Abstract
High-resolution structure determination of small proteins in solution is one of the big assets of NMR spectroscopy in structural biology. Improvements in efficiency of NMR structure determination by advances in NMR experiments and automation of data handling therefore attracts continued interest. Here, non-uniform sampling (NUS) of 3D heteronuclear-resolved [1H,1H]-NOESY data yielded two- to three-fold savings of instrument time for structure determinations of soluble proteins. With the 152-residue protein NP_372339.1 from Staphylococcus aureus and the 71-residue protein NP_346341.1 from Streptococcus pneumonia we show that high-quality structures can be obtained with NUS NMR data, which are equally well amenable to robust automated analysis as the corresponding uniformly sampled data.
Keywords: NMR spectroscopy, Proteins, Compressed sensing, Automated data analysis, J-UNIO structure determination protocol
INTRODUCTION
NMR spectroscopy in solution enables a wide range of different investigations, including de novo protein structure determination, and studies of intramolecular dynamics and intermolecular interactions with small and large ligand molecules.[1-8] Among these, structure determination of small proteins, in the size range up to about 25 kD, is particularly attractive. Due to the large percentage of surface-exposed amino acid residues, a significant part of the molecular structure is dynamically disordered and potentially available for interactions with the solvent and with small or macromolecular ligands. As a consequence, small proteins tend to be more difficult to crystallize, whereas they can readily be studied with standard solution NMR structure determination approaches.[9-11] The efficiency of these protocols has recently been significantly improved, based primarily on the use of more sophisticated correlation experiments,[12-16] and on the introduction of automation for discrete steps of NMR data analysis and structural interpretation.[17-41] Here, we explore the use of non-uniform sampling (NUS) of NMR data sets to further reduce the requirements for NMR spectrometer time. We thus achieved a 2- to 3-fold reduction of the overall measurement time needed per structure determination, which represents an impressive progress resulting from basic physical chemistry research.[42-57]
NMR data acquisition with non-uniform sampling has in recent years attracted renewed interest for the recording of high-dimensional data sets.[58-64] Applications have primarily been proposed for obtaining improved sensitivity in studies of large proteins or multimolecular complexes, specifically membrane proteins reconstituted in detergent micelles.[56, 65-67] In this paper, we explore applications of NUS for improved efficiency of routine structure determinations of soluble proteins, by reducing the use of instrument time. A novel aspect results also from the requirement that the non-uniformly sampled NMR data sets must be amenable to automated spectral analysis and structural interpretation.
In our laboratory we use the J-UNIO protocol for protein structure determination by NMR in solution, which affords extensive automation of this process.[68] A structure determination with the J-UNIO protocol typically requires a set of three APSY-NMR (“Automated Projection SpectroscopY”) experiments[13, 69] and three 3D heteronuclear-resolved [1H,1H]-NOESY data sets[70] for chemical shift assignment and collection of the input for the structure calculation.[9] Using the UNIO software, peaks in the NMR spectra are automatically picked[23] and used as input for automated chemical shift assignment routines.[26, 29] Following interactive validation of the chemical shift assignments, additional automated routines[33] are used to prepare the input of NOE distance restraints for the protein structure calculation, for which the torsion angle molecular dynamics algorithm in the program CYANA[9] is used.
With uniform sampling, about 8 days of measurement time are needed for the three 3D heteronuclear-resolved [1H,1H]-NOESY experiments used in protein structure determinations with J-UNIO, whereas the required APSY-NMR measurements typically take less than 30 hours of instrument time per protein, and in many cases have been obtained in less than 10 hours.[71] The 3D heteronuclear-resolved [1H,1H]-NOESY measurements were thus the experiments where significant savings of the overall instrument time could be achieved with NUS. Here, this was implemented with reconstruction of the undersampled data with the compressed sensing algorithm.[57]
An important goal of the present paper was to study how reconstructed NUS spectra would be amenable for the automated steps of the J-UNIO protocol. We validated the NUS approach through comparison with results obtained from corresponding uniformly sampled NMR data on the 152-residue protein NP_372339.1 (PDB: 2MQB, BMRB: 25026) and the 71-residue protein NP_346341.1 (PDB: 2M7O, BMRB: 19198), i.e., statistics on the NOESY-based automated side chain assignments and inputs of conformational constraints, and the results of the structure determinations.
RESULTS AND DISCUSSION
Structure determination of the 152-residue protein NP_372339.1 with uniformly and non-uniformly sampled NMR data
Uniformly and non-uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY spectra of the [13C,15N]-labeled 152-residue protein NP_372339.1 from Staphylococcus aureus were acquired using the parameters listed in Table 1. Sparsely sampled data were obtained with 25% of the initial number of indirect hypercomplex points sampled for the 15N- and 13Caro-resolved [1H,1H]-NOESY experiments, and with 50% sampling for the 13Cali-resolved [1H,1H]-NOESY (Table 1). The spectra were reconstructed using the compressed sensing algorithm IRLS.[57] As a first result, we noticed that the reconstructed spectra contained nearly as many peaks as the conventionally acquired data, with similar spectral resolution, peak positions and signal intensities (Figure 1).
Table 1.
Acquisition parameters used for the 3D heteronuclear-resolved [1H,1H]-NOESY data sets of the proteins NP_372339.1 and NP_345341.1.
| 3D X-resolved [1H,1H]-NOESY[a] | NP_372339.1 152 residues | NP_346341.1 71 residues | ||||
|---|---|---|---|---|---|---|
| X | 15N | 13Cali | 13Caro | 15N | 13Cali | 13Caro |
| t1 max (ms) | 13 | 10.4 | 8.3 | 11 | 11 | 11 |
| t2 max (ms) | 28 | 6.2 | 9.7 | 28 | 5.8 | 11 |
| t3 max (ms) | 85 | 85 | 85 | 85 | 85 | 85 |
| τm (ms) | 65 | 65 | 65 | 90 | 90 | 90 |
| d (s) | 1 | 1 | 1 | 1 | 1 | 1 |
|
Number of sampled points (US[b]) | ||||||
| n(t1) | 128 | 128 | 128 | 118 | 100 | 100 |
| n(t2) | 64 | 48 | 64 | 48 | 45 | 45 |
| n(t3) | 1024 | 1024 | 1024 | 1024 | 1024 | 1024 |
|
Number of sampled points (NUS[c]) | ||||||
| % | 25 | 50 | 25 | 25 | 25 | 25 |
| n | 2048 | 3072 | 2048 | 1416 | 1125 | 1125 |
| n(t3) | 1024 | 1024 | 1024 | 1024 | 1024 | 1024 |
|
Measurement Time (hours) | ||||||
| US[b] | 85 | 64 | 85 | 60 | 48 | 48 |
| NUS[c] | 21 | 32 | 21 | 15 | 12 | 12 |
d is the delay time between subsequent recordings. The other parameters are defined in the experimental schemes of Figure S1.
US: Uniformly sampled 3D X-resolved [1H,1H]-NOESY experimental set-up. n(t1) and n(t2) indicate the number of increments in the t1 and t2 indirect dimensions, and n(t3) is the number of directly recorded complex points.
NUS: Non-uniformly sampled 3D X-resolved [1H,1H]-NOESY experimental set-ups. % and n indicate, respectively, the percentage and the total number of sampled indirect hypercomplex points, and n(t3) is the number of directly recorded complex points.
Figure 1.
Comparison of [ω1(1H), ω3(1H)] strips from uniformly sampled (blue) and non-uniformly sampled (NUS) 3D heteronuclear-resolved [1H,1H]-NOESY data sets of the 152-residue protein NP_372339.1. For NUS, 25 % sampling is shown in red, and in (b), 50 % sampling (green) has been added (see text). (a) 3D 15N-resolved [1H,1H]-NOESY. (b) 3D 13Cali-resolved [1H,1H]-NOESY. (c) 3D 13Caro-resolved [1H,1H]-NOESY.
Complete polypeptide backbone assignments had been obtained from APSY-NMR data.[69] After the recording of the NMR spectra (Figure 1), the present work continued with the preparation of the input for the program UNIO-ATNOS/ASCAN to assign the amino acid side chains of the protein NP_372339.1. This input contained the three 3D X-resolved [1H,1H]-NOESY spectra of Table 1, and the complete polypeptide backbone chemical shift assignments.[71] Two independent computations were performed, using, respectively, the uniformly and non-uniformly sampled three 3D X-resolved [1H,1H]-NOESY data sets. As a second result, the percentage of automated side chain assignments made using the two different NOESY data sets was found to be comparable (Table 2). For both data sets, the automatically determined amino acid side chain chemical shift assignments were interactively validated and completed using the respective experimental NOESY data. The extent of these final side chain chemical shift assignments was again nearly identical for the two approaches.
Table 2.
Results of automated amino acid side chain chemical shift assignments with UNIO-ATNOS/ASCAN[23, 26] for the proteins NP_372339.1 and NP_346341.1, when using either uniformly or non-uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY data sets (Table 1, Figure 1).
| Protein | NOESY data sampling[a] | Automated assignments | |
|---|---|---|---|
| number[b] | %[b] | ||
| NP_372339.1, 152 aa | Uniform | 1195 (89) | 73 (7) |
| NUS 25/50/25 | 1210 (67) | 74 (6) | |
| NP_346341.1, 71 aa | Uniform | 563 (63) | 72 (11) |
| NUS 25/25/25 | 556 (64) | 71 (12) | |
For the non-uniformly sampled spectra (NUS), the percentage of hypercomplex points sampled in the indirect dimensions are indicated in the order of the 3D 15N- / 13Cali- / 13Caro- resolved [1H,1H]-NOESY spectra.
The total number of assignments corresponds to the number of heavy atom and hydrogen atom assignments made automatically by ASCAN. In parentheses, the extent of erroneous assignments by the automated procedure is indicated.[26] The percentages are relative to the number of assignments expected from the amino acid sequence. Prior to the start of the structure calculation, the assignments obtained with ASCAN were interactively validated. This resulted in the elimination of all the erroneous assignments and to increasing the extent of the correct assignments to 89% for NP_372339.1, and 86% for NP_346341.1, respectively.[68]
The chemical shift assignments provided the basis for automated identification of NOE distance restraints with the software UNIO-ATNOS/CANDID,[23, 33] which was used in combination with the torsion angle dynamics algorithm in the program CYANA.[9] As a third result, comparison of the NOE upper distance constraints obtained based either on the uniformly or the non-uniformly sampled NOESY data showed that the distance constraints used as input for the final round of the structure calculation were nearly identically distributed along the amino acid sequence (Figure 2), and that the number of constraints per residue correlates with r = 0.94 between the uniformly and non-uniformly sampled NOESY data.
Figure 2.
Plots of the number, N, of medium-range and long-range NOE upper distance limits per residue contained in the input for the final cycle of the structure calculation of the 152-residue protein NP_372339.1 with CYANA 3.0. (a) Data obtained from the three uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY spectra. (b) Data obtained from the non-uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY with 25% (15N) / 50% (13Cali) / 25% (13Caro) sampling of hypercomplex points in the indirect dimensions. The correlation of the number of constraints per residue between the uniformly and the non-uniformly sampled data is r = 0.94.
The final, conclusive result comes from comparison of the NMR structures of NP_372339.1 obtained with the two NOESY data sets. The two structures are of similar quality (Figure 3a, Table 3), and they could be superimposed with backbone and all-heavy-atom RMSD values calculated between the mean structures of 1.10 Å and 1.54 Å, respectively.
Figure 3.
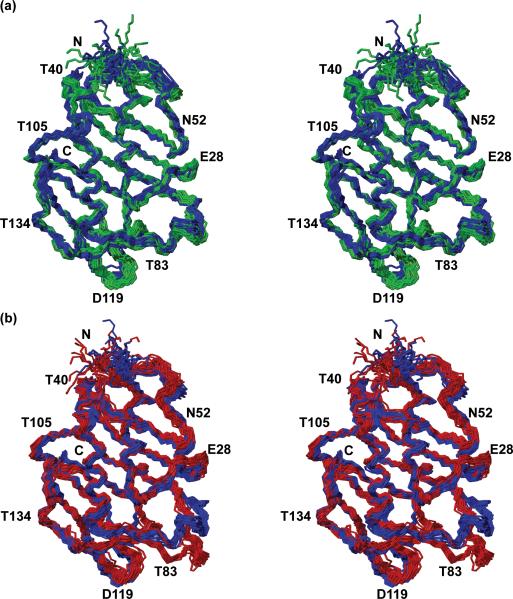
Superposition of the NMR structures of the 152-residue protein NP_372339.1 obtained from uniformly sampled (blue) and non-uniformly sampled (NUS) 3D heteronuclear-resolved [1H,1H]-NOESY data sets. Stereo views are shown of bundles of 20 NMR conformers. In (a), the structure obtained from 25% (15N) / 50% (13Cali) / 25% (13Caro) NUS is shown in green. In (b) the structure from 25% (15N) / 25% (13Cali) / 25% (13Caro) NUS is shown in red. The chain ends and selected amino acid residues are identified to guide the eye.
Table 3.
Input data and calculation statistics for the structure determination of the 152-residue protein NP_372339.1 based on uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY data and on two different non-uniformly sampled NOESY data sets.
| Quantity | Uniform | NUS[a] 25 / 50 / 25 | NUS[a] 25 / 25 / 25 |
|---|---|---|---|
| NOE upper distance limits[b] | 2521 | 2577 | 2319 |
| Intraresidual (|i – j| = 0) | 640 | 685 | 611 |
| Short-range (|i – j| = 1) | 687 | 645 | 630 |
| Medium-range (1 < i – j| < 5) | 431 | 466 | 398 |
| Long-range (|i – j| ≥ 5) | 763 | 781 | 680 |
| Dihedral angle constraints | 830 | 816 | 828 |
| Residual target function value (Å2) | 3.74 ± 0.53 | 3.98 ± 0.89 | 3.39 ± 0.83 |
| Residual NOE Violations | |||
| Number ≥ 0.1 Å | 18 ± 4 | 22 ± 4 | 18 ± 4 |
| Maximum (Å) | 0.14 ± 0.01 | 0.22 ± 0.16 | 0.14 ± 0.02 |
| Residual dihedral angle violations | |||
| Number ≥ 2.5° | 0 ± 0 | 0 ± 1 | 0 ± 1 |
| Maximum (°) | 2.17 ± 1.05 | 2.06 ± 0.95 | 2.32 ± 1.36 |
| Amber energies (kcal/mol) | |||
| Total | −5842 ± 512 | −5725 ± 739 | −5907 ± 625 |
| Van der Waals | −379 ± 223 | −263 ± 355 | −314 ± 280 |
| Electrostatic | −6623 ± 286 | −6655 ± 365 | −6740 ± 375 |
| RMSD from ideal geometry | |||
| Bond lengths (Å) | 0.0078 ± 0.0001 | 0.0081 ± 0.0002 | 0.0079 ± 0.0001 |
| Bond Angles (°) | 1.952 ± 0.046 | 2.012 ± 0.043 | 1.957 ± 0.049 |
| RMSD to the mean coordinates (Å)[c] | |||
| bb (5-54,61-151) | 0.58 ± 0.08 | 0.67 ± 0.16 | 0.70 ± 0.18 |
| ha (5-54,61-151) | 0.96 ± 0.09 | 1.03 ± 0.17 | 1.05 ± 0.15 |
| Ramachandran plot statistics (%)[d] | |||
| Most favored regions | 72.4 | 72.3 | 67.9 |
| Additional allowed regions | 26.9 | 27.0 | 30.7 |
| Generously allowed regions | 0.7 | 0.6 | 1.2 |
| Disallowed regions | 0.0 | 0.0 | 0.2 |
For the non-uniformly sampled data (NUS), the percentages of hypercomplex points sampled in the indirect dimensions for the 3D 15N- / 13Cali- / 13Caro-resolved [1H,1H]-NOESY data sets are indicated.
Except for the top six entries, which describe the input generated in the final cycle of the ATNOS/CANDID/CYANA calculation, the entries refer to the 20 energy-minimized CYANA conformers selected to represent the NMR structure. Where applicable, the average value for the bundle of 20 conformers and the standard deviation are given.
bb indicates the backbone atoms N, Cα, and C’; ha stands for all heavy atoms. Numbers in parentheses indicate the polypeptide segments for which the RMSD was calculated.
As determined by PROCHECK. The averages of the 20 conformers representing the NMR structure are given.
The 25/50/25 protocol for NUS in NMR structure determination of proteins
From the experience with the structure determination of the protein NP_3722339.1, we recommend to use non-uniform sampling in the extent of 25% for 3D 15N- and 13Caro-resolved NOESY experiments, and 50% for the 13Cali-resolved NOESY. This yields between two- and three-fold savings in the overall instrument time needed for a protein structure determination, when compared to using uniformly sampled NOESY experiments. The NMR data thus obtained were amenable to spectral analysis and structural interpretation by the automated steps of the J-UNIO protocol, and hence the use of NUS resulted in genuinely improved efficiency of the NMR structure determination. The 25/50/25 protocol should be applicable for proteins with up to about 200 amino acid residues, provided that a structure-quality protein solution[68, 72] is available.
Since non-uniformly sampled reconstructed data contain non-Gaussian noise[73] and artifactual peak shapes,[74] it was particularly intriguing to explore the use of automated peak picking with NUS data. The J-UNIO protocol employs the automated NOE peak picking algorithm ATNOS.[23] Remarkably, the use of ATNOS with the non-uniformly sampled heteronuclear-resolved [1H,1H]-NOESY data yielded results of similar quality to those obtained with the corresponding uniformly sampled data.
Robustness of the 25/50/25 NUS protocol
We arrived at the aforementioned protocol by detailed investigation of the minimal extent of sampling that would be consistent with robust structure determination. To this end, sampling was varied in discrete steps over the range 25% to 50%. The critical issue turned out to be the sampling of the 3D 13Cali-resolved [1H,1H]-NOESY spectrum. As an illustration, we present the results obtained with 25% and 50% non-uniform sampling of this spectrum for the 152-residue protein NP_372339.1 (Figs. 1 and 3, Table 3). When using 25% sampling for all three 3D heteronuclear-resolved [1H,1H]-NOESY data sets, we observed a 10 % decrease in the overall number of NOE distance constraints, which includes a 14% decrease of medium-range and long-range restraints (Table 3). As a result, the NMR structure showed significant differences to the structure obtained with uniform sampling, which is particularly pronounced in the loop containing T 83 (Figure 3b).
For a quantitative assessment, we randomly picked 2D [1H,1H] strips (Figure 1) from the uniformly sampled 3D 15N-, 13Caro- and 13Cali-resolved NOESY spectra, and calculated the intensities of all peaks present in each of them. The corresponding strips in the non-uniformly sampled spectra were similarly analyzed. Peak intensities of the NOE signals in the 25% sampled 3D 15N- and 13Caro-resolved NOESY spectra of the 152-residue protein NP_372339.1 correlated well with the corresponding peaks in the uniformly sampled data, but in the 25% sampled 3D 13Cali-resolved NOESY, 27% of the signals failed to be reconstructed (Figure 1). From comparison with the other two data sets, we tentatively rationalize the poor quality of the reconstructed 25% sampled 3D 13Cali-resolved [1H,1H]-NOESY spectrum by having sampled an insufficient number of points, rather than by a low signal-to-noise ratio due to the short measurement time. 25% of sampling of the 3D 13Cali-resolved [1H,1H]-NOESY experiment corresponds to collecting 1536 hypercomplex points (Table 1), while the number of NOE signals in this spectrum is about 3000. Sampling of 50% of the hypercomplex points correlated well with the number of expected signals, and it increased the number of reconstructed signals to 98%. This positive result is in line with the theorem that the required minimal number of sampling points is dictated primarily by the number of signals to be reconstructed.[75]
Structure determination of the 71-residue protein NP_346341.1 with NUS NMR data
For an experimental test of the above explanation of the minimal sampling required for the 3D 13Cali-resolved [1H,1H]-NOESY experiment, we performed a structure determination of a smaller protein, for which about 1500 NOE peaks are expected in this spectrum. The results showed that for NP_346341.1, NUS in the extent of 25% / 25% / 25% of the hypercomplex points in the indirect dimensions of the 3D 15N- / 13Cali- / 13Caro-resolved [1H,1H]-NOESY spectra (Table 1) yields a high-quality result. The uniformly and non-uniform sampled NOESY data sets provided nearly identical statistics for automated side chain assignment with ATNOS/ASCAN (Table 2), and the numbers of NOE distance restraints (Table 4) and their distribution per residue along the sequence (Figure 4) were also closely similar. The NMR structures from uniformly sampled and 25% NUS NOESY data can be near-identically superimposed, with backbone and all-heavy atom RMSD values of 0.98 Å and 1.52 Å, respectively (Figure 5).
Table 4.
Input data and calculation statistics for the structure determination of the 71-residue protein NP_346341.1 based on uniformly sampled and NUS 3D heteronuclear resolved [1H,1H]-NOESY data sets.
| Quantity | Uniform sampling | NUS[a] 25 / 25 / 25 |
|---|---|---|
| NOE upper distance limits[b] | 1265 | 1195 |
| Intraresidual (|i – j| = 0) | 333 | 308 |
| Short-range (|i – j| = 1) | 355 | 348 |
| Medium-range (1 < |i – j| < 5) | 268 | 247 |
| Long-range (|i – j| ≥ 5) | 309 | 292 |
| Dihedral angle constraints | 303 | 304 |
| Residual target function value (Å2) | 1.07 ± 0.10 | 0.98 ± 0.33 |
| Residual NOE Violations | ||
| Number ≥ 0.1 Å | 8 ± 3 | 10 ± 3 |
| Maximum (Å) | 0.16 | 0.15 |
| Residual dihedral angle violations | ||
| Number ≥ 2.5° | 0 ± 1 | 0 ± 1 |
| Maximum (°) | 2.04 | 2.53 |
| Amber energies (kcal/mol) | ||
| Total | −2087 ± 57 | −2216 ± 67 |
| Van der Waals | −178 ± 9 | −175 ± 10 |
| Electrostatic | −2455 ± 61 | −2582 ± 70 |
| RMSD from ideal geometry | ||
| Bond lengths (Å) | 0.0076 ± 0.0002 | 0.0076 ± 0.0002 |
| Bond Angles (°) | 1.91 ± 0.06 | 1.91 ± 0.05 |
| RMSD to the mean coordinates (Å)[c] | ||
| bb (6-70) | 0.55 ± 0.17 | 0.64 ± 0.12 |
| ha (6-70) | 0.98 ± 0.13 | 1.11 ± 0.15 |
| Ramachandran plot statistics (%)[d] | ||
| Most favored regions | 76.6 | 73.4 |
| Additional allowed regions | 21.8 | 21.9 |
| Generously allowed regions | 0.0 | 1.6 |
| Disallowed regions | 1.6 | 3.1 |
For the non-uniformly sampled data (NUS), the percentages of hypercomplex points sampled in the indirect dimensions for the 3D 15N- / 13Cali- / 13Caro-resolved [1H,1H]-NOESY data sets are indicated.
Except for the top six entries, which describe the input generated in the final cycle of the ATNOS/CANDID/CYANA calculation, the entries refer to the 20 energy-minimized CYANA conformers selected to represent the NMR structure. Where applicable, the average value for the bundle of 20 conformers and the standard deviation are given.
bb indicates the backbone atoms N, Cα, and C’; ha stands for all heavy atoms. Numbers in parentheses indicate the polypeptide segments for which the RMSD was calculated.
As determined by PROCHECK. The averages of the 20 conformers representing the NMR structure are given.
Figure 4.
Plots of the number, N, of medium-range and long-range NOE upper distance limits per residue contained in the input for the final cycle of the structure calculation of the 71-residue protein NP_346341.1 with CYANA 3.0. (a) Data obtained from the three uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY spectra. (b) Data obtained from the three 3D heteronuclear-resolved [1H,1H]-NOESY spectra with 25% (15N) / 25% (13Cali) / 25% (13Caro) NUS. The correlation of the number of constraints per residue between the uniformly and the non-uniformly sampled data is r = 0.94.
Figure 5.
Superposition of the NMR structures of the 71-residue protein NP_346341.1 obtained from uniformly sampled (blue) and 25% (15N) / 25% (13Cali) / 25% (13Caro) NUS (red) 3D heteronuclear-resolved [1H,1H]-NOESY data. Stereo views are shown of bundles of 20 NMR conformers. The chain ends and selected amino acid residues are identified to guide the eye.
CONCLUSIONS
Non-uniform sampling of the 3D heteronuclear-resolved [1H,1H]-NOESY spectra used for NMR structure determination of soluble proteins with sizes of up to about 200 amino acid residues results in 2- to 3-fold reduction of the overall instrument time needed for a structure determination, i.e., from about eight days of high-field spectrometer time to about two to three days. The NUS data can be similarly used for automated analysis by J-UNIO routines68 as corresponding uniformly sampled data. Whereas 25% NUS is uncritical for the 3D 15N- and 13Caro-resolved [1H,1H]-NOESY experiments with proteins in the aforementioned size range, the 13Cali-resolved experiment requires 50% NUS for a robust protocol of structure determination. Depending on the protein size, this percentage can be fine-tuned, as shown by the two structure determinations described in this paper, so that a maximal reduction of the instrument time can be achieved for smaller proteins. The comparison of the work with different protein sizes may provide guidance for optimizing NUS for use with smaller chemical compounds with much lower signal densities, where improved sensitivity might also be of interest, for example, when studying dilute solutions of organic compounds with natural isotope abundance.
EXPERIMENTAL SECTION
Solutions of the 13C, 15N-labeled proteins NP_372339.1 and NP_346341.1 were produced using a standard purification protocol.[68] For the NMR experiments, 1.2 mM solutions of the proteins were prepared in 95% H2O/5% 2H2O (v/v) containing 20 mM sodium phosphate at pH 6.0, 50 mM sodium chloride and 5 mM sodium azide. Prior to the structure determination, high quality of the protein solutions was documented with NMR-profiles[72] generated from 2D [15N,1H]-HSQC spectra recorded on a Bruker Avance 700 MHz spectrometer equipped with a 1.7 mm TXI z-gradient microcoil probe.
3D heteronuclear-resolved [1H,1H]-NOESY spectra of [13C,15N]-NP_372339.1 and [13C,15N]-NP_346341.1 were recorded at 298 K on a Bruker Avance 800 MHz spectrometer equipped with a 5 mm TXI probe, using the experimental schemes of Figure S1 with the parameters of Table 1. Quadrature detection in all experiments was achieved by following the States–TPPI method.[76] Non-uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY data sets, with otherwise the same experimental parameters, were recorded with variable undersampling in the range 25% to 50% of the hypercomplex points in the indirect dimensions. We applied an exponentially weighted random sampling schedule in the indirect heteronuclear dimension,[77] and a random sampling schedule in the indirect proton dimension. The sampling schedules were generated using the Topspin 3.1 software (Bruker™), with the same random seed for all the experiments.
The uniformly sampled NOESY spectra were processed with the Topspin 3.1 software (Bruker™). Prior to Fourier transformation, the time domain data were multiplied with a sine-squared window[78] and zero-filled to 512 and 256 points in the t1 and t2 dimensions, respectively (Figure S1). To reconstruct the non-uniformly sampled data, we employed the Iterative Reweighted Least Squares (IRLS) algorithm implemented in the MDDNMR software.[45, 57] Spectra were reconstructed by 25 to 35 iterations, with 8 to 16 parallel calculations. Using an Intel Core™ 3.40 GHz processor and 16 GB of memory, this required, per spectrum, about 3 hours of cpu time for 25% sampled data, and about 15 hours for 50% sampled data. Reconstructed NUS spectra were identically processed as the uniformly sampled data, using the NMRPipe software.[79]
Structures were determined either from the uniformly or the non-uniformly sampled 3D heteronuclear-resolved [1H,1H]-NOESY data sets, using the J-UNIO protocol[68] with the torsion angle dynamics algorithm in the program CYANA 3.0.[9]
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIH Protein Structure Initiative (PSI) grants U540-GM074898 (Joint Center for Structural Genomics) and U54-GM094618 (GPCR-Network) from the National Institute of General Medical Sciences (www.nigms.nih.gov), and by the NIH Common Fund grant P50-GM073197 (Joint Center for Innovative Membrane Protein Technologies). Kurt Wüthrich is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute.
REFERENCES
- 1.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 2.Wüthrich K. Angew. Chem. Int. Ed. Engl. 2003;42:3340–3363. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- 3.Göbl C, Madl T, Simon B, Sattler M. Prog. Nucl. Magn. Reson. Spectrosc. 2014;80:26–63. doi: 10.1016/j.pnmrs.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Frueh DP, Goodrich AC, Mishra SH, Nichols SR. Curr. Opin. Struct. Biol. 2013;23:734–739. doi: 10.1016/j.sbi.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osawa M, Takeuchi K, Ueda T, Nishida N, Shimada I. Curr. Opin. Struct. Biol. 2012;22:660–669. doi: 10.1016/j.sbi.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Smith AE, Zhang Z, Pielak GJ, Li C. Curr. Opin. Struct. Biol. 2015;30:7–16. doi: 10.1016/j.sbi.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Williamson MP. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Billeter M, Wagner G, Wüthrich K. J. Biomol. NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Güntert P, Mumenthaler C, Wüthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 10.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS. Acta Crystallogr. Sect. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 11.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 12.Malmodin D, Billeter M. J. Am. Chem. Soc. 2005;127:13486–13487. doi: 10.1021/ja0545822. [DOI] [PubMed] [Google Scholar]
- 13.Hiller S, Fiorito F, Wüthrich K, Wider G. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10876–10881. doi: 10.1073/pnas.0504818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Szyperski T. J. Am. Chem. Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 15.Eghbalnia HR, Bahrami A, Tonelli M, Hallenga K, Markley JL. J. Am. Chem. Soc. 2005;127:12528–12536. doi: 10.1021/ja052120i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lescop E, Schanda P, Brutscher B. J. Magn. Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Orekhov VY, Ibraghimov IV, Billeter M. J. Biomol. NMR. 2001;20:49–60. doi: 10.1023/a:1011234126930. [DOI] [PubMed] [Google Scholar]
- 18.Fredriksson J, Bermel W, Staykova D, Billeter M. J. Biomol. NMR. 2012;54:43–51. doi: 10.1007/s10858-012-9649-y. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt E, Güntert P. J. Am. Chem. Soc. 2012;134:12817–12829. doi: 10.1021/ja305091n. [DOI] [PubMed] [Google Scholar]
- 20.Atreya H, Sahu S, Chary K, Govil G. J. Biomol. NMR. 2006;17:125–136. doi: 10.1023/a:1008315111278. [DOI] [PubMed] [Google Scholar]
- 21.Coggins BE, Zhou P. J. Biomol. NMR. 2003;26:93–111. doi: 10.1023/a:1023589029301. [DOI] [PubMed] [Google Scholar]
- 22.Hyberts SG, Wagner G. J. Biomol. NMR. 2003;26:335–344. doi: 10.1023/a:1024078926886. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann T, Güntert P, Wüthrich K. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 24.Koradi R, Billeter M, Engeli M, Güntert P, Wüthrich K. J. Magn. Reson. 1998;135:288–297. doi: 10.1006/jmre.1998.1570. [DOI] [PubMed] [Google Scholar]
- 25.Shen Y, Delaglio F, Cornilescu G, Bax A. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiorito F, Herrmann T, Damberger FF, Wüthrich K. J. Biomol. NMR. 2008;42:23–33. doi: 10.1007/s10858-008-9259-x. [DOI] [PubMed] [Google Scholar]
- 27.Moseley HN, Monleon D, Montelione GT, Montelione G. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 28.Bartels C, Billeter M, Güntert P, Wüthrich K. J. Biomol. NMR. 1996;7:207–213. doi: 10.1007/BF00202037. [DOI] [PubMed] [Google Scholar]
- 29.Volk J, Herrmann T, Wüthrich K. J. Biomol. NMR. 2008;41:127–138. doi: 10.1007/s10858-008-9243-5. [DOI] [PubMed] [Google Scholar]
- 30.Jung Y-S, Zweckstetter M. J. Biomol. NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 31.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. PLoS Comp. Biol. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann T, Güntert P, Wüthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 34.Gronwald W, Moussa S, Elsner R, Jung A, Ganslmeier B, Trenner J, Kremer W, Neidig K-P, Kalbitzer HR. J. Biomol. NMR. 2002;23:271–287. doi: 10.1023/a:1020279503261. [DOI] [PubMed] [Google Scholar]
- 35.Grishaev A, Llinás M. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6707–6712. doi: 10.1073/pnas.082114199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moseley HN, Montelione GT. Curr. Opin. Struct. Biol. 1999;9:635–642. doi: 10.1016/s0959-440x(99)00019-6. [DOI] [PubMed] [Google Scholar]
- 37.Huang YJ, Tejero R, Powers R, Montelione GT. Proteins: Struct. Funct. Bioinform. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- 38.Crippen GM, Rousaki A, Revington M, Zhang Y, Zuiderweg ER. J. Biomol. NMR. 2010;46:281–298. doi: 10.1007/s10858-010-9403-2. [DOI] [PubMed] [Google Scholar]
- 39.Lescop E, Brutscher B. J. Biomol. NMR. 2009;44:43–57. doi: 10.1007/s10858-009-9314-2. [DOI] [PubMed] [Google Scholar]
- 40.Leutner M, Gschwind RM, Liermann J, Schwarz C, Gemmecker G, Kessler H. J. Biomol. NMR. 1998;11:31–43. doi: 10.1023/a:1008298226961. [DOI] [PubMed] [Google Scholar]
- 41.Schmucki R, Yokoyama S, Güntert P. J. Biomol. NMR. 2009;43:97–109. doi: 10.1007/s10858-008-9291-x. [DOI] [PubMed] [Google Scholar]
- 42.Hoch JC. Methods Enzymol. 1989;176:216–241. doi: 10.1016/0076-6879(89)76014-6. [DOI] [PubMed] [Google Scholar]
- 43.Schmieder P, Stern AS, Wagner G, Hoch JC. J. Biomol. NMR. 1993;3:569–576. doi: 10.1007/BF00174610. [DOI] [PubMed] [Google Scholar]
- 44.Schmieder P, Stern AS, Wagner G, Hoch JC. J. Biomol. NMR. 1994;4:483–490. doi: 10.1007/BF00156615. [DOI] [PubMed] [Google Scholar]
- 45.Orekhov VY, Jaravine VA. Prog. Nucl. Magn. Reson. Spectrosc. 2011;59:271–292. doi: 10.1016/j.pnmrs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Rovnyak D, Frueh DP, Sastry M, Sun Z-YJ, Stern AS, Hoch JC, Wagner G. J. Magn. Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Luan T, Jaravine V, Yee A, Arrowsmith C, Orekhov VY. J. Biomol. NMR. 2005;33:1–14. doi: 10.1007/s10858-005-1363-6. [DOI] [PubMed] [Google Scholar]
- 48.Tugarinov V, Kay LE, Ibraghimov I, Orekhov VY. J. Am. Chem. Soc. 2005;127:2767–2775. doi: 10.1021/ja044032o. [DOI] [PubMed] [Google Scholar]
- 49.Frueh DP, Sun Z-YJ, Vosburg DA, Walsh CT, Hoch JC, Wagner G. J. Am. Chem. Soc. 2006;128:5757–5763. doi: 10.1021/ja0584222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiller S, Ibraghimov I, Wagner G, Orekhov VY. J. Am. Chem. Soc. 2009;131:12970–12978. doi: 10.1021/ja902012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyberts SG, Frueh DP, Arthanari H, Wagner G. J. Biomol. NMR. 2009;45:283–294. doi: 10.1007/s10858-009-9368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyberts SG, Takeuchi K, Wagner G. J. Am. Chem. Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mobli M, Stern AS, Bermel W, King GF, Hoch JC. J. Magn. Reson. 2010;204:160–164. doi: 10.1016/j.jmr.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holland DJ, Bostock MJ, Gladden LF, Nietlispach D. Angew. Chem. 2011;123:6678–6681. doi: 10.1002/anie.201100440. [DOI] [PubMed] [Google Scholar]
- 56.Bostock MJ, Holland DJ, Nietlispach D. J. Biomol. NMR. 2012;54:15–32. doi: 10.1007/s10858-012-9643-4. [DOI] [PubMed] [Google Scholar]
- 57.Kazimierczuk K, Orekhov VY. Angew. Chem. Int. Ed. 2011;50:5556–5559. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 58.Hyberts SG, Arthanari H, Robson SA, Wagner G. J. Magn. Reson. 2014;241:60–73. doi: 10.1016/j.jmr.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arthanari H, Billeter M, Orekhov V. Novel sampling approaches in higher dimensional NMR. Springer Science & Business Media; 2012. [Google Scholar]
- 60.Mobli M, Hoch JC. Prog. Nucl. Magn. Reson. Spectrosc. 2014;83:21–41. doi: 10.1016/j.pnmrs.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isaksson L, Mayzel M, Saline M, Pedersen A, Rosenlöw J, Brutscher B, Karlsson BG, Orekhov VY. PLoS One. 2013;8:e62947. doi: 10.1371/journal.pone.0062947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayzel M, Rosenlöw J, Isaksson L, Orekhov VY. J. Biomol. NMR. 2014;58:129–139. doi: 10.1007/s10858-013-9811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saxena S, Stanek J, Cevec M, Plavec J, Koźmiński W. J. Biomol. NMR. 2014;60:91–98. doi: 10.1007/s10858-014-9861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shigemitsu Y, Ikeya T, Yamamoto A, Tsuchie Y, Mishima M, Smith BO, Güntert P, Ito Y. Biochem. Biophys. Res. Commun. 2015;457:200–205. doi: 10.1016/j.bbrc.2014.12.088. [DOI] [PubMed] [Google Scholar]
- 65.Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen MK, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J. Angew. Chem. Int. Ed. 2011;50:11942–11946. doi: 10.1002/anie.201105648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gautier A, Kirkpatrick JP, Nietlispach D. Angew. Chem. 2008;120:7407–7410. doi: 10.1002/anie.200802783. [DOI] [PubMed] [Google Scholar]
- 67.Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D. Nat. Struct. Mol. Biol. 2010;17:768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serrano P, Pedrini B, Mohanty B, Geralt M, Herrmann T, Wüthrich K. J. Biomol. NMR. 2012;53:341–354. doi: 10.1007/s10858-012-9645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiller S, Wider G, Wüthrich K. J. Biomol. NMR. 2008;42:179–195. doi: 10.1007/s10858-008-9266-y. [DOI] [PubMed] [Google Scholar]
- 70.Fesik SW, Zuiderweg ER. J. Magn. Reson. 1988;78:588–593. [Google Scholar]
- 71.Dutta SK, Serrano P, Proudfoot A, Geralt M, Pedrini B, Herrmann T, Wüthrich K. J. Biomol. NMR. 2014:1–7. doi: 10.1007/s10858-014-9881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedrini B, Serrano P, Mohanty B, Geralt M, Wüthrich K. Biopolymers. 2013;99:825–831. doi: 10.1002/bip.22348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hyberts SG, Robson SA, Wagner G. J. Biomol. NMR. 2013;55:167–178. doi: 10.1007/s10858-012-9698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I. J. Magn. Reson. 2007;188:344–356. doi: 10.1016/j.jmr.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 75.Candès EJ, Romberg J, Tao T. IEEE Trans. Inf. Theory. 2006;52:489–509. [Google Scholar]
- 76.Marion D, Ikura M, Tschudin R, Bax A. J. Magn. Reson. 1989;85:393–399. [Google Scholar]
- 77.Rovnyak D, Sarcone M, Jiang Z. Magn. Reson. Chem. 2011;49:483–491. doi: 10.1002/mrc.2775. [DOI] [PubMed] [Google Scholar]
- 78.De Marco A, Wüthrich K. J. Magn. Reson. 1976;24:201–204. [Google Scholar]
- 79.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.