Abstract
Natural product discovery arises through a unique interplay between chromatographic purification and biological assays. Currently, most techniques used for natural product purification deliver leads without a defined biological action. We now describe a technique, referred to herein as functional chromatography, that deploys biological affinity as the matrix for compound isolation.
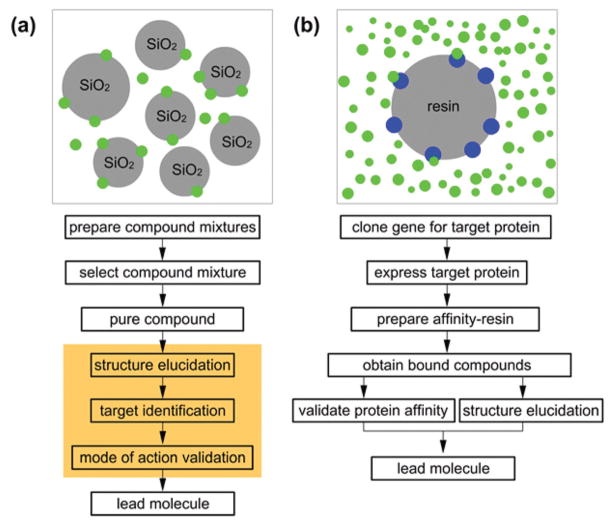
To date, a large portion of natural product discovery has focused its first step on purifying materials based on their affinity to silica gel. While not the target of the discovery program, silica gel and related supports are often the primary tool for discovery efforts with methods including flash chromatography,1 vacuum dry column chromatography,2 and ultra-high performance liquid chromatography (UPLC)3 leading the effort. As outlined in Fig. 1a, this approach occurs without an input from a targeted biological response. While secondary assays can often support this effort through bioactivity guidance,4 the outcome of this approach often becomes restricted by bottlenecks such as target identification or associated mode of action (MOA) validation efforts (orange shaded region of Fig. 1a).
Fig. 1.
A comparison between (a) flash chromatography and (b) functional chromatography. Blue spheres indicate a biological target and green spheres, small molecules. The orange region depicts steps that typically require complicated studies and often reduce the speed of the discovery process.
Alternatively, one can reverse this process by adapting a biological target as the vector for purification. Here the biological function is used as the tool to lead the purification scheme (Fig. 1b). This so called ‘functional chromatography’ approach offers several key advantages that are not available by conventional methods such as flash chromatography (Fig. 1a). We now report on the development of a practical protocol for using functional chromatography to isolate compounds based on their affinity to recombinantly-expressed and purified target proteins.
Over recent years, we have examined the use of reverse affinity strategies as a tool to expedite mode of action studies.5 In these efforts whole or fractionated proteomes presented on resin were used as tools to identify lead molecules in concert with their molecular targets. Perhaps, the first example in target-guided purification was reported by Corti and Cassani in 1985,6 and further developed by teams at Smith, Kline and French Laboratories.7 From their studies, agarose linked-D-Ala-D-Ala resins have become a common tool for the purification of glycopeptide antibiotics such as vancomycin.8 Given the success of this work, we wondered if simple extension to full length purified proteins would provide a logical next step. To this end, we developed functional chromatography by using protein-loaded resins as a tool to isolate small molecules.9
After evaluation, we were able to generate a process that required five-steps over two stages. As shown in Fig. 2, the first stage (Steps 1–2) involved the preparation of protein-coated resins, a process that has been well defined for agarose (i.e. Affi-Gel) and PEGA resins.10 The latter stage (Steps 3–5) applied these resins for purification by the sequential presentation of an extract or crude compound mixture (Step 3), washing and removal of unbound ligands (Step 4), and isolation of the bound ligands by eluting with organic solvents (Step 5).
Fig. 2.
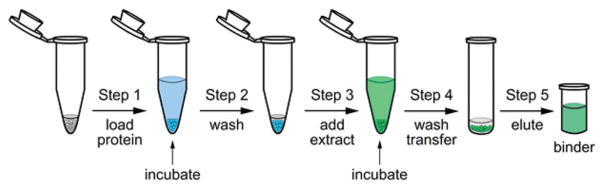
Functional chromatography arises through a 5-step procedure that can be completed in 6–12 h using conventional Eppendorf tubes and glass vials. (Step 1) The process begins by coupling a purified protein to a resin. Protein loading typically requires 1–2 h of incubation at 4 °C to complete. (Step 2) The protein-charged resin is capped to block reactive sites and washed to remove any unbound protein or blocking agent. (Step 3) A natural product extract is added in an aqueous buffer. (Step 4) After incubation, the resin is washed and transferred to a glass vial. The bound materials are eluted using 95% EtOH. Use of a glass vial at the elution stage was key to avoiding background from contaminants in plastic.
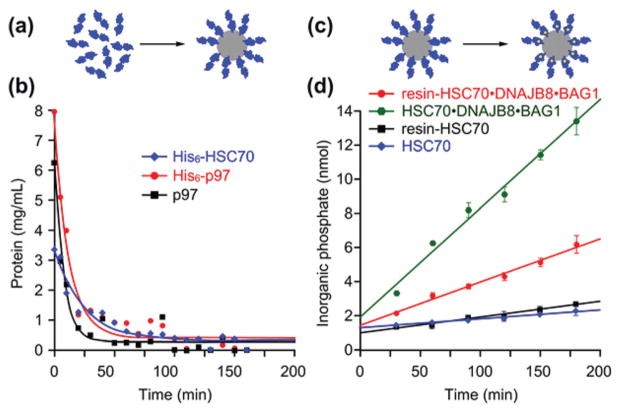
For the first stage, we applied a combination of parallel analyses for protein loading (Fig. 3a and b) and protein activity (Fig. 3c and d) to guide the selection of resin and associated media. Shown in Fig. 3 are three proteins that were investigated in the present study: p97 (also known as valosin containing protein (VCP) or cdc48),11 His6-p97, and His6-HSC70.12 In addition to these we also investigated HSPA1A13 and commercially available malate dehydrogenase (MDH)14 (ESI†). We selected these proteins due to our interest in the kinetics of loading using oligomeric proteins (p97, Fig. 3b and MDH, ESI†) or monomeric proteins (HSC70, Fig. 3b and HSPA1A, ESI†) and effects of changes to the N-termini (His6-p97 and p97, Fig. 3b). We were also interested in how these parameters might affect biochemical function in a number of contexts including oligomeric assembly (p975d), cofactor function (p97;5d HSC70, Fig. 3d; and HSPA1A, ESI†), and a multi-reactant, dimeric enzyme (MDH, ESI†). The kinetics of loading were largely independent of protein identification, but for reasons yet unclear the biochemical function of HSPA1A was compromised when loaded on either Affi-Gel 10 or Affi-Gel 15 (data not shown).
Fig. 3.
Protein loading. (a) Schematic representation of proteins (blue) being coupled to a resin (grey). (b) Plots depicting the amount of unloaded protein remaining as a function of time. The loading efficiencies for three proteins, His6-HSC70, His6-p97 and untagged p97 are shown. (c) Schematic representation of the biochemical validation of a resin as given by confirmation of active site function (yellow dot) within a protein (blue) on the given resin (grey). (d) ATPase activity of resin-loaded and solution samples of HSC70 and the HSC70·DNAJB8·BAG1 complex using a Malachite green assay to measure the release of inorganic phosphate from ATP. The function of the p97 resins has already been reported.5d
As illustrated in Fig. 3, both HSC70 and p97 were effectively coupled and active when loaded on Affi-Gel 10 or Affi-Gel 15. During these investigations, we did find slight differences in rates and activity as a function of conditions or resin used. For instance, p97 did not couple efficiently to Affi-Gel 10 and the optimal coupling conditions (Affi-Gel 15 in 20 mM HEPES pH 8.0, 150 mM KCl, 1 mM MgCl2, 2 mM BME), were slightly different than for the other proteins: HSC70 (Affi-Gel 10 in 20 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 2 mM BME), HSPA1A (Affi-Gel 15 in 20 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 2 mM BME), and MDH (Affi-Gel 15 in 20 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 2 mM BME), as described in Fig. S1, ESI.† Except for the p97-Affi-Gel 10 resin, these differences were modest and do not seem to present a major impediment to the technology. To verify uncompromised function of the resin bound proteins, we used an ATPase assay (p97;5d HSPA1A, ESI,† and HSC70, Fig. 3d) or NADH reduction of ketomalonic acid (MDH; ESI†). We also were able to use an ATPase assay to evaluate the proteins temporal stability on resin and found that in the case of p97, activity was lost only after ~9 days at 4 °C (Fig. S2, ESI†), indicating that our resins would remain active within the 6–12 h experimental period.
We then evaluated these resins to identify protein selective binders (Steps 3–5, Fig. 2). First, we standardized our p97 resins using the commercial inhibitor DBeQ.15 We were consistently able to use 50 μL of resin bearing 5 mg mL−1 of p97 (theoretically capable of isolating 956 ng of DBeQ) to return ~952 ng of DBeQ from 500 μL of a 10 μM solution of DBeQ in buffer (20 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 2 mM BME), indicating that nearly all of p97 remained functional for ligand isolation. Based on this, we calibrated our experiments to 200 μL of resin (theoretical maximum yield of 1.12 mg of ligand), an application that was at least 3000 fold larger than the detection limit of our HPLC-MS analysis (based on the HPLC-MS limits for DBeQ).
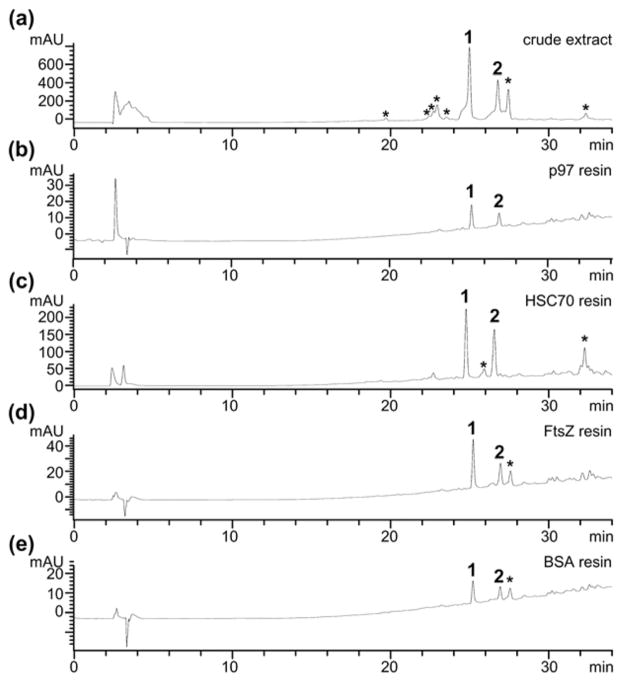
Using this information, we set out to examine application of this technique to an extract derived from the fungus, Lecythophora sp. (Fig. 4a).16 We incubated 200 μL of a 2 mg mL−1 solution of extract in buffer (20 mM HEPES pH 7.4, 150 mM KCl, 1 mM MgCl2, 2 mM BME) with 200 μL of p97 resin for 12 h at 4 °C (Step 3, Fig. 2). After this period, the resin was developed using the procedures in Steps 4–5, Fig. 2 (protocol provided in the ESI†). The resulting bound fraction was dried and evaluated by HPLC analysis. As illustrated in Fig. 4b, incubating p97 returned two peaks at 25.1 min and 26.8 min, referred to as compounds 1 and 2, respectively.
Fig. 4.
Application of functional chromatography. (a) An exemplary HPLC trace of the crude extract of Lecythophora sp. HPLC traces depicting the eluted fraction from application of resins loaded with: (b) p97; (c) HSC70; (d) FtsZ; or (e) bovine serum albumin (BSA) to the extract of Lecythophora sp. All studies used the same concentration of extract and buffers. A 400 μL total volume (200 μL of resin and 200 μL of media containing extract) was used for the p97, FtsZ and BSA studies while 1 mL (500 μL of resin and 500 μL of media) was used for the HSC70 study. Each resin contained 5 mg mL−1 of protein. An asterisk (*) denotes compounds that were not eluted from the p97 resin. HPLC traces were collected by UV absorption at 254 nm.
We then examined the resins bearing HSC70. As shown in Fig. 4c, we observed peaks that corresponded to the isolation of 1 and 2 but also contained a third, minor, peak at 32.2 min that was not observed in experiments with the p97 resin. As a means of control, we applied the same procedure in parallel using FtsZ, a bacterial equivalent to actin,17 and bovine serum albumin (BSA), a protein with high affinity to small molecules.18 As depicted in Fig. 4d and e, HPLC/MS analysis indicated that application of the FtsZ and BSA resin under identical conditions returned three compounds with retention times at 25.2, 26.9 and 27.5 min, which corresponded to the major components within the extract (Fig. 4a). While the p97 resin (Fig. 4b) displayed selective isolation of 1 and 2, FtsZ (Fig. 4d) and BSA (Fig. 4e) lacked this selectivity. While the data indicate that 1 and 2 display a general lack in protein selectivity, as indicated by their isolation by all four protein resins, only the p97 resin delivered pure 1 and 2, without other components of the extract, therein demonstrating the applicability of functional chromatography.
We then turned to conventional isolation methods to obtain compounds 1 and 2 from the Lecythophora sp. extract (ESI†) for their activity validation and structure elucidation efforts.16 Excitingly, we determined that purified samples of 2 displayed modest p97 ATPase inhibition (IC50 value of 31.2 ± 3.0 μM), but 1 showed no inhibition. This difference in activity was also observed in cellular assays using the p97 reporter constructs UbG76VGFP (Fig. S3, ESI†) and TCRα-GFP (Fig. S4, ESI†).
We then turned to spectroscopic methods to elucidate the structures of 1 and 2. Comparison of spectroscopic (MS, 1H and 13C NMR) data19 with those reported suggested that 1 and 2 may be identical with oxaspirol C and oxaspirol B, respectively. However, the isolation and the planar structures of these have exclusively appeared in the Japanese patent literature with incomplete and unassigned NMR spectroscopic data and meagre information on details of their structure elucidation. We have therefore acquired 2D NMR (COSY, HSQC, HMBC, and ROESY) data for 1 (Fig. S5–S10, ESI†) and 2 (Fig. S11–S17, ESI†) interpretation of which allowed us to assign all their 1H and 13C NMR data and their relative stereochemistry (Fig. 5). As reported before,19 oxaspirol B (2) was encountered as an inseparable mixture of two isomers and our ROESY data suggested that they are epimeric at the spiro-carbon (C-5).
Fig. 5.
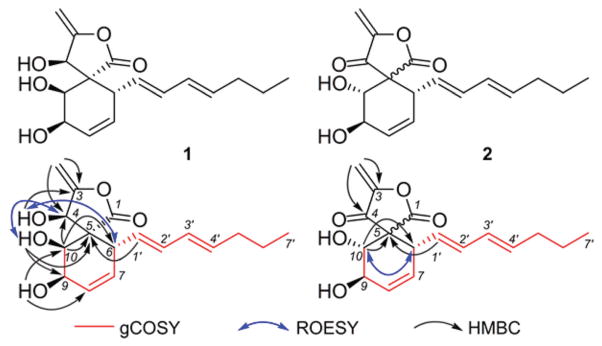
Structures of oxaspirols C (1) and B (2) along with select 1H–1H COSY, HMBC, and 1H–1H ROESY correlations.
Oxaspirols belong to a rare class of natural products, represented thus far by oxaspirols A–C encountered in the fungus, Rhodotorula glutinis, and reported to have weak antibacterial activity.19,20 It is intriguing that the resin-bound p97 was able to retain both oxaspirol B (2) and C (1), but only 2 was capable of inhibiting p97 activity. This prompted us to hypothesize that structural features common to both 1 and 2 are responsible for their binding to p97 but the Michael-acceptor enone moiety present in 2 may be essential for its p97 inhibitory activity.
Conclusions
Here, we describe a procedure, called functional chromatography21 that offers a cost-effective means to purify compounds based on their binding to a recombinantly-expressed and purified protein. Given structural genomics fueled advances in rapid cloning and expression of proteins on large scales, we believe this protocol offers an efficient tool to rapidly address small molecule libraries (either natural product extracts or mixtures of synthetically prepared materials) for novel protein binders.
We now find that functional chromatography offers a practical parallel tool to identify protein binding and more importantly protein specificity. As illustrated by the example in Fig. 4, this method is not a replacement for conventional silica gel based chromatography. Rather, this tool provides a parallel use by allowing a user to rapidly identify ligands to a protein of their interest and hence the term functional chromatography. As shown by comparing the traces in Fig. 4b to Fig. 4a, compounds 1 and 2 were isolated from an extract in one step by their binding to p97. While not selective to p97 as illustrated by the traces in Fig. 4c and e, this study demonstrates how one can rapidly interrogate an extract using resin-bound proteins to identify ligands with differing degrees of selectivity.
In this study, this method is capable of finding materials that are not required to alter an established catalytic site. Both 1 and 2 were discovered in the same extract using the p97-resin, however, only compound 2 showed biochemical activity against p97 ATPase activity (Fig. S3, ESI†) and in cellular studies (Fig. S4, ESI†). While compound 1 bound to p97, it targeted a site that did not alter the ATPase activity of p97. We are presently investigating the mode of binding of oxaspirols B (2) and C (1) and structure–activity relationships of 2 with the aim of enhancing p97 inhibitory activity with compound 2.
Given the low cost and practical efficacy, we envision that this functional chromatographic method22 will have utility for lead discovery in general with immediate applications to natural products programs,22 high-content screening programs,23 or related fragment-based24 or natural-product like25 discovery initiatives. We also see that this tool could be fused directly inline with the steps of these procedures such as natural product isolation or synthetic efforts associated with optimizing small molecule libraries or fragments.
We dedicate this manuscript to the impact that Flash Chromatography published over 35 years ago by W. Clark Still and his team at Columbia University. We thank Abimael Rodriguez (University of Puerto Rico, Rio Piedas), Martin Lear (Tohoku University), and Phil Crews (UC Santa Cruz) for suggestions during the development of this method. This work was supported by a grant (RO1 ES023875) from the National Institute of Environmental Health Sciences (E.C.), a grant (R01 CA90265) from the National Cancer Institute (A.A.L.G.), and a grant (ES006694) from the National Institute of Environmental Health Sciences (a center grant).
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: Methods, procedures as well as copies of spectroscopic data for 1 and 2. See DOI: 10.1039/c4ob02292k
Contributor Information
James J. La Clair, Email: i@xenobe.org.
Eli Chapman, Email: chapman@pharmacy.arizona.edu.
Notes and references
- 1.Still WC, Kahn M, Mitra A. J Org Chem. 1978;43:2923. [Google Scholar]
- 2.Leopold EJ. J Org Chem. 1982;47:4592. [Google Scholar]
- 3.(a) Ali I, Al-Othman ZA, Nagae N, Gaitonde VD, Dutta KK. J Sep Sci. 2012;35:3235. doi: 10.1002/jssc.201200454. [DOI] [PubMed] [Google Scholar]; (b) Aubry AF. Bioanalysis. 2011;3:1819. doi: 10.4155/bio.11.166. [DOI] [PubMed] [Google Scholar]; (c) Wang X, Sun H, Zhang A, Wang P, Han Y. J Sep Sci. 2011;34:3451. doi: 10.1002/jssc.201100333. [DOI] [PubMed] [Google Scholar]
- 4.(a) Agarwal A, D’Souza P, Johnson TS, Dethe SM, Chandrasekaran C. Curr Opin Biotechnol. 2014;25:39. doi: 10.1016/j.copbio.2013.08.010. [DOI] [PubMed] [Google Scholar]; (b) Cheng Z, Wu T. Comb Chem High Throughput Screening. 2013;16:531. doi: 10.2174/1386207311316070004. [DOI] [PubMed] [Google Scholar]; (c) Peach KC, Bray WM, Winslow D, Linington PF, Linington RG. Mol Bio Syst. 2013;9:1837. doi: 10.1039/c3mb70027e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Houssen WE, Jaspars M. Methods Mol Biol. 2012;864:367l. doi: 10.1007/978-1-61779-624-1_14. [DOI] [PubMed] [Google Scholar]; (e) Peach KC, Linington RG. Future Med Chem. 2009;4:593. doi: 10.4155/fmc.09.56. [DOI] [PubMed] [Google Scholar]
- 5.(a) Vera B, Rodríguez AD, La Clair JJ. Angew Chem, Int Ed. 2011;50:8134. doi: 10.1002/anie.201102546. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rodríguez AD, Lear MJ, La Clair JJ. J Am Chem Soc. 2008;130:7257. doi: 10.1021/ja7114019. [DOI] [PubMed] [Google Scholar]; (c) La Clair JJ, Rodríguez AD. Bioorg Med Chem. 2011;19:6645. doi: 10.1016/j.bmc.2011.06.051. [DOI] [PubMed] [Google Scholar]; (d) Kang MJ, Wu T, Wijeratne EMK, Lau EC, Mason DJ, Mesa C, Tillotson J, Zhang DD, Gunatilaka AAL, La Clair JJ, Chapman E. Chem Bio Chem. 2014;15:2125. doi: 10.1002/cbic.201402258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti A, Soffientini A, Cassani G. J Appl Biochem. 1985;7:133. [PubMed] [Google Scholar]
- 7.(a) Folena-Wasserman G, Sitrin RD, Chapin F, Snader KM. J Chromatogr. 1987;392:225. doi: 10.1016/s0021-9673(01)94268-2. [DOI] [PubMed] [Google Scholar]; (b) Dingerdissen JJ, Sitrin RD, DePhillips PA, Giovenella AJ, Grappel SF, Mheta RJ, Oh YC, Pan CH, Roberts GD, Sheare MC, Nisbet LJ. J Antibiot. 1987;40:165. doi: 10.7164/antibiotics.40.165. [DOI] [PubMed] [Google Scholar]
- 8.(a) Chen YY, He WY, Wu Y, Niu CQ, Liu G. J Comb Chem. 2008;10:914. doi: 10.1021/cc800104b. [DOI] [PubMed] [Google Scholar]; (b) Yao N, Wu CY, Xiao W, Lam KS. Biopolymers. 2008;90:421. doi: 10.1002/bip.20949. [DOI] [PubMed] [Google Scholar]
- 9.(a) Zhang Y, Shi S, Guo J, You Q, Feng D. J Chromatogr, A. 2013;1293:92. doi: 10.1016/j.chroma.2013.04.015. [DOI] [PubMed] [Google Scholar]; (b) Yang Z, Zhang Y, Sun L, Wang Y, Gao X, Chen Y. Anal Chim Acta. 2012;719:87. doi: 10.1016/j.aca.2012.01.018. [DOI] [PubMed] [Google Scholar]; (c) Wan H, Rehngren M. J Chromatogr, A. 2006;1102:125. doi: 10.1016/j.chroma.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 10.(a) Kaboord B, Perr M. Methods Mol Biol. 2008;424:349. doi: 10.1007/978-1-60327-064-9_27. [DOI] [PubMed] [Google Scholar]; (b) Kuramochi K, Miyano Y, Enomoto Y, Takeuchi R, Ishi K, Takakusagi Y, Saitoh T, Fukudome K, Manita D, Takeda Y, Kobayashi S, Sakaguchi K, Sugawara F. Bioconjugate Chem. 2008;19:2417. doi: 10.1021/bc8002716. [DOI] [PubMed] [Google Scholar]; (c) Takahashi T, Shiyama T, Mori T, Hosoya K, Tanaka A. Anal Bioanal Chem. 2006;385:122. doi: 10.1007/s00216-006-0335-3. [DOI] [PubMed] [Google Scholar]
- 11.(a) Baek GH, Cheng H, Choe V, Bao X, Shao J, Luo S, Rao H. Amino Acids. 2013;2013:183421. doi: 10.1155/2013/183421. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu Y, Ye Y. Curr Protein Pept Sci. 2012;13:436. doi: 10.2174/138920312802430608. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bug M, Meyer H. J Struct Biol. 2012;179:78. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]; (d) Meyer H, Bug M, Bremer S. Nat Cell Biol. 2012;14:117. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 12.(a) Liu T, Daniels CK, Cao S. Pharmacol Ther. 2012;136:354. doi: 10.1016/j.pharmthera.2012.08.014. [DOI] [PubMed] [Google Scholar]; (b) Witt SN. Curr Protein Pept Sci. 2009;10:424. doi: 10.2174/138920309789352047. [DOI] [PubMed] [Google Scholar]
- 13.(a) Yamashima T. J Neurochem. 2012;120:477. doi: 10.1111/j.1471-4159.2011.07596.x. [DOI] [PubMed] [Google Scholar]; (b) Khalouei S, Chow AM, Brown IR. Cell Stress Chaperones. 2014;19:321. doi: 10.1007/s12192-013-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sasi BK, Sonawane PJ, Gupta V, Sahu BS, Mahapatra NR. J Mol Biol. 2014;426:116. doi: 10.1016/j.jmb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 14.(a) Danis P, Farkas R. Endocr Regul. 2009;43:39. doi: 10.4149/endo_2009_01_39. [DOI] [PubMed] [Google Scholar]; (b) Minárik P, Tomásková N, Kollárová M, Antalík M. Gen Physiol Biophys. 2002;21:257. [PubMed] [Google Scholar]
- 15.(a) Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ. Proc Natl Acad Sci U S A. 2011;108:4834. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chou TF, Bulfer SL, Weihl CC, Li K, Lis LG, Walters MA, Schoenen FJ, Lin HJ, Deshaies RJ, Arkin MR. J Mol Biol. 2014;246:2886. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Details of the culturing, extraction, bioactivity-guided isolation and identification of compounds 1 and 2, and characterization of other metabolites from Lecythophora sp. will be communicated elsewhere.
- 17.FtsZ was chosen due to ready access in our laboratory and its well-documented exploration as a drug target, see: Ma S, Ma S. Chem Med Chem. 2012;7:1161.Kapoor S, Panda D. Expert Opin Ther Targets. 2009;13:1037. doi: 10.1517/14728220903173257.
- 18.(a) Bujacz A, Zielinski K, Sekula B. Proteins. 2014;82:2199. doi: 10.1002/prot.24583. [DOI] [PubMed] [Google Scholar]; (b) Peters T. Adv Protein Chem. 1985;37:61. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]; (c) Kragh-Hansen U. Pharmacol Rev. 1981;33:17. [PubMed] [Google Scholar]
- 19.Isogai A, Nakagawa S, Hirota A, Doi A, Nakanishi O. JP 62164679 A 19870721. Jpn Kokkai Tokkyo Koho. 1987; Chem Abstr. 1987;108:185624. [Google Scholar]
- 20.Doi J, Hirota A, Nakagawa M, Sakai H, Isogai A. Agric Biol Chem. 1985;49:2247. [Google Scholar]
- 21.The term functional chromatography describes a chromatographic method that links a biological function to the matrix for purification. In this manuscript, we demonstrate how reversing conventional protein ligand affinity can be used to isolate select protein binders. Typically small molecules on resins are used to isolate proteins or labelled proteins (i.e. Ni-NTA resins and His-tagged proteins) and here the reversal allows one to isolate natural products based on their protein binding. Hence, the term “functional” chromatography as the method allows one to rapidly identify small molecules based on their function. For the example in Fig. 4, there function was binding to p97, an event that was not that selective as illustrated by the binding to HSC70, FtsZ and BSA. While not a good function in terms of drug discovery, this is a function. We also refer readers to ref. 5d.
- 22.(a) Basmadjian C, Zhao Q, Bentouhami E, Djehal A, Nebigil CG, Johnson RA, Serova M, de Gramont A, Faivre S, Raymond E, Désaubry LG. Front Chem. 2014;2:20. doi: 10.3389/fchem.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Butler MS, Blaskovich MA, Cooper MA. J Antibiot. 2013;66:571. doi: 10.1038/ja.2013.86. [DOI] [PubMed] [Google Scholar]; (c) Bologa CG, Ursu O, Oprea TI, Melançon CE, 3rd, Tegos GP. Curr Opin Pharmacol. 2013;13:678. doi: 10.1016/j.coph.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Huang L, Su T, Li X. Curr Top Med Chem. 2013;13:1864. doi: 10.2174/15680266113139990142. [DOI] [PubMed] [Google Scholar]
- 23.(a) Caliandro R, Belviso DB, Aresta BM, de Candia M, Altomare CD. Future Med Chem. 2013;5:1121. doi: 10.4155/fmc.13.84. [DOI] [PubMed] [Google Scholar]; (b) Harner MJ, Frank AO, Fesik SW. J Biomol NMR. 2013;56:65. doi: 10.1007/s10858-013-9740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wilde F, Link A. Expert Opin Drug Discovery. 2013;8:597. doi: 10.1517/17460441.2013.780022. [DOI] [PubMed] [Google Scholar]
- 24.(a) Abet V, Mariani A, Truscott FR, Britton S, Rodriguez R. Bioorg Med Chem. 2014;22:4474. doi: 10.1016/j.bmc.2014.04.019. [DOI] [PubMed] [Google Scholar]; (b) Martell RE, Brooks DG, Wang Y, Wilcoxen K. Clin Ther. 2013;35:1271. doi: 10.1016/j.clinthera.2013.08.005. [DOI] [PubMed] [Google Scholar]; (c) Fang Y. Front Pharmacol. 2014;27:52. doi: 10.3389/fphar.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Singh S, Carpenter AE, Genovesio A. J Biomol Screening. 2014;19:640. doi: 10.1177/1087057114528537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.(a) Thomas GL, Johannes CW. Curr Opin Chem Biol. 2011;145:516. doi: 10.1016/j.cbpa.2011.05.022. [DOI] [PubMed] [Google Scholar]; (b) Bauer RA, Wurst JM, Tan DS. Curr Opin Chem Biol. 2010;14:308. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.