaining high activity, the trade-off seems to be a modest decrease in stability as we showed here. Such a trade-off is consistent with results from numerous studies indicating that the evolution of new functions is often accompanied by a decrease in thermodynamic stability [26–28]. In support of this notion, when an identical antigen-independent approach was used to enhance scFv stability in vivo, the binding activity of selected clones was significantly reduced [25].
The implementation of a secondary screen based on CMCase activity was crucial to the discovery of highly active clones, and provided an important proof-of-concept for our two-tiered directed evolution strategy. However, we recognize that screening on soluble substrates such as CMC may be a suboptimal choice for designing enzymes that catalyze cellulose conversion on pretreated biomass. Therefore, in the future, refinements to the secondary screen could include relevant pretreated substrates, synergistic cellulose cocktails, and more practical reaction conditions [1, 29]. Furthermore, the advent of robotic screening platforms [43] aims to simultaneously optimize many of these parameters to create high efficiency enzyme cocktails. Such screens could be easily adapted to include our genetic selection for folding and would be highly beneficial for future engineering of cellulolytic enzymes. It should also be pointed out that increased stringency in the secondary screening thresholds could allow for enhancement of properties such as conversion in ionic liquid buffers, heat tolerance, low pH activity, or synergism with other cellulytic enzymes to improve the approach prescribed above. Regardless, current cellulase engineering efforts towards activity or other enzyme enhancements will not be biotechnologically relevant if they result in poor host production, necessitating a multi-tiered approach as prescribed here.
While adaption of the two-tiered strategy for cellulase production is a logical progression, the true power of QC-based evolution lies in remodeling heterologous enzymes for in vivo functions. Indeed, optimizing the in vivo properties of two heterologous enzymes by adaptive evolution had a profound effect on the productivity of a constructed metabolic pathway [44]. We anticipate that combining the Tat QC selection with rational protein design and established secondary screens will provide a pipeline of optimized enzymes for functional construction (or reconstruction) of synthetic biological pathways. Such a pipeline could enable intracellular chemistries that have been difficult to achieve due to poor expression of heterologous enzymes. Additionally, the use of selective pressure could be used for tuning the stoichiometry of pathway enzymes in a manner that eliminates metabolic bottlenecks and minimizes the burden on the cell that results from gross overexpression.
Materials and Methods
Bacterial strains and plasmids
E. coli strain DH5α was used for all cloning including library construction, strain MC4100 was used for genetic selection and spot plating experiments, strain BL21(DE3) and Origami B(DE3) (Novagen) were used for cytoplasmic expression analysis, and strain SHuffle T7 Express (New England Biolabs) was used for purification. For plate-based selection experiments, the plasmid pSALect was used [22]. The Fg_cel5A gene from F. graminearum (BROAD locus FG03795; GenBank locus NT_086532) was codon optimized and synthesized by GenScript. The resulting cel5A gene was PCR amplified without its native signal peptide and cloned between the XbaI and SalI sites in pSALect, yielding pSALect-FgCel5A. An error prone DNA library of the entire 1,098 base-pair long cel5A gene was created with a theoretical DNA mutation rate of 0.15% mutations per base pair as described [45]. Library size was determined by plating the transformed library on Luria-Bertani (LB) agar plates supplemented with 25 μg/mL chloramphenicol (Cm), and the error rates were confirmed by sequencing the DNA of naïve library members. The library size for the first round of directed evolution was determined to be 1.7 × 106 members and for the second round it was 6.7×104 members. Error rates in both libraries were experimentally confirmed to be 0.3% mutations per base pair, which corresponds to approximately 3.3 nucleotide or 2.5 non-synonymous amino acid substitutions per gene. For cytoplasmic expression analysis, plasmid pET28a (Novagen) was modified to contain an N-terminal 6x poly-histidine tag followed by FLAG-epitope tag. The wt Fg_cel5A gene (or isolated hits) was then cloned between the NdeI and SalI sites of the resulting pET28a plasmid, yielding pET28-FgCel5A. A similar strategy was followed to create plasmid pET22b-FgCel5A but with FLAG and 6x-His tags at the C-terminus of FgCel5A. All plasmids were confirmed by sequencing.
Selective growth and enzyme activity screening
MC4100 cells carrying pSALect-FgCel5A (or pSALect encoding isolated variants) were grown overnight in LB medium containing 25 μg/mL Cm. Antibiotic sensitivity of cells was assayed by spotting 5 μL of 10-fold diluted overnight cells directly onto LB agar plates supplemented with 100 μg/mL Carb or 25 μg/mL Cm and growing overnight at 30°C. Library selections were performed by electroporating pSALect plasmids containing the Fg_cel5A library into MC4100 cells followed by direct plating on LB agar plates supplemented the desired selective concentration of Carb to ensure cells grew as single colonies [46]. Plated bacteria were cultured overnight at 30°C and individual colonies were picked into 96-well plates containing LB media supplemented with Cm. The following day, 10 μL of the overnight culture was subcultured in a 96-deep-well culture plate containing 1 mL of LB supplemented with 25 μg/mL Cm. The deep-well cultures were grown for 2 h at 37°C and then transferred to 30°C for an additional 8 h. Cultures were centrifuged at 4,000×g for 10 min to pellet cells and pellets were frozen at −20°C overnight. To lyse cells, 50 μL of BugBuster was added to each well followed by shaking of the plates at room temperature for 30 min. The soluble cell lysate was recovered by centrifugation at 4,000×g for 30 min. The activity of each sample was determined by performing an assay using CMC, which contained 10 μL soluble lysate, 10 μL water and 20 μL of 2% CMC, and was carried out for 24 h at 37°C. Extent of reaction was quantified as described below. The LB-agar plate-based secondary screen for CMC hydrolysis of selected colonies was performed exactly as described elsewhere [47].
Protein production and purification
BL21(DE3) or Origami B(DE3) cells were freshly transformed with pET28-FgCel5A, pET28a encoding isolated hits, or pET22b-FgCel5A. Overnight cells were subcultured in baffled flasks containing LB supplemented with kanamycin (Kan). Cultures were grown to an absorbance at 600 nm (Abs600) of ~0.5 at 37°C. Protein production was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 2 h at 30°C. Cells were harvested by centrifugation at 16,000×g for 3 min and lysed using BugBuster. The soluble fraction was recovered by centrifugation at 16,000×g for 30 min and total protein determined by Bradford assay with BSA as standard. The insoluble pellet was washed twice with 50 mM Tris (pH 8), 1 mM EDTA followed by extraction of the insoluble sample in phosphate-buffered saline (PBS) with 2% SDS at 100°C for 10 min. Protein samples were normalized by total soluble protein, separated by SDS-PAGE, transferred to a PVDF membrane, and detected using an anti-FLAG primary antibody (Sigma) and appropriate secondary antibodies.
To determine the amount of soluble cellulase produced, CMC hydrolysis was measured. Cells were cultured exactly as described above and then harvested by centrifugation at 16,000×g for 3 minutes. The resulting cell pellet was resuspended in 50 mM sodium acetate buffer (pH 5). Cells were lysed by sonication and the soluble fraction was recovered by centrifugation at 16,000×g for 30 min. CMC hydrolysis was carried out as described below for purified samples except the reaction was carried out for 13 h at 37°C. Lysate derived from BL21(DE3) cells that did not contain a plasmid was used as a blank for each amount of lysate tested.
To purify enzymes, SHuffle T7 Express cells were freshly transformed with pET28-FgCel5A (or pET28a encoding isolated hits). An overnight culture of cells was subcultured in shake flasks containing 1 L of terrific broth supplemented with Kan. Cultures were grown to an Abs600 of ~0.7 at 37°C. Protein production was induced with 0.1 mM IPTG for 24 h at room temperature. Cells were pelleted by centrifugation at 4,000×g for 10 min and pellets were frozen at −80°C. Cell pellets were resuspended in 50 mM sodium phosphate (pH 7.4), 0.5 M NaCl and 20 mM imidazole and lysed using a cell homogenizer (Avestin). Lysates were cleared of cell debris by centrifugation at 30,000×g for 30 min and filtered through a 0.22-μm filter. Protein purification was carried out using an FPLC by Ni2+-affinity chromatography followed by desalting into 50 mM sodium acetate buffer (pH 5). Proteins were further concentrated using 3-kDa molecular weight cutoff centrifuge columns made of polyethylsulfone (Sartorius). Protein concentration was determined by BCA assay using a BSA standard curve. Size exclusion chromatography was performed using an FPLC with Superdex 75 column (GE Health Sciences). Protein purity was determined by separating the enzymes with SDS-PAGE gels and staining the resulting gels with Coomassie blue.
Enzyme activity assays
Extents of reaction and specific activity on low-viscosity CMC (Sigma) were measured according to IUPAC standard protocols for endoglucanases [37]. For CMC hydrolysis, various amounts of FgCel5A were incubated with 1% CMC at 50°C or 37°C in 50 mM sodium acetate buffer (pH 5). Following reaction, samples were mixed in a 1:3 ratio with DNS to detect the number of reducing ends formed during the reaction. The DNS reaction was carried out at 95°C or 100°C for 5 min. For each set of samples tested, glucose standards (0.25 to 2 mg/mL) and appropriate blank reactions (1% CMC in sodium acetate buffer) were included. DNS-reacted samples were cooled and diluted 1:3 in water prior to measuring the Abs540. Glucose standards were used to create a standard for converting Abs540 to concentration of glucose.
To determine the extent of reaction on insoluble substrates, Avicel and BMCC, specific enzyme loadings per gram substrate were used. For Avicel reactions, between 133–340 nmol enzyme per gram Avicel was tested. Prior to setting up the BMCC reaction, the mass per volume of BMCC was measured by drying the substrate. For BMCC reactions, between 1,250–5,000 nmol enzyme per gram BMCC was tested. To analyze a simple synergy between cellulases, a β-glucosidase (Sigma) was added to 30 cellobiase units (CBU)/g. Reactions were carried out for 17.5 h at 50°C using screw capped tubes and a rotating platform. The samples were centrifuged at 16,000×g for 5 min to separate soluble sugars from substrate that remained insoluble. The amount of reducing ends formed in the soluble sugar fraction were measured using DNS as described above, and the extent of reaction calculated based on the theoretical maximum number of ends formed for each substrate loading.
SAXS data collection and envelope construction
Proteins were purified as previously described. Extinction coefficients were calculated using known concentrations as measured by BCA assay and measured absorbance values (A280). A dilution series of FgC5A.3 was created between 40 mg/mL to 0.1 mg/mL. The Cornell High Energy Synchrotron Source (CHESS) was used as the source of high energy X-rays and hutch G1 was set up with a robotic sampling unit as described in [48]. For each sample, 10 spectra were taken by exposing for 2 seconds and these spectra were averaged. Blank samples of 50 mM sodium acetate buffer were subtracted from each sample and scattering was collected while oscillating samples to reduce X-ray damage. After each sample was measured, the A280 was collected to give an accurate concentration of the sample used to collect the SAXS data. Using the measured concentration and a Guinier fit (I(q) vs q2), it was possible to determine aggregation and estimate the radius of gyration for each sample. The distance distribution was calculated using the Primus program from the ATSAS package (EMBL Hamburg) [49]. The Dmax was set to 106 Å. The DAMMIF algorithm was used to predict ten envelope structures for FgC5A.3 [39]. The predicted envelopes were reoriented to overlap using the DAMSUP algorithm and were compared using the DAMSEL program to test the similarity of the structures [50]. Of the 10 envelopes predicted for FgC5A.3, 9 were predicted to be similar. Likely position of the homology-modeled CD and CBM were predicted individually using the SUPCOMB program.
Supplementary Material
Acknowledgments
This material is based upon work supported by a USDA NIFA Grant # 2009–02202 (to D.M.G., L.P.W. and M.P.D.), a DOE Emerging Opportunities Program Grant 115K555 through the Grant Lakes Bioenergy Research Center (to M.P.D.), and an NSF GK-12 “Grass Roots” Fellowship under Grant # DGE-1045513 (to J.T.B.). This work is also based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF and the NIH/NIGMS under NSF award DMR-1332208, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award GM-103485 from the NIGMS, NIH.
References
- 1.Garvey M, Klose H, Fischer R, Lambertz C, Commandeur U. Cellulases for biomass degradation: comparing recombinant cellulase expression platforms. Trends Biotechnol. 2013;31:581–93. doi: 10.1016/j.tibtech.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Assenberg R, Wan PT, Geisse S, Mayr LM. Advances in recombinant protein expression for use in pharmaceutical research. Curr Opin Struct Biol. 2013;23:393–402. doi: 10.1016/j.sbi.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Almo SC, Garforth SJ, Hillerich BS, Love JD, Seidel RD, Burley SK. Protein production from the structural genomics perspective: achievements and future needs. Curr Opin Struct Biol. 2013;23:335–44. doi: 10.1016/j.sbi.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 5.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–38. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact. 2011;10:32. doi: 10.1186/1475-2859-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eijsink VG, Bjork A, Gaseidnes S, Sirevag R, Synstad B, van den Burg B, et al. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–20. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci U S A. 1987;84:6663–7. doi: 10.1073/pnas.84.19.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumura M, Signor G, Matthews BW. Substantial increase of protein stability by multiple disulphide bonds. Nature. 1989;342:291–3. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson H, Becktel WJ, Matthews BW. Enhanced protein thermostability from designed mutations that interact with alpha-helix dipoles. Nature. 1988;336:651–6. doi: 10.1038/336651a0. [DOI] [PubMed] [Google Scholar]
- 11.Serrano L, Fersht AR. Capping and alpha-helix stability. Nature. 1989;342:296–9. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- 12.Malakauskas SM, Mayo SL. Design, structure and stability of a hyperthermophilic protein variant. Nat Struct Biol. 1998;5:470–5. doi: 10.1038/nsb0698-470. [DOI] [PubMed] [Google Scholar]
- 13.Dale GE, Broger C, Langen H, D’Arcy A, Stuber D. Improving protein solubility through rationally designed amino acid replacements: solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Eng. 1994;7:933–9. doi: 10.1093/protein/7.7.933. [DOI] [PubMed] [Google Scholar]
- 14.Roodveldt C, Aharoni A, Tawfik DS. Directed evolution of proteins for heterologous expression and stability. Curr Opin Struct Biol. 2005;15:50–6. doi: 10.1016/j.sbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Waldo GS. Genetic screens and directed evolution for protein solubility. Curr Opin Chem Biol. 2003;7:33–8. doi: 10.1016/s1367-5931(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 16.Magliery TJ, Regan L. Combinatorial approaches to protein stability and structure. Eur J Biochem. 2004;271:1595–608. doi: 10.1111/j.1432-1033.2004.04075.x. [DOI] [PubMed] [Google Scholar]
- 17.Martineau P, Jones P, Winter G. Expression of an antibody fragment at high levels in the bacterial cytoplasm. J Mol Biol. 1998;280:117–27. doi: 10.1006/jmbi.1998.1840. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki T, Yasugi M, Arisaka F, Yamagishi A, Oshima T. Adaptation of a thermophilic enzyme, 3-isopropylmalate dehydrogenase, to low temperatures. Protein Eng. 2001;14:85–91. doi: 10.1093/protein/14.2.85. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci U S A. 2008;105:14808–13. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17:691–5. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell KL, Mittermaier AK, Forman-Kay JD, Davidson AR. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–11. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006;15:449–58. doi: 10.1110/ps.051902606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, et al. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–71. doi: 10.1016/j.molcel.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson AJ, Lim HK, Xu H, Rocco MA, Bratkowski MA, Ke A, et al. Engineering antibody fitness and function using membrane-anchored display of correctly folded proteins. J Mol Biol. 2012;416:94–107. doi: 10.1016/j.jmb.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher AC, DeLisa MP. Efficient isolation of soluble intracellular single-chain antibodies using the twin-arginine translocation machinery. J Mol Biol. 2009;385:299–311. doi: 10.1016/j.jmb.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beadle BM, Shoichet BK. Structural basis of stability-function tradeoffs in enzymes. J Mol Biol. 2002;321:285–96. doi: 10.1016/s0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 27.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225:585–9. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 28.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20:295–9. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 30.King BC, Waxman KD, Nenni NV, Walker LP, Bergstrom GC, Gibson DM. Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol Biofuels. 2011;4:4. doi: 10.1186/1754-6834-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong J, Tamaki H, Akiba S, Yamamoto K, Kumagai H. Cloning of a gene encoding a highly stable endo-beta-1,4-glucanase from Aspergillus niger and its expression in yeast. J Biosci Bioeng. 2001;92:434–41. doi: 10.1263/jbb.92.434. [DOI] [PubMed] [Google Scholar]
- 32.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci U S A. 1999;96:13703–8. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocco MA, Waraho-Zhmayev D, DeLisa MP. Twin-arginine translocase mutations that suppress folding quality control and permit export of misfolded substrate proteins. Proc Natl Acad Sci U S A. 2012;109:13392–7. doi: 10.1073/pnas.1210140109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLisa MP, Tullman D, Georgiou G. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci U S A. 2003;100:6115–20. doi: 10.1073/pnas.0937838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King BC, Donnelly MK, Bergstrom GC, Walker LP, Gibson DM. An optimized microplate assay system for quantitative evaluation of plant cell wall-degrading enzyme activity of fungal culture extracts. Biotechnol Bioeng. 2009;102:1033–44. doi: 10.1002/bit.22151. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Z, Storms R, Tsang A. Microplate-based carboxymethylcellulose assay for endoglucanase activity. Anal Biochem. 2005;342:176–8. doi: 10.1016/j.ab.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 37.Ghose TK. Measurement of Cellulase Activities. Pure and Applied Chemistry. 1987;59:257–68. [Google Scholar]
- 38.Watson DL, Wilson DB, Walker LP. Synergism in binary mixtures of Thermobifida fusca cellulases Cel6B, Cel9A, and Cel5A on BMCC and Avicel. Appl Biochem Biotechnol. 2002;101:97–111. doi: 10.1385/abab:101:2:097. [DOI] [PubMed] [Google Scholar]
- 39.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42:342–6. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Receveur V, Czjzek M, Schulein M, Panine P, Henrissat B. Dimension, shape, and conformational flexibility of a two domain fungal cellulase in solution probed by small angle X-ray scattering. J Biol Chem. 2002;277:40887–92. doi: 10.1074/jbc.M205404200. [DOI] [PubMed] [Google Scholar]
- 41.Bianchetti CM, Brumm P, Smith RW, Dyer K, Hura GL, Rutkoski TJ, et al. Structure, dynamics, and specificity of endoglucanase D from Clostridium cellulovorans. J Mol Biol. 2013;425:4267–85. doi: 10.1016/j.jmb.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim DS, Song HN, Nam HJ, Kim SG, Park YS, Park JC, et al. Directed evolution of human heavy chain variable domain (VH) using in vivo protein fitness filter. PLoS One. 2014;9:e98178. doi: 10.1371/journal.pone.0098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walton J, Banerjee G, Car S. GENPLAT: an automated platform for biomass enzyme discovery and cocktail optimization. J Vis Exp. 2011 doi: 10.3791/3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshikuni Y, Dietrich JA, Nowroozi FF, Babbitt PC, Keasling JD. Redesigning enzymes based on adaptive evolution for optimal function in synthetic metabolic pathways. Chem Biol. 2008;15:607–18. doi: 10.1016/j.chembiol.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–53. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 46.Fisher AC, Rocco MA, DeLisa MP. Genetic selection of solubility-enhanced proteins using the twin-arginine translocation system. Methods Mol Biol. 2011;705:53–67. doi: 10.1007/978-1-61737-967-3_4. [DOI] [PubMed] [Google Scholar]
- 47.Farrow MF, Arnold FH. High throughput screening of fungal endoglucanase activity in Escherichia coli. J Vis Exp. 2011 doi: 10.3791/2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen SS, Moller M, Gillilan RE. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J Appl Crystallogr. 2012;45:213–23. doi: 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semenyuk AV, Svergun DI. Gnom - a Program Package for Small-Angle Scattering Data-Processing. J Appl Crystallogr. 1991;24:537–40. [Google Scholar]
- 50.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–4. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.