Rab-coupling protein–mediated integrin trafficking promotes filopodia formation via RhoA-ROCK-FHOD3, generating non-lamellipodial actin spike protrusions that drive cancer cell migration in 3D extracellular matrix and in vivo.
Abstract
Invasive migration in 3D extracellular matrix (ECM) is crucial to cancer metastasis, yet little is known of the molecular mechanisms that drive reorganization of the cytoskeleton as cancer cells disseminate in vivo. 2D Rac-driven lamellipodial migration is well understood, but how these features apply to 3D migration is not clear. We find that lamellipodia-like protrusions and retrograde actin flow are indeed observed in cells moving in 3D ECM. However, Rab-coupling protein (RCP)-driven endocytic recycling of α5β1 integrin enhances invasive migration of cancer cells into fibronectin-rich 3D ECM, driven by RhoA and filopodial spike-based protrusions, not lamellipodia. Furthermore, we show that actin spike protrusions are Arp2/3-independent. Dynamic actin spike assembly in cells invading in vitro and in vivo is regulated by Formin homology-2 domain containing 3 (FHOD3), which is activated by RhoA/ROCK, establishing a novel mechanism through which the RCP–α5β1 pathway reprograms the actin cytoskeleton to promote invasive migration and local invasion in vivo.
Introduction
Malignant transformation and metastatic spread is the main cause of death in cancer patients. To metastasize, cells must acquire the ability to migrate and invade in 3D matrices, requiring dynamic reorganization of the actin cytoskeleton to alter morphology and provide protrusive force (Bravo-Cordero et al., 2012). Cancer cells are understood to adopt a range of migratory strategies, from collective to single cell invasion, and the mechanisms that drive protrusion are thought to be dictated by Rho GTPases (Sanz-Moreno et al., 2008). For example, the leader cells in collective invasion and single “mesenchymal” cells migrate in a Rac-dependent manner (Friedl and Alexander, 2011; Friedl et al., 2012; Bravo-Cordero et al., 2012; Theveneau and Mayor, 2013), with the mechanisms of actin polymerization, protrusion, and force generation thought to be reliant on Arp2/3, analogous to lamellipodial migration in 2D (Law et al., 2013; Giri et al., 2013; Krause and Gautreau, 2014). Lamellipodium-independent 3D migration strategies have also been described. Single cells can adopt an “amoeboid” migration strategy, similar to the movement of leukocytes, whereby RhoA/ROCK activity promotes actomyosin contractility and membrane blebbing to provide protrusive force (Friedl and Alexander, 2011), and lobopodial migration is driven by RhoA/ROCK-mediated contractility, providing the force to drive nuclear pistoning (Petrie et al., 2012, 2014). Both of these mechanisms require actomyosin contractility at the rear of the cell to drive an increase in hydrostatic pressure and forward movement of the cell in the absence of actin polymerization–dependent protrusive structures. We have recently shown that Rab-coupling protein (RCP)-mediated α5β1 integrin recycling locally activates RhoA at the cell front to promote formation of pseudopodial protrusions tipped by actin spikes (Jacquemet et al., 2013a). However, an understanding of how the molecular mechanisms underlying lamellipodial protrusion in 2D are reflected in 3D, and how nonlamellipodial actin-based protrusions are dynamically regulated in 3D, is lacking.
Integrins are α/β heterodimeric receptors that mediate communication between the cell and the ECM, capable of eliciting a plethora of signaling responses to effect a host of functional outcomes (Hynes, 2002; Legate et al., 2009; Ivaska and Heino, 2011). Although integrins alone are not oncogenic, dysregulation of integrin signaling is frequently a prognostic indicator of tumor progression (Desgrosellier and Cheresh, 2010). For example, in high-grade ovarian tumors, αvβ3 integrin expression is down-regulated (Maubant et al., 2005) and patients with high β3 integrin expression have an improved prognosis (Kaur et al., 2009), whereas high expression of α5β1 integrin is an indicator of a poor outcome (Sawada et al., 2008).
The endocytic trafficking of integrins plays an important role in regulating integrin function during cell division and migration (Caswell and Norman, 2006; Pellinen and Ivaska, 2006; Caswell et al., 2009; Bridgewater et al., 2012; Jacquemet et al., 2013b). In particular, the recycling of the fibronectin (FN) receptor α5β1 promotes invasive migration in 3D ECM (Caswell et al., 2007, 2008; Caswell and Norman, 2008; Muller et al., 2009; Dozynkiewicz et al., 2012). Rab coupling protein (RCP, Rab11-FIP1) can interact with α5β1 to control its recycling, and inhibition of αvβ3 integrin (with small-molecule inhibitors, e.g., cilengitide, cRGDfV; or soluble ligands, e.g., osteopontin) or expression of gain-of-function mutant p53 (e.g., R273H, R175H) promotes the association of RCP with α5β1 and leads to rapid recycling of this integrin (Caswell et al., 2008; Muller et al., 2009). RCP–α5β1 vesicles accumulate in protrusive pseudopods in 3D matrix, driving their extension and resulting in invasive migration (Caswell et al., 2008; Rainero et al., 2012). Rather than directly influence the adhesive capacity of the cell, RCP-driven α5β1 recycling coordinates signaling of receptor tyrosine kinases (RTKs, including EGFR1 and c-Met; Caswell et al., 2008; Muller et al., 2009) to drive polarized signaling within the tips of invasive pseudopods through the RacGAP1–IQGAP1 complex. This leads to local suppression of activity in the small GTPase Rac1 and increased activity of RhoA, which drives extension of long pseudopodial processes tipped with actin spikes at the cell front, as opposed to formation of wave-like structures, enabling subsequent migration and invasion in 3D ECM (Jacquemet et al., 2013a).
Reorganization of the actin cytoskeleton to promote actin-based protrusion requires actin filament elongation, catalyzed by actin assembly factors that promote nucleation and/or elongation of actin filaments (Nürnberg et al., 2011; Krause and Gautreau, 2014). The Arp2/3 complex polymerizes actin filaments as branches from existing filaments, generating a complex, dense, and highly branched network of actin filaments, such as that seen within the lamellipodium of cells migrating in 2D (Krause and Gautreau, 2014). Actin polymerization within the lamellipodium pushes filaments against the membrane at the leading edge, and this force, combined with actomyosin contractility, generates retrograde flow of actin filaments and complexes associated with the actin network (Lai et al., 2008). Some of these rearward-sliding actin filaments are captured by integrin adhesion complexes that form within the lamellipodium; this acts as a “molecular clutch” to transfer retrograde actin flow into protrusive traction force, driving the lamellipodium forward (Gardel et al., 2008; Thievessen et al., 2013). Filopodia are an alternate class of protrusion found at the leading edge of migrating cells that can be generated through Arp2/3-dependent (convergent elongation) and -independent (tip nucleation) means of actin polymerization (Mattila and Lappalainen, 2008; Mellor, 2010). Filopodia contain more stable F-actin bundles, and rates of protrusion and retrograde flow are inversely correlated (Mallavarapu and Mitchison, 1999). Formins promote the formation of unbranched actin filaments by nucleating actin polymerization at the barbed end processively and promoting elongation by resisting capping (Mattila and Lappalainen, 2008; Mellor, 2010; Nürnberg et al., 2011). Diaphanous-related formins (DRFs) are effectors for Rho GTPases, which bind to DRFs to relieve the autoinhibitory interaction between the diaphanous inhibitory domain (DID) and the diaphanous autoregulatory domain (DAD), and have been reported to play a role in filopodia formation and contribute to lamellipodial extension (Peng et al., 2003; Pellegrin and Mellor, 2005; Block et al., 2012). The DRFs FMNL2 and FMNL3 have further been shown to play a role in migration in 3D matrix (Kitzing et al., 2010; Vega et al., 2011), although how they contribute to actin-based protrusion is currently unclear.
In the present study, we have interrogated the dynamics of actin-based protrusion in migrating and invading carcinoma cells. We find evidence of retrograde flow in wave-like lamellipodial protrusions in 3D matrix; however, upon increasing the invasive capacity of cells by promoting RCP–α5β1 trafficking, we observe more stable long filopodia that form actin spike protrusions. RCP–α5β1 trafficking increased local invasion of cancer cells in vivo, where cells exhibit dynamic actin spike protrusions. Furthermore, we show that formation of actin spikes is Arp2/3-independent, but requires ROCK-mediated phosphorylation and activation of the DRF Formin homology domain-2 containing 3 (FHOD3). Finally, FHOD3 is required for local invasion in vivo, downstream of RCP–α5β1 trafficking, providing evidence that integrin trafficking dictates a novel actin polymerization program to promote actin spike protrusion formation and invasion in 3D microenvironments.
Results
RCP–α5β1 integrin trafficking alters F-actin dynamics in cell protrusions
Retrograde flow of actin filaments is a feature of lamellipodial migration, which, when coupled to integrin adhesion complexes, provides a powerful force driving forward movement of cells (Gardel et al., 2008; Lai et al., 2008; Thievessen et al., 2013). We used a photoactivation approach in cells expressing photoactivatable GFP (paGFP)-actin and Lifeact-mRFPmars in combination with spinning disk confocal microscopy to analyze retrograde actin flow in carcinoma cell lines migrating on 2D surfaces. Upon activation of paGFP-actin within a small region (5 µm2) at the leading edge of the lamellipodium, evidence of retrograde flow could be seen in A2780 ovarian cancer cells and H1299 non-small cell lung cancer cells, as a pool of photoactivated paGFP-actin dissipated from the photoactivated region at a slower rate than freely diffusible paGFP and passed through a region of interest (ROI) immediately behind that activated within ∼30 s (Fig. S1, A and B).
We have previously shown that RCP–α5β1-dependent trafficking, induced by inhibition of αvβ3 integrin (using cRGDfV, a selective cyclic peptide inhibitor of αvβ3 integrin) or mutant p53 expression promotes the formation of dynamic actin-rich protrusions that actively ruffle to promote rapid but less directionally persistent migration in 2D (Caswell et al., 2008; Muller et al., 2009). Upon activation of paGFP-actin within dynamically ruffling protrusions, the lower rate of diffusion of paGFP-actin away from the photoactivated region suggested that these structures were more stable, and retrograde flow of actin filaments was also slowed as a lower level of photoactivated paGFP-actin dispersed through the region immediately behind that photoactivated (Fig. S1, A and B).
While there is evidence that Rac and downstream effector pathways leading to activation of Arp2/3 (Sanz-Moreno et al., 2008; Giri et al., 2013; Law et al., 2013) play a role in 3D migration, there is little evidence to suggest that the mechanism that drives lamellipodial traction force in 3D ECM is linked to retrograde actin flow. We seeded carcinoma cells onto cell-derived matrix (CDM), a linearly elastic 3D matrix made up of fibrillar collagen and FN (Cukierman et al., 2001; Petrie et al., 2012, 2014). Under basal conditions, A2780 cells formed wave-like protrusions resembling small lamellipodia toward the cell front (reflecting Rac activity localized in this subcellular region; Jacquemet et al., 2013a), structures that have been noted previously in different cell lines in 3D ECM (Petrie et al., 2012; Giri et al., 2013; Jacquemet et al., 2013a). Upon activation of paGFP-actin at the leading edge of these protrusive structures, retrograde flow of actin was clearly observable within the region immediately behind that activated (Fig. 1, A–C; and Video 1), with similar kinetics to those observed in 2D. Interestingly, when RCP–α5β1 trafficking was induced with cRGDfV, protrusions again appeared more stable, and F-actin structures showed slower rates of retrograde flow (Fig. 1, A–C; and Video 1). Moreover, the dynamics of paGFP-actin in MDA-MB-231 cells, which express an endogenous gain-of-function mutant p53, suggest that these cells generate F-actin protrusions that are not subjected to rapid retrograde flow in FN-rich ECM (Fig. S1 C). These data suggest that RCP–α5β1 integrin-driven actin-based protrusions are relatively stable and exhibit low rates of retrograde flow.
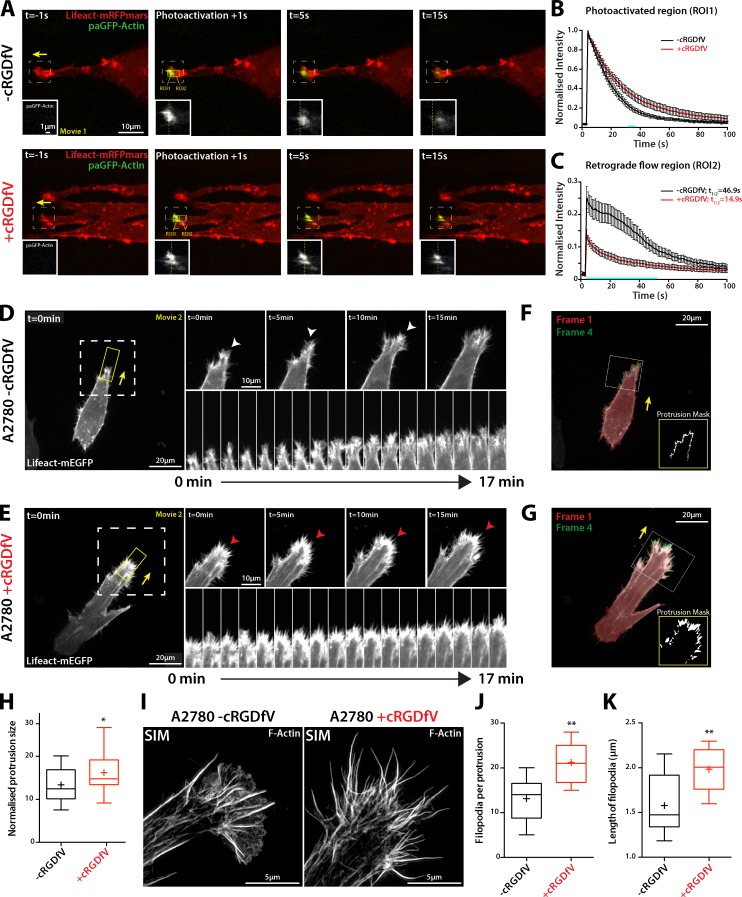
Figure 1.
RCP–α5β1 integrin trafficking promotes actin spike formation. (A) A2780 cells transiently transfected with Lifeact-mRFPmars and paGFP-Actin were seeded onto 3D CDM 4 h before imaging and treated with cRGDfV (2.5 µM, 2 h) as indicated. Yellow broken lines mark the center of the photoactivated region. The boxed regions are enlarged in the insets. (B and C) ROI1 was photoactivated and cells were imaged every second for 100 s. Normalized intensity was analyzed for ROI1 (B) and ROI2 (C; n = 15 [−cRGDfV], n = 16 [+cRGDfV]). Blue bars indicate time points at which datasets are significantly different (P < 0.01; Student’s t test). Yellow arrows indicate the direction of protrusion. Error bars represent the SEM. (D and E) A2780 cells transiently transfected with Lifeact-mEGFP were seeded as in A and z stacks were captured (7 sections, 0.2 µm intervals) on a spinning disk confocal microscope every minute for >25 min. Z projections are shown. White arrowheads indicate lamellipodia-like actin veils, red arrowheads indicate actin spikes. The white boxed regions are enlarged on the right, and the kymograph images are taken from the yellow boxed regions. (F and G) Quantification of Lifeact-mEGFP protrusions. Protrusions were identified by overlapping thresholded video frames with a lag of 3 min. Insets show the boxed regions. (H) Normalized protrusion size was measured using image masks as represented in F and G. n = 28 cells per condition (n > 25 frames per cell). (I) A2780 cells were seeded as in A before fixation and Alexa Fluor 488 Phalloidin staining. SIM was used to generate super-resolution images; maximum intensity projections are shown. The number (J) and length (K) of filopodia per protrusion was quantified from SIM images; n = 10 cells per condition. All data represent at least three independent experiments; statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test (H, J, and K). +, mean; *, P < 0.05; **, P < 0.01.
Promoting RCP–α5β1 trafficking leads to the formation of F-actin spikes and increased invasion in vivo
RCP–α5β1 integrin trafficking leads to an increase in F-actin density at protrusions of cells migrating in 3D CDM or high-concentration collagen/FN hydrogels (Jacquemet et al., 2013a). Live-cell spinning disk imaging, capturing z-stacks of A2780 cells transfected with Lifeact-mEGFP migrating in CDM over time courses that expose protrusive events, revealed small, wave-like protrusions under basal conditions, which, like 2D lamellipodia, were made up of F-actin veils between filopodia (Fig. 1 D and Video 2). Upon addition of cRGDfV to promote RCP–α5β1 recycling, cells migrating in 3D CDM formed large actin spike–based protrusions (Fig. 1 E and Video 2). Using an unbiased analysis to mask protrusions at the front of migrating cells over many frames (Fig. 1, F and G), we found that RCP–α5β1 trafficking generates larger protrusive areas (Fig. 1 H). H1299-Vec cells, which move with broad lamellipodia in 2D (Fig. S1 B; Muller et al., 2009), possess wavelike lamellipodia in 3D CDM; however, upon expression of mutant p53 in H1299 cells, dynamic bursts of spikelike structures were observed (Fig. S2, A and B). Similarly, MDA-MB-231 cells also displayed intense clusters of actin spikes, which were concentrated at cell protrusions in the direction of cell migration (Fig. S2 C). Structured illumination microscopy (SIM) enabled the resolution of bundled F-actin filaments within protrusions as cells move in 3D CDM, and revealed a switch from predominantly lamellipodial protrusion to an increase in the number and length of filopodia at the tips of invasive pseudopods upon induction of RCP–α5β1 trafficking with cRGDfV (Fig. 1, I–K). Live imaging further demonstrated that filopodial lifetime was also increased in cRGDfV-treated cells (Fig. S2 D). These data suggest that, in several carcinoma cells lines, signaling downstream of RCP–α5β1 trafficking suppresses lamellipodia formation and promotes formation of filopodia that are arranged into F-actin spikes in the direction of migration in 3D CDM.
We have previously demonstrated that RCP-driven recycling of α5β1 and co-cargo RTKs, induced by inhibition of αvβ3 integrin or mutant p53 expression, leads to increased invasion into FN-rich ECM (Caswell et al., 2008; Muller et al., 2009, 2013), and gain-of-function mutant p53 promotes metastasis in genetically engineered mouse models (Caulin et al., 2007; Adorno et al., 2009; Muller et al., 2009; Doyle et al., 2010; Arjonen et al., 2014). We sought to determine whether manipulation of α5β1 integrin trafficking could influence local invasion in a physiologically relevant in vivo environment, a zebrafish xenograft model in which melanoma cells have been shown to disseminate in association with collagen and FN (Chapman et al., 2014). Under basal conditions, cells formed a xenograft tumor mass in the pericardial cavity, but showed negligible signs of dissemination through surrounding tissue (Fig. 2, A, C, D, and F). Upon addition of cRGDfV (A2780 cells) or expression of mutant p53 R273H (H1299 cells), a significant increase in the ability of cells to locally invade into the jaw region of the embryo was observed (Fig. 2, B, C, E, and F), suggesting that promoting RCP–α5β1 trafficking increases the capacity of cells to invade an in vivo tissue environment.
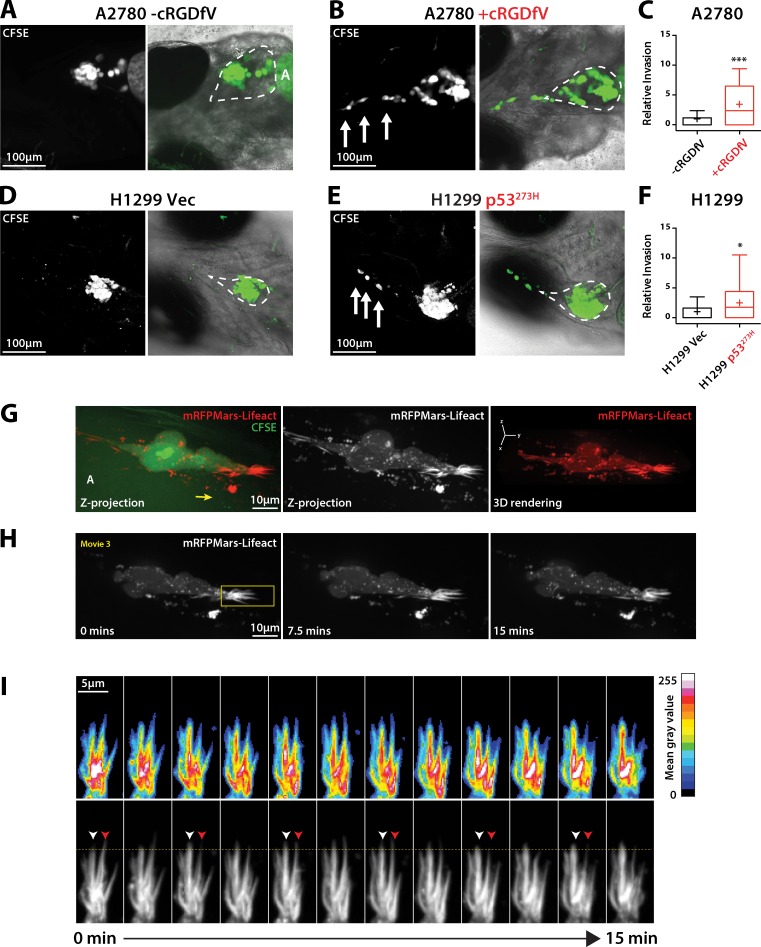
Figure 2.
Dynamic actin spikes form protrusions in cells invading locally in vivo. (A, B, D, and E) CFSE-labeled A2780 cells (A and B) or H1299 cells (D and E) were injected into the pericardial cavity of zebrafish embryos. cRGDfV (2.5 µM) was added 1 dpi as appropriate. Embryos were imaged at 4 dpi to determine invasion. Broken lines indicate the pericardial cavity. A, autofluorescence from the yolk sac. White arrows indicate invading cells. (C and F) Quantification of relative invasion. n = 28–33 fish per condition. (G) CFSE-labeled A2780 cells stably expressing Lifeact-mRFPmars were injected into zebrafish embryos as in A and B, and treated with cRGDfV (2.5 µM). Imaging was performed 3 dpi using a spinning disk confocal microscope. Images represent z projections and 3D rendering. The yellow arrow indicates the direction of migration. (H) Time lapse of Lifeact-mRFPmars–expressing cells over limited z range (2.5 µm). (I) Kymograph of protrusive area highlighted in H (yellow box). White arrowheads indicate an actin spike actively protruding; red arrowheads indicate retraction. The yellow horizontal line provides a reference for protrusion/retraction events. Mean gray value indicates intensity of Lifeact fluorescence. All data represent at least three independent experiments; statistical significance was evaluated using a Student’s t test with Welch’s correction. +, mean; *, P < 0.05; ***, P < 0.001.
Casper strain zebrafish are amenable to imaging, so we next investigated the dynamics of F-actin protrusions in cells invading in vivo. A2780 cells stably expressing Lifeact-mRFPmars were injected into zebrafish embryos and allowed to invade for 48 h before live spinning disk confocal imaging. Invading cells possessed large spike-like F-actin protrusions in the direction of invasion (Fig. 2, G–I), which appeared to be larger and thicker structures than those observed in cells migrating in vitro, but clearly showed dynamic actin polymerization and depolymerization (Fig. 2 I and Video 3). Together these data reveal that RCP-driven α5β1 integrin trafficking leads to the formation of filopodia and actin spikes (but not lamellipodia or membrane blebs) and local invasion in vivo.
The Arp2/3 complex is not required for RCP–α5β1-driven actin polymerization and invasive migration
We next sought to identify potential downstream effectors of the RCP–α5β1 integrin pathway that led to altered morphology and dynamics of actin-based protrusion. CK-666, an Arp2/3 inhibitor (Nolen et al., 2009), caused lamellipodia to collapse under basal conditions in 2D, whereas the inactive analogue CK-689 had no effect (Fig. S3 A). However, CK-666–treated cells were able to form protrusions in the direction of migration, similar to actin-rich ruffling protrusions that form in response to RCP–α5β1 trafficking (Fig. S1 A; Caswell et al., 2008) and knockdown of ArpC2 or ArpC3 (Fig. 3 A). CK-666–treated A2780 cells moved in a rapid but less directionally persistent manner (Fig. S3, B–E), similar to the 2D migratory phenotype observed upon induction of RCP-driven α5β1 recycling (+cRGDfV), while migration of mouse embryonic fibroblasts was impaired by these concentrations of CK-666 (Fig. S3 F), as previously observed (Wu et al., 2012). Moreover, Arp2/3 inhibition had no significant influence on 2D migration in the presence of cRGDfV (Fig. S3, B–E). RCP–α5β1-driven morphology and migration in 3D CDM was unaffected by ArpC2 and ArpC3 knockdown (Fig. 3, B–D; and Fig. S3, H–J) or Arp2/3 inhibition (Fig. 3, F and G; and Fig. S3 G), and under basal conditions these interventions did not influence migration (Fig. 3, C, F, and G; and Fig. S3, G and I), but ArpC2 knockdown and CK-666–treated A2780 cells appeared to extend longer pseudopodial protrusions (Fig. 3, D and H; and Fig. S3 J). Interestingly, Arp2/3 inhibition did not abrogate the recycling of α5β1, and in fact induced a modest increase in recycling under basal conditions (Fig. S3 K). Furthermore, inhibition of Arp2/3 promoted the accumulation of RCP-containing vesicles to pseudopod tips (Fig. S3, L and M), suggesting that Arp2/3 could oppose RCP–α5β1 trafficking. These data indicate that in cancer cells Arp2/3 is not always required for migration, and may even suppress RCP–α5β1-driven signaling to the cytoskeleton and filopodial actin spike formation.
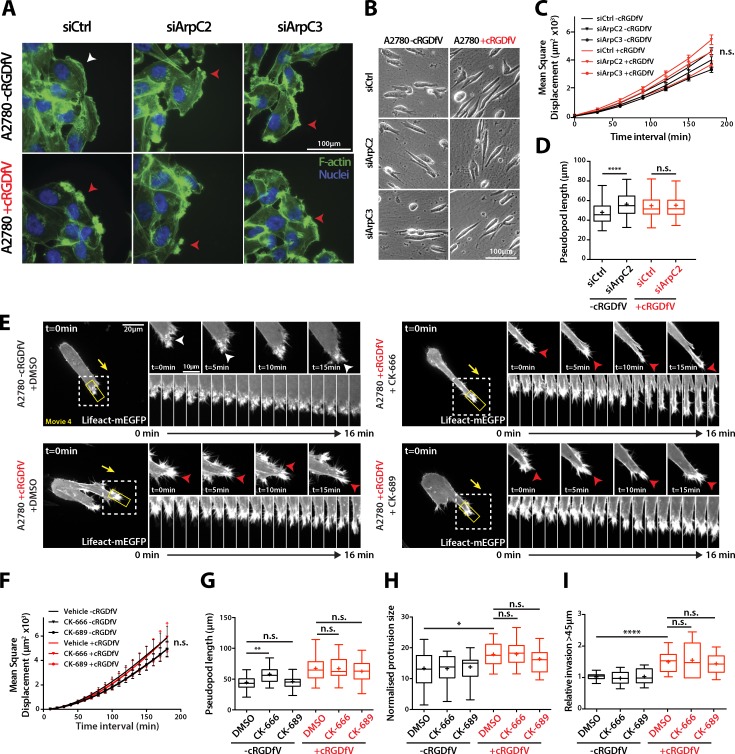
Figure 3.
RCP–α5β1-driven migration and invasion is Arp2/3-independent. (A) siControl, ArpC2 knockdown, or ArpC3 knockdown A2780 cells were seeded onto glass coverslips, wounded after 24 h, and treated with cRGDfV (2.5 µM) as indicated before fixation. F-actin was stained with FITC-phalloidin and nuclei with DAPI. The white arrowhead indicates lamellipodia; red arrowheads indicate nonlamellipodial ruffling protrusions. (B) siControl, ArpC2 knockdown, or ArpC3 knockdown A2780 cells were seeded onto 3D CDM for 4 h and treated with cRGDfV (2.5 µM) where indicated before time-lapse imaging. (C) MSD measurement of siCtrl, siArpC2, and siArpC3 A2780 cells migrating in CDM. n ≥ 99 cells per condition. (D) Pseudopod length of siCtrl and siArpC2 A2780 cells migrating in CDM. n = 100 cells per condition. (E) A2780 cells transiently transfected with Lifeact-mEGFP were seeded onto 3D CDM for 4 h, and treated with cRGDfV (2.5 µM, >2 h) and the indicated inhibitors (50 µM) or vehicle control immediately before imaging. Maximum z projections are shown. White arrowheads indicate lamellipodia; red arrowheads indicate ruffling actin spikes. The white boxed regions are enlarged on the right, and the kymograph images are taken from the yellow boxed regions. Yellow arrows indicate the direction of migration. (F and G) MSD (F; n = 20) and pseudopod length (G; n = 50) of DMSO, CK-666, and CK-689–treated A2780 cells migrating in CDM. (H) Normalized protrusion size of A2780 cells expressing Lifeact-mEGFP treated with the indicated inhibitor (50 µM) as in E. n = 13–21 cells per condition with n ≥ 25 frames quantified per cell. (I) A2780 cells were seeded into an inverted invasion assay in the presence of cRGDfV (2.5 µM) where indicated for 72 h. DMSO, CK-666 (50 µM), and CK-689 (50 µM) were added 24 h after seeding and imaged 48 h later to quantify invasion. All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. +, mean; n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars represent the SEM.
Live imaging of A2780 cells expressing Lifeact-mEGFP showed that cells treated with cRGDfV were still able to generate large, filopodia-based protrusions upon addition of CK-666 and CK-689 (Fig. 3, E and H; and Video 4). Furthermore, Arp2/3 inhibition had no significant influence on the very low basal level of A2780 cell invasion, nor did it affect RCP–α5β1-driven invasion into plugs of FN-rich fibrillar collagen (Fig. 3 I). Together, these data suggest that RCP–α5β1-driven protrusion, migration, and invasion are not reliant on Arp2/3-mediated actin polymerization, suggesting that other pathways influence actin dynamics in this context.
Formin inhibition abrogates invasive migration
Formins are a family of 15 actin nucleators that drive processive polymerization of actin filaments from the barbed end, some of which can act as Rho GTPase effectors (DRFs; Mellor, 2010; Nürnberg et al., 2011). The pan-formin inhibitor SMIFH2 (Rizvi et al., 2009) profoundly abrogated protrusion formation in cells migrating in 3D CDM (Fig. 4, A and B) and invasion into collagen/FN-rich ECM (Fig. 4 D), and cells instead formed membrane blebs (Fig. 4, A and C), suggesting that formins mediate actin reorganization within protrusions as cells migrate in 3D ECM.
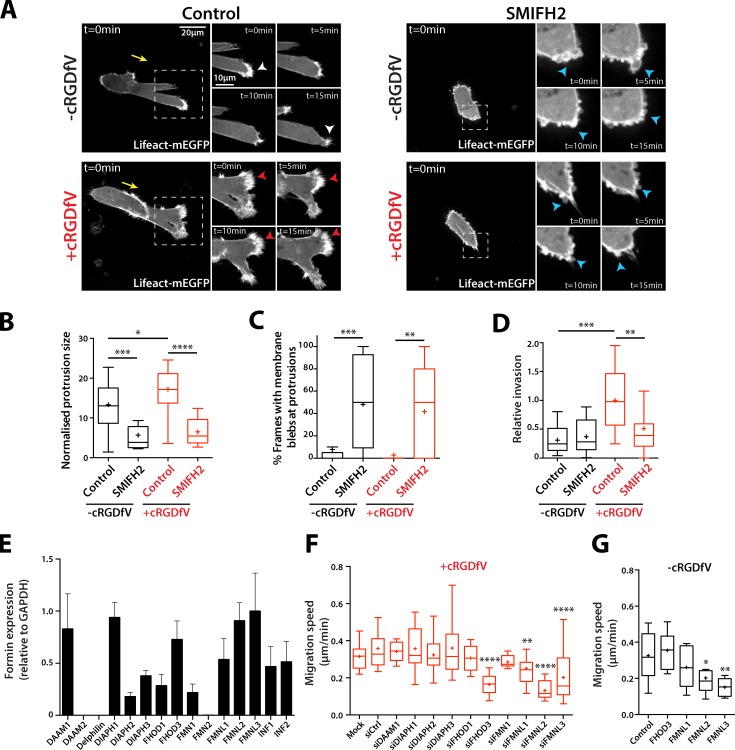
Figure 4.
Formin function is required for protrusion formation. (A) A2780 cells transiently transfected with Lifeact-mEGFP were seeded onto 3D CDM for 4 h, and treated with cRGDfV (2.5 µM, 2 h). Cells were treated with SMIFH2 (25 µM) or vehicle control immediately before imaging. Maximum z projections are shown. White arrowheads indicate lamellipodia-like protrusions; red arrowheads indicate actin spikes; blue arrowheads indicate blebs. Yellow arrows indicate direction of migration. The white boxed regions are enlarged on the right. (B) Normalized protrusion size of cells as in A. n = 12–20 cells per condition with n ≥ 25 frames quantified per cell. (C) Percentage of movie frames with bleb protrusions. n = 17 cells per condition. (D) A2780 cells were seeded into inverted invasion assays in the presence of cRGDfV (2.5 µM) where indicated. SMIFH2 (25 µM) and vehicle control were added 24 h after seeding, and invasion assays were imaged for quantification 48 h later. n = 15 per condition. (E) Formin expression in A2780 cells was measured by qRT-PCR. Gene expression is shown relative to GAPDH expression. n = 3. Error bars represent the SEM. (F and G) siRNA miniscreen of formins in A2780 cells. Migration speed on CDM in the presence of cRGDfV (2.5 µM; F) or under basal conditions (G) is shown. n = 10–30 cells per condition. All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. n.s., not significant; +, mean; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
FHOD3 is specifically required for RCP–α5β1 integrin-driven actin dynamics and migration
Of the 15 mammalian formins, 12 were found to be expressed at relatively high levels in A2780 cells (Fig. 4 E). Because the function of inverted formins (INF1/2) in actin polymerization is less well defined, we knocked down the remaining 10 formins (Fig. S4 A) and measured migration speed of cells in 3D CDM. In cells migrating in an RCP–α5β1-dependent manner, knockdown of the formin-like family (FMNL, in particular FMNL2 and FMNL3) and FHOD3 significantly decreased migration speed (Fig. 4 F). However, FHOD3 knockdown did not affect migration speed under basal conditions (Fig. 4 G), suggesting that this DRF could function specifically downstream of the RCP–α5β1 integrin pathway.
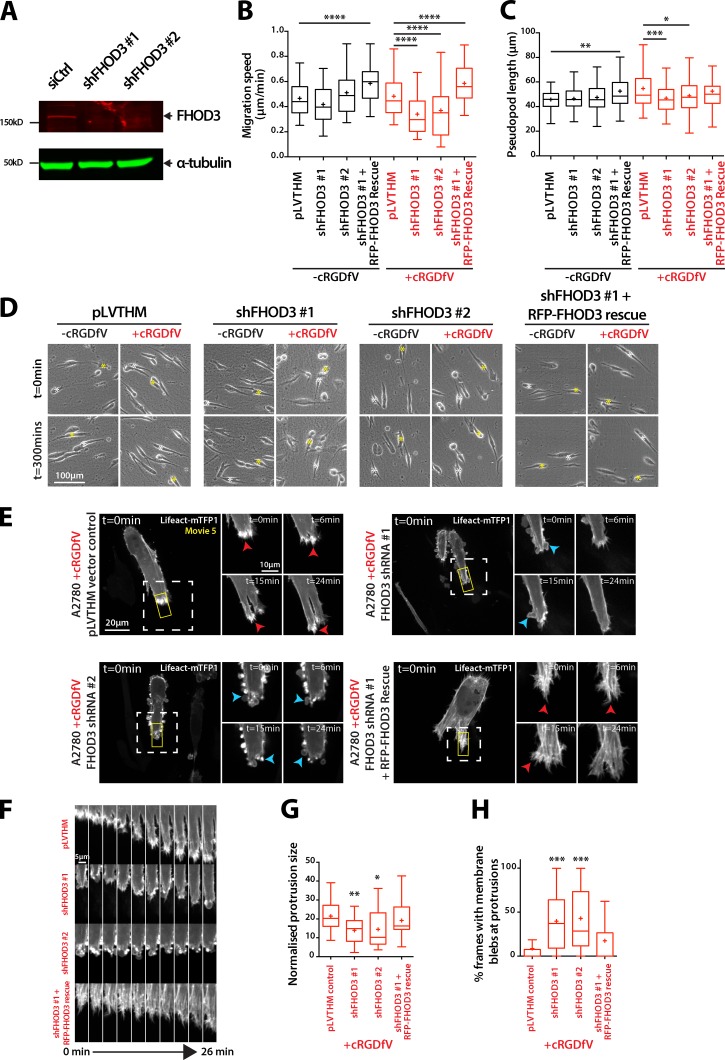
Confirming the siRNA miniscreen, stable knockdown of FHOD3 reduced RCP–α5β1-driven (but not basal) migration and suppressed pseudopod extension in A2780 cells in 3D CDM (Fig. 5, A–D; and Fig. S4 B), but had no discernible influence on cell migration in 2D (Fig. S4, C–E), and the effect of FHOD3 knockdown on 3D migration was rescued by reexpression of shRNA-resistant RFP-FHOD3; in fact, RFP-FHOD3-expressing cells migrated significantly faster (Figs. 5 B and S4 F). The cell lines used in this study expressed only the nonmuscle-specific isoform of FHOD3 (Fig. S4 G), and this isoform was used for rescue experiments.
Figure 5.
FHOD3 drives actin spike formation and migration in 3D matrix. (A) A2780 cells stably transfected with control or FHOD3 targeting shRNA constructs were lysed, then lysates were subjected to SDS-PAGE and Western blotting using antibodies specific for FHOD3 and α-tubulin, and analyzed using Odyssey. (B) A2780 cells stably expressing shFHOD3 #1, shFHOD3 #2, pLVTHM vector control, or RFP-FHOD3 rescue were seeded onto CDM and treated with cRGDfV (2.5 µM) where indicated before time-lapse imaging, and migration speed (B; n = 96–134 cells per condition) and pseudopod length (C; n = 100 cells per condition) were quantified. (D) Representative images of cells from B and C. Asterisks indicate the position of individual cells at different time points. (E) A2780 cells stably transfected as in B were transiently transfected with Lifeact-mTFP1, seeded onto CDM for 4 h, and treated with cRGDfV (2.5 µM) before spinning disk confocal imaging. Maximum z projections are shown. Red arrowheads indicate actin spikes; blue arrowheads indicate blebs. White boxed regions are enlarged on the right. (F) Kymographs of yellow boxes in E. (G and H) Normalized protrusion size of cells (G; n = 23–28 cells per condition) and percentage of movie frames with bleb protrusions (H; n = 30 cells per condition). All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
RFP-FHOD3 showed a punctate distribution throughout the cytoplasm of cells in 3D CDM, sometimes decorating F-actin bundles under basal conditions but with little localization at the leading edge (Fig. S5 A). Upon induction of RCP–α5β1 trafficking, cells generated numerous filopodial protrusions at the leading edge, and RFP-FHOD3 puncta could be seen within these actin spike protrusions, whereas RFP alone was diffuse (Fig. S5 A). Interestingly, a constitutively active mutant of FHOD3, which lacks the DAD domain (GFP-FHOD3ΔDAD), promoted formation of spike-like actin protrusions, and accumulated at these structures (Fig. S5, B and C), suggesting that FHOD3 could directly influence the formation of filopodia and actin spikes in invading cells. Live-cell imaging of Lifeact-mTFP1–expressing FHOD3 knockdown cells revealed nonapoptotic membrane blebs at the cell periphery, and a complete loss of intense, filopodia-based protrusions (Fig. 5, E–H; and Video 5). The knockdown phenotype was rescued by reexpression of shRNA-resistant RFP-FHOD3, which in fact increased actin polymerization to form actin spike protrusions and stress fiber-like structures behind the leading edge (Fig. 5 E and Video 5). Together, these data indicate that FHOD3 is required for actin polymerization at the tips of invasive pseudopods downstream of RCP–α5β1 integrin to form filopodia and actin spike–based protrusions that drive invasive migration in 3D ECM.
ROCK-dependent phosphorylation and activation of FHOD3 is required for RCP–α5β1 integrin-driven migration
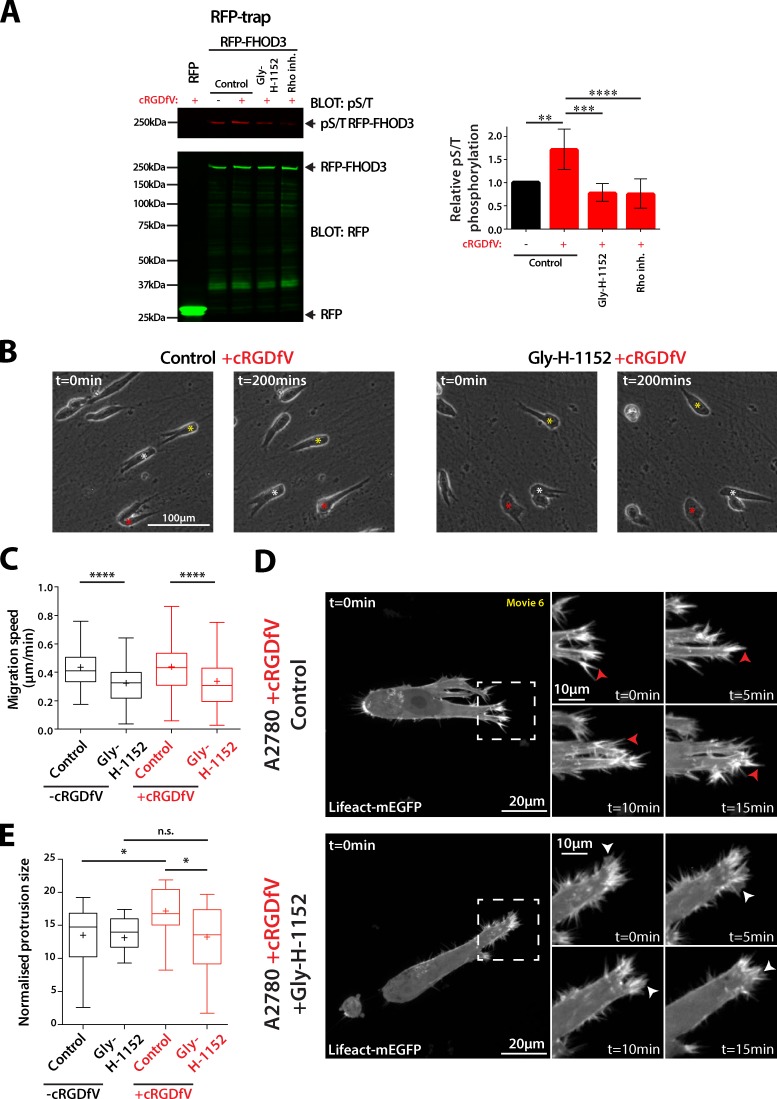
FHOD3 has no known GTPase interactor/activator, but, like FHOD1, can be activated by ROCK-dependent phosphorylation of Ser and Thr residues close to the core DAD motif (Takeya et al., 2008; Iskratsch et al., 2013a). Given that RCP–α5β1 integrin-driven rapid, random migration in 2D is regulated by ROCK (White et al., 2007; Rainero et al., 2012), and that RhoA activity is elevated at the tips of invasive pseudopods in this context (Jacquemet et al., 2013a), we investigated the possibility that ROCK might mediate FHOD3 phosphorylation and activation downstream of RCP–α5β1 trafficking.
RFP-FHOD3 immunoprecipitated from A2780 cells demonstrated a low level of constitutive Ser/Thr phosphorylation, which was significantly increased upon induction of RCP–α5β1 trafficking with cRGDfV (Fig. 6 A). Pretreatment of cells with a Rho inhibitor, or the highly specific ROCK inhibitor Gly-H1152, opposed cRGDfV-induced FHOD3 phosphorylation (Fig. 6 A), suggesting that RhoA-ROCK activity promotes FHOD3 phosphorylation. ROCK inhibition also abrogated migration on 3D CDM (Fig. 6, B and C), and prevented RCP–α5β1 trafficking–induced actin spike formation and protrusive area (Fig. 6, D and E). Interestingly, upon ROCK inhibition, protrusions appeared more wavelike, and membrane blebs were not observed (Fig. 6 D).
Figure 6.
ROCK phosphorylates FHOD3 and promotes formation of actin spike protrusions. (A) A2780 cells stably expressing RFP or RFP-FHOD3/shFHOD3#1 were untreated or treated with cRDGfV (2.5 µM, 30 min) and ROCK inhibitor Gly-H1152 (100 nM, 30 min) or Rho inhibitor I (4 µg/ml, 4 h) as indicated. RFP trap was performed from cell lysates, and subjected to SDS-PAGE and Western blotting using RFP and pS/T antibodies. Fluorescence intensity of pS/T bands was measured using ImageJ, and expressed relative to untreated (n > 4 per condition). Error bars represent the SEM. (B) A2780 cells were seeded onto CDMs and treated with cRGDfV (2.5 µM) and ROCK inhibitor Gly-H1152 (100 nM) where indicated before time-lapse imaging. Asterisks indicate the position of individual cells at different time points. (C) Migration speed was measured for cells treated as in B; n = 150 cells per condition. (D and E) A2780 cells transiently transfected with Lifeact-mEGFP were seeded onto 3D CDM for 4 h, and cRGDfV (2.5 µM, 2 h) and Gly-H1152 were added as indicated. Z stacks were captured on a spinning disk confocal microscope every minute for ≥25 min. Z projections are shown. White arrows indicate lamellipodia-like actin veils; red arrows indicate actin spikes. Boxed regions are enlarged on the right. (E) Normalized protrusion size was measured for cells in D; n = 21–27 cells per condition. All data represent at least three independent experiments; statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. +, mean; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
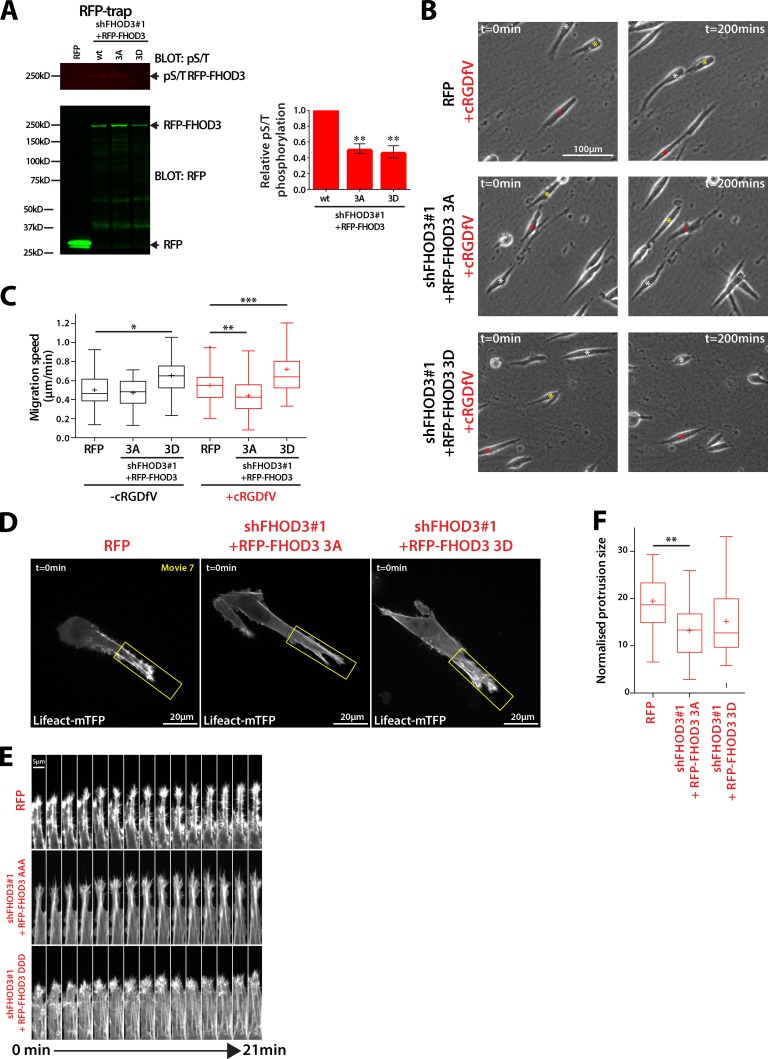
Because FHOD3 is phosphorylated by ROCK on three conserved residues (Iskratsch et al., 2013a), we mutated each to nonphosphorylatable Ala (3A) or phosphomimetic Asp (3D) within full-length shFHOD3#1-resistant RFP-FHOD3 and expressed these in A2780 shFHOD3#1 knockdown cells. Both RFP-FHOD3-3A and -3D showed reduced levels of Ser/Thr phosphorylation compared with wild-type RFP-FHOD3 (Fig. 7 A), and RFP-FHOD3-3A–expressing cells moved more slowly in 3D CDM, whereas expression of RFP-FHOD3-3D increased migration speed (Fig. 7, B and C). Furthermore, RFP-FHOD3-3A–expressing cells showed fewer intense F-actin spike protrusions and decreased protrusion size (Fig. 7 D). Collectively, these data suggest that, downstream of RCP–α5β1 trafficking, localized RhoA activity promotes ROCK-dependent phosphorylation and activation of FHOD3 to promote actin polymerization and filopodia formation at the tips of invasive pseudopods.
Figure 7.
FHOD3 phosphorylation is required for actin spike protrusions and migration in 3D CDM. (A) A2780 cells stably expressing RFP or RFP-FHOD3 wt/3A/3D were treated with cRDGfV (2.5 mM, 30 min) and RFP trap as in Fig. 6 A. Fluorescence intensity of pS/T bands was measured using ImageJ, and expressed relative to untreated (n = 4 per condition). (B) A2780 cells were seeded onto CDM and treated with cRGDfV (2.5 µM) and ROCK inhibitor Gly-H1152 (100nM) where indicated before time-lapse imaging. Asterisks indicate the position of individual cells at different time points. (C) Migration speed was determined for cells treated as in B; n > 60 cells per condition. (D) A2780 cells stably expressing RFP/pLVTHM, RFP-FHOD3 3A/shFHOD3#1, or RFP-FHOD3 3D/shFHOD3#1 were transiently transfected with Lifeact-mTFP1, seeded onto CDM for 4 h, and treated with cRGDfV (2.5 µM) before spinning disk confocal imaging. Maximum z projections are shown. (E) Kymograph analysis of D, taken from the areas in yellow boxes. (F) Normalized protrusion size of cells in D. n = 22–27 cells per condition. All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. +, mean; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FHOD3 is required for RCP–α5β1 integrin-driven invasion in vitro and in vivo
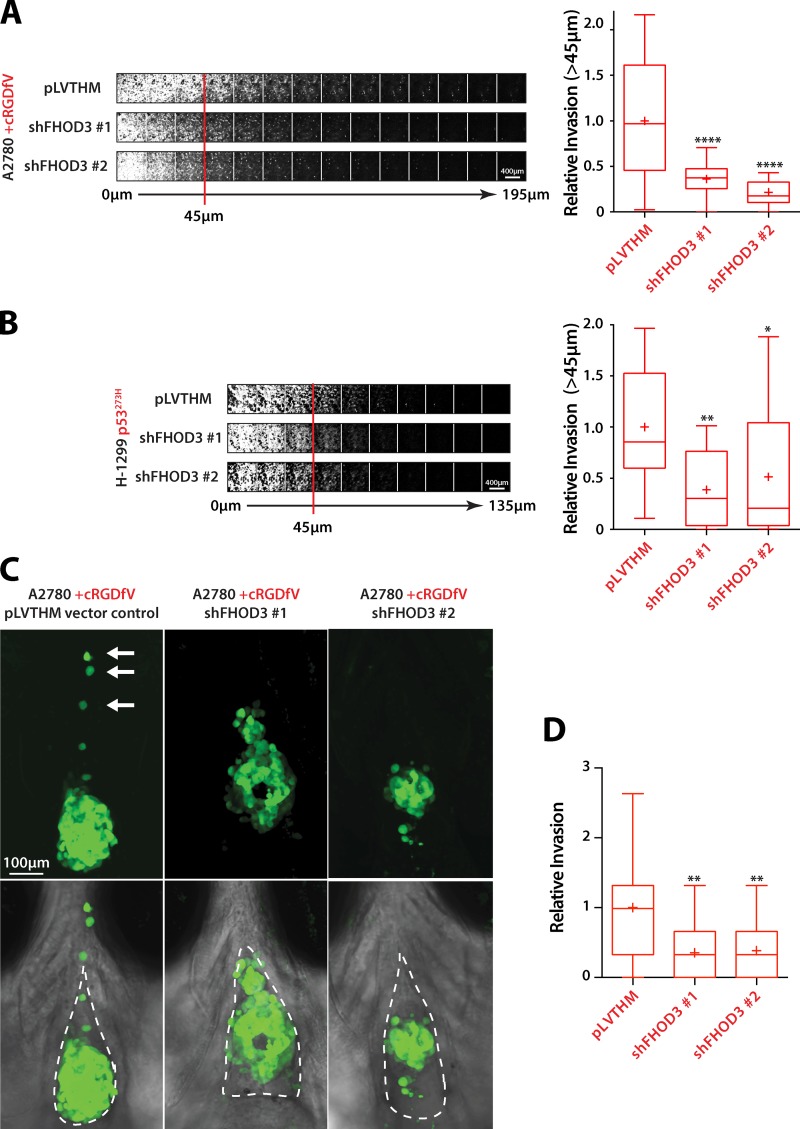
To assess the role of FHOD3 in invasive migration, we introduced FHOD3 knockdown A2780 or H1299-p53273H cells into inverted invasion assays. FHOD3 knockdown abrogated RCP–α5β1–driven invasion into FN-rich collagen hydrogels (Fig. 8, A and B; and Fig. S5 D), confirming its requirement in 3D invasive migration. We also injected stable FHOD3 knockdown cells into zebrafish embryos. Whereas control A2780 cells were able to invade into the surrounding stromal environment when RCP–α5β1 recycling was induced, FHOD3 knockdown cells showed little evidence of dissemination (Fig. 8, C and D). These data suggest that FHOD3 is an actin nucleator that operates downstream of RCP–α5β1 integrin trafficking to promote invasive migration in a physiologically relevant in vivo tissue environment.
Figure 8.
FHOD3 is required for invasive migration in vitro and in vivo. (A) A2780 cells stably expressing pLVTHM vector control, shFHOD3 #1, or shFHOD3 #2 were seeded into inverted invasion assays and allowed to invade into Collagen-I/FN matrix for 72 h. Cells were visualized with calcein-AM and serial confocal sections were captured at 15-µm intervals. (B) Invasion of H1299-p53-273H cells stably expressing pLVTHM vector control, shFHOD3 #1, or shFHOD3 #2 was determined as in A. (C) A2780 cells stably expressing pLVTHM vector control, shFHOD3 #1, or shFHOD3 #2 were injected into the pericardial cavity of zebrafish embryos and treated with cRGDfV (2.5 µM). White arrows indicate invading cells. The broken lines indicate the pericardial cavity. (D) Relative invasion quantification of C. n = 24–27 fish per condition. All data represent at least three independent experiments. Statistical significance was evaluated using ANOVA/Tukey’s multiple comparison test. +, mean; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Discussion
We have shown that lamellipodia formation and retrograde actin flow, which form a major mechanism that drives forward movement of cells in 2D, is observed in cells migrating in 3D matrix. However, promoting RCP–α5β1 trafficking, which increases invasive migration into FN-rich ECM and local invasion in vivo, leads to generation of filopodia to form actin spike–based protrusions that extend in the direction of invasion, rather than lamellipodia. Filopodial actin spike protrusions formed in this context require the formin FHOD3, and not Arp2/3-mediated actin polymerization. This novel form of actin-based protrusion provides the driving force for invasion of cancer cells in 3D ECM in vitro and in vivo.
In 3D collagen, there is some evidence that lamellipodia can facilitate migration (Giri et al., 2013), and αvβ3 integrin controls lamellipodial migration in 2D (White et al., 2007; Caswell et al., 2008) and promotes invasive migration in low FN microenvironments (Christoforides et al., 2012). Our demonstration of retrograde actin flow in unstimulated cancer cells moving in 3D, within protrusions made up of lamellipodial F-actin veils, provides further evidence to support the notion that the mechanisms that support lamellipodial migration in 2D can facilitate invasive migration in 3D (Fig. 1, D–F). However, the RCP–α5β1 pathway drives invasion into FN-rich ECM in a manner that does not rely on lamellipodial protrusion, suggesting that alternative mechanisms can drive rapid migration in 3D ECM to contribute to cancer cell invasion and metastasis depending on the composition of the microenvironment. Loss of the Rac activator Tiam1, or the downstream WAVE complex component CYFIP1, can contribute to cell migration, invasion, and metastasis (Malliri et al., 2002; Silva et al., 2009; Vaughan et al., 2015), suggesting that suppression of major regulators of lamellipodia formation can lead to increased metastatic potential.
Nonlamellipodial migratory mechanisms control migration in 3D ECM microenvironments (Friedl and Alexander, 2011; Petrie et al., 2012, 2014); however the RCP–α5β1 pathway promotes migration of cancer cells driven by filopodial actin spike–based protrusions that are morphologically distinct from protrusions seen in amoeboid or lobopodial 3D migration (Fig. 1 and Fig. 2). RCP–α5β1-driven filopodial actin-based protrusions extend beyond the cortex, appear rapidly in collective bursts (Fig. 1, E–H; and Fig. S2), show dynamic protrusion and retraction (Fig. 1, E–H; Fig. 2; and Fig. S2), and have a rate of retrograde actin flow that is lower than in lamellipodial-based protrusions (Fig. 1, A–C; and Fig. S1). Filopodia have been shown to exhibit retrograde F-actin flow (Okabe and Hirokawa, 1991; Lin et al., 1996; Anderson et al., 2008), and, interestingly, retrograde flow may be reduced in actively protruding filopodia (Mallavarapu and Mitchison, 1999), a notion supported by our data.
Filopodia can arise via at least two broad mechanistic routes: convergent elongation of F-actin derived from the lamellipodial network and tip nucleation through formins (Mattila and Lappalainen, 2008; Mellor, 2010). ArpC3 knockout MEFs retain some ability to migrate in 2D, extending filopodial-like protrusions (FLPs; Suraneni et al., 2012, 2015), confirming that formins can nucleate actin filaments to generate filopodia in the absence of a dendritic lamellipodial actin network, and here we have shown that filopodia in invasive cells can form in an Arp2/3-independent, FHOD3-driven manner. Long-lived FLPs, formed as a consequence of Rif/mDia2 and integrin signaling, facilitate metastatic colonization of breast cancer cells (Shibue et al., 2012, 2013). The dynamics of these structures differs from FHOD3-induced filopodia, suggesting that while in general integrins may be linked with formin function and filopodial extension, the properties of these filopodial subtypes may be very different. The DRF Rho GTPase effectors have previously been linked to cell migration in 2D and 3D (Kitzing et al., 2010; Vega et al., 2011; Block et al., 2012; Breitsprecher and Goode, 2013). Indeed, we and others find that members of the formin-like (FMNL) family contribute to basal migration in 3D CDM (Fig. 5 I; Kitzing et al., 2010; Vega et al., 2011). However, our data suggest that the DRF FHOD3 is specifically required for RCP–α5β1-driven filopodial actin spike formation, migration, and local invasion in vivo.
FHOD3 shows relatively restricted expression (heart, kidney, and brain; Katoh and Katoh, 2004; Kanaya et al., 2005; Iskratsch et al., 2010), and contributes to sarcomeric organization in cardiomyocytes by promoting actin polymerization and contributing to myofibril formation (Taniguchi et al., 2009; Iskratsch et al., 2010). FHOD3 can be activated by ROCK-mediated phosphorylation of Ser/Thr residues within the DAD domain (Iskratsch et al., 2013a), and we now show that FHOD3 is the target of localized RhoA-ROCK signaling downstream of RCP–α5β1 trafficking in invasive cancer cells. However, while knockdown of FHOD3 has a profound influence on the morphology of cells in 3D CDM, promoting membrane blebbing (Fig. 5 E), inhibition of ROCK has a more subtle influence (Fig. 6 D). ROCK is known to oppose Rac activity through FilGAP (Ohta et al., 2006), and it is therefore possible that ROCK inhibition leads to an increase in Rac activity and formation of lamellipodial protrusions, as observed (Fig. 6 D). Similarly, expression of nonphosphorylatable FHOD3 induces a more subtle phenotype than knockdown of FHOD3 (Fig. 7 D). While no Rho GTPase interactor has been described for FHOD3, its closest relative FHOD1 can be activated by Rac binding and/or ROCK phosphorylation, and integrin signaling controls its targeting (Gasteier et al., 2003; Hannemann et al., 2008; Takeya et al., 2008; Iskratsch et al., 2013b). Together with our data, this could suggest that numerous signals converge to control the precise local activation of DRFs.
The recycling of α5β1 via the RCP pathway is critical for the establishment of polarized signaling to activate RhoA at the front of migrating and invading cells (Jacquemet et al., 2013a). WASH is a nucleation-promoting factor (NPF) for Arp2/3, which is required for RCP-mediated α5β1 recycling (Zech et al., 2011), and it is therefore surprising that RCP–α5β1-driven recycling, migration, and invasion is unperturbed when Arp2/3 activity is impaired (Figs. 3 and S3). However, WASH has been reported to interact with the F-actin nucleator Spire (Liu et al., 2009) and V-ATPase (Carnell et al., 2011), which acidifies vesicular compartments, and it is possible that WASH could contribute to α5β1 trafficking in an Arp2/3-independent fashion. The fact that interfering with Arp2/3 function promotes pseudopod extension (Fig. 3 D, G), promotes accumulation of RCP vesicles at the tips of pseudopods (Fig. S3, L and M), and is dispensable for α5β1 recycling (Fig. S3 K) suggests that Arp2/3-mediated actin polymerization might restrain vesicular cargo, and remodeling of actin (or its disassembly) might be required to facilitate trafficking that leads to downstream signaling to promote migration and invasion.
We have shown that the RCP–α5β1 pathway drives local invasion in vitro and in vivo by controlling the activation of FHOD3 to induce filopodial actin spike protrusions in the direction of invasive migration (Fig. 3 and Fig. 7) similar to those seen in FN-rich ECM in vitro (Fig. 2; Jacquemet et al., 2013a). Breast cancer cell lines expressing GFP-Lifeact implanted into the mammary fat pad of mice in a collagen-rich xenograft environment have been shown to have the ability to generate actin-based protrusions using intravital imaging, but in this context blebbing amoeboid-type migration predominates (Tozluoğlu et al., 2013). In the zebrafish xenograft model, carcinoma cells may migrate in a similar manner to invasive melanoma cells, utilizing collagen I and FN ECM tracks that facilitate invasion (Chapman et al., 2014). This suggests that, as we find in vitro, the composition (presence of FN and soluble αvβ3 ligands e.g., osteopontin) and topology of the ECM combine with cell-intrinsic factors (e.g., expression of gain-of-function mutant p53) to produce a migratory response for invasion of stromal environments in vivo.
Materials and methods
Cell culture and transient transfection
A2780 cells were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FCS. H1299, MDA-MB-231, telomerase-immortalized fibroblast (TIF), MEF, and 293T cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FCS. All cells were maintained at 37°C and 5% CO2. Transient transfections and siRNA knockdowns were performed using nucleofector (Solution T; 3 µg plasmid DNA or 1 µM siRNA; program A-23; Amaxa; Lonza), according to the manufacturer’s instructions. Formin knockdown was performed with one round of transfection, whereas ArpC2 and ArpC3 knockdown was performed with two rounds of transfection.
CDMs were prepared as described previously (Cukierman et al., 2001; Caswell et al., 2008). In brief, plastic tissue-culture plates or glass bottom dishes (for high-resolution imaging) were gelatin coated, cross-linked with glutaraldehyde, quenched with glycine, and equilibrated in DMEM containing 10% FCS. Human TIFs were seeded at near confluence and grown for 8–10 d in DMEM containing 10% FCS and 50 µg/ml ascorbic acid. Matrices were denuded of live cells by incubation with PBS containing 20 mM NH4OH and 0.5% Triton X-100, and DNA residue was removed by incubation with DNase I (Lonza).
Plasmids and reagents
Rabbit anti-ArpC2 and ArpC3 antibodies were purchased from Proteintech. Mouse anti–α-tubulin (DM1A) was purchased from Sigma-Aldrich. Rat anti-RFP antibody and RFP-trap beads were from ChromoTek. Rabbit anti-phospho S/T antibody was purchased from ECM Biosciences. Affinity purified rabbit anti-FHOD3 (antigen His-FHOD31-339 in pEt3b) has been described previously (Iskratsch et al., 2010). DAPI and Alexa Fluor 488/555/647 phalloidin were purchased from Life Technologies. Arp2/3 inhibitor CK-666, CK-689 (inactive analogue of CK-666), and ROCK inhibitor Glycyl-H-1152 were purchased from EMD Millipore. SMIFH2 was purchased from Sigma-Aldrich. cRGDfV was purchased from Bachem.
FHOD3 (lacking muscle-specific exon 26) and delta-DAD FHOD3 constructs in pEGFP-C style vectors were as described previously (Iskratsch et al., 2010, 2013a). RFP-FHOD3 was generated by digesting HA-FHOD3-C1 with NheI and PvuI to remove the HA tag, and ligating an NheI–PvuI fragment in its place. paGFP-actin was a gift from K. Anderson (Beatson Institute for Cancer Research, Glasgow, Scotland, UK). Lifeact-mEGFP and Lifeact-mRFPmars were gifts from R. Wedlich-Soeldner (University of Münster, Münster, Germany; Riedl et al., 2008), and mTFP1-LifeAct-7 was from AddGene (plasmid #54749; all in EGFP-N1-style vectors).
Knockdown of formins and ArpC2/ArpC3 was performed using siRNA pools from an siARRAY (#M-012081-00 and M-005284-00, respectively; GE Healthcare). Stable FHOD3 shRNA was generated by cloning the following sequences into pLVTHM (Addgene) using Mlu1 and Cla1 restriction sites: FHOD3 shRNA #1, 5′-CGCGTCCCCATAGACCAGTTGGAGAACAATATTCAAGAGATATTGTTCTCCAACTGGTCTATTTTTTGGAAAT-3′ (adapted from Iskratsch et al., 2010); and FHOD3 shRNA #2, 5′-CGCGTCCCCATACGGAGCTACTGGTTTATTCAAGAGATAAACCAGTAGCTCCGTATTTTTTGGAAAT-3′. shRNA-resistant FHOD3 was generated using the Q5 site-directed mutagenesis kit (New England Biolabs, Inc.) using the following primers: forward, 5′-AGAAAATAACAAAACCTTGGGCTT-TATCC-3′; and reverse, 5′-AATTGATCGATTCCTTCCTTCAGGTCCAG-3′. 3A and 3D mutant RFP-FHOD3 were generated using the following primers: 3A forward, 5′-AAGCTTTGCGAAGAGCCCTGAAGAGCGGCCTGAC-3′; reverse, 5′-TCCGGTTGGCCCGGGCTCGTTTCCTCTCCCTCGG-3′; and 3D forward, 5′-AAGATTTGCGAAGAGACCTGAAGAGCGGCCTGAC-3′; and reverse, 5′-TCCGGTTGGCCCGGTCTCGTTTCCTCTCCCTCGG-3′. Lifeact-mRFPmars was subcloned into pCDH (gift from A. Gilmore, University of Manchester, Manchester, England, UK) using restriction sites EcoRI and NotI, and shRNA-resistant RFP-FHOD3 was cloned into pCDH using Nhe1 and Not1.
Lentiviral production and transduction
Lentiviruses were produced by transfecting 293T cells with psPAX2 and pMD2.G (Addgene) and pLVTHM or pCDH viral vectors. Conditioned medium containing viruses was collected after 5 d and then used immediately to infect cells or stored at −80°C. Infected cells were sorted for EGFP or mRFPMars expression using FACS ARIA Fusion (BD) with excitation at 488 nm and 561 nm, respectively. FACS data were processed and analyzed using DIVA8 software (BD).
SDS-PAGE and quantitative Western blotting
Cells were lysed in lysis buffer (200 mM NaCl, 75 mM Tris-HCl, pH 7.4, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% [vol/vol] Triton X-100, 0.75% [vol/vol] NP-40, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride). Lysates were clarified by centrifugation at 10,000 g for 10 min at 4°C.
Cell lysates were resolved under denaturing conditions by SDS-PAGE (4–12% Bis-Tris gels; Invitrogen) and transferred to nitrocellulose membrane. Membranes were blocked and incubated overnight at 4°C with the appropriate primary antibody and then at room temperature for 1 h with the appropriate fluorophore-conjugated secondary antibody. Membranes were scanned using an infrared imaging system (Odyssey; LI-COR Biosciences). Band intensity was determined by digital densitometric analysis using Image Studio (version 4.0). Western blots shown are representative of at least three independent experiments.
RNA extraction and PCRs
RNA was extracted from cells using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions.
For real-time qPCR, reactions were performed using the Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems) with 1 µg RNA template and run using a StepOnePlus Real-Time PCR System and StepOnePlus software (Applied Biosystems). Fold change in gene expression was calculated using the comparative CT method (Schmittgen and Livak, 2008). Primers used in the reactions are listed in Supplementary Table 2 in Kitzing et al. (2010). GAPDH expression was used as an endogenous control.
For reverse-transcription PCR, reactions were performed using the OneStep RT-PCR kit (QIAGEN) with 1 µg RNA template and either pan-FHOD3 forward primer, 5′-CTCCTTTTTACTCTTTATGGGCCATCC-3′, or muscle-specific FHOD3 forward primer, 5′-ACTGATGAGGAGGAGGAAGTTGAG-3′, with pan-FHOD3 reverse primer, 5′-CTCGCTGACTGGGCACTTGGGTGG-3′.
Microscopy
All images were taken with a constant exposure time between all the conditions of the same experiment. Images were processed using ImageJ v1.49b (National Institutes of Health) and only subjected to signal rescaling using linear transformation for display in the figures.
For 2D scratch wound assays, cells were seeded onto tissue culture plates to confluency, and after 24 h, cell monolayers were wounded with a plastic pipette tip. For 3D migration assays, cells were plated onto CDM for >4 h. For long-term time lapse, cells were imaged in the presence or absence of cRGDfV and maintained at 37°C and 5% CO2. Phase-contrast images were captured on an inverted microscope system (AS-MDW; Leica) using a 20× objective lens (HC Plan Fluotar Ph2, NA 0.50) every 10 min using a charge-coupled device (CCD) camera (CoolSNAP HQ; Photometrics) and the Micro-Manager plugin for ImageJ (version 1.4.17; Edelstein et al., 2010, 2014). Point visiting mode was used to allow multiple positions to be imaged within the same time course. Images and videos were analyzed using ImageJ with the MTrackJ plugin (Meijering et al., 2012). Analysis of speed, endpoint directional persistence, direction autocorrelation, and mean square displacement (MSD) was performed using DiPer (Gorelik and Gautreau, 2014). Endpoint directional persistence and autocorrelation (a measure of directionality less influenced by speed) were used in 2D to accurately reflect a cell’s ability to migrate in a persistent manner, but this is less appropriate in 3D CDM where cells generally follow the fibrillar nature of the ECM. We therefore used MSD (a measure of the area explored) to better define migration capacity in 3D CDM. To obtain a measure of pseudopod length, the distance between the center of the nucleus and the cell front (with respect to the direction of migration) was measured using ImageJ.
Wounded cells fixed with 4% paraformaldehyde were stained with FITC-phalloidin and DAPI and imaged with a restoration microscope (Delta Vision RT; Applied Precision) using a 40×/NA 1.30 Uplan FLN objective lens and a Sedat filter set (86000v2; Chroma Technology Corp.). Images were collected using a CoolSNAP HQ camera (Photometrics).
For photoactivation, cells transiently cotransfected with Lifeact-mRFPmars and paGFP-Actin were seeded onto glass-bottom dishes or CDMs, and media was replaced with Ham’s F12 (Gibco) containing 25 mM Hepes and 10% FCS before addition of cRGDfV (>2 h) where appropriate for image acquisition at 37°C. Using a spinning disk confocal inverted microscope (Marianas; 3i), paGFP-Actin was photoactivated at cell protrusions within a square ROI (5 µm2) for 10 ms using a 405 nm laser. paGFP and mRFPMars fluorescence emission was captured every second for 100 s using a 63× objective lens (NA 1.4 Plan Apochromat) and SlideBook 6.0 software. Images analysis was performed in ImageJ: ROIs were positioned over the point of photoactivation, immediately behind the photoactivated region (relative to the direction of protrusion) or outside the area of the cell (used for background subtraction). Fluorescence intensity was normalized to the intensity of the photoactivated region in the first frame after photoactivation.
For live-cell imaging of actin dynamics during invasive migration, A2780, H1299, and MDA-MB-231 cells transiently transfected with Lifeact-mEGFP or Lifeact-mTFP1 were plated on CDMs for 4 h and normal culture medium was replaced with Ham’s F12 containing 25 mM Hepes and 10% FCS before addition of cRGDfV (>2 h) where appropriate before image acquisition. Image stacks (seven positions every 0.2 µm) were collected every 1 min (Lifeact-mEGFP) or 3 min (Lifeact-mTFP1) at 37°C on a spinning disk confocal inverted microscope (Marianas) using a 63× objective lens (NA 1.4 Plan Apochromat) and SlideBook 6.0 software.
RFP- or RFP-FHOD3–expressing cells were seeded onto CDMs 4 h before treatment with cRGDfV for 2 h as appropriate. Cells were fixed with 4% paraformaldehyde and stained with rat anti-RFP primary antibody (ChromoTek), Rhodamine Red-X–conjugated secondary antibody (Jackson ImmunoResearch), and Alexa Fluor 647–phalloidin (Life Technologies). Cells were imaged with a Delta Vision RT (Applied Precision) restoration microscope using a 60×/NA 1.42 Plan-Apochromat objective lens and the Sedat filter set (86000v2; Chroma Technology Corp.). Images were collected using a CoolSNAP HQ (Photometrics) camera with a Z optical spacing of 0.2 µm. Raw images were then deconvolved (aggressive mode, 10 pass) using SoftWoRx software (Applied Precision) and maximum intensity projections of these deconvolved images are shown.
SIM was performed on Alexa Fluor 488 phalloidin–stained A2780 cells on CDM. Images were acquired on a ELYRA S.1 system (Carl Zeiss) with a pco.edge 5.5 scientific CMOS camera using a 63×/1.4 NA oil immersion objective lens. For optimal lateral spatial sampling, a 1.6× intermediate magnification lens was used, resulting in a pixel size of 64 nm. A z interval of 110 nm was used to ensure adequate spatial sampling in the axial dimension. Excitation was achieved with a 488-nm laser and emission was filtered with a 495–550-nm band-pass filter. Modulation of the illumination light was achieved using a physical grating with 28 µm spacing. For each z plane, this patterned illumination was shifted through five phases at each of five rotational angles. Raw data were processed using ZEN software and analyzed using ImageJ and the Matlab application CellGeo (Tsygankov et al., 2014) to determine filopodia number and length, respectively.
Quantification of actin protrusions in 3D CDM was performed using maximum z projections of cells imaged for 30–60 min. Using custom software written in Python and NumPy, images in the time-lapse sequence were individually band-pass filtered (à trous wavelet, linear 3 × 3 filter, keeping scales 2–8) to remove both high-frequency noise and stationary background. Images were thresholded using a fixed threshold (pixel gray value >100) and all the objects in the resulting binary image were identified using an 8-connected component labeling. All but the largest object in the image were discarded. The difference between binary images with a lag of 3 min between frames was used to identify protrusive areas. To limit the analysis to the front of the cell only, the following automated method was devised: the axis aligned minimum bounding box was calculated for each cell at each time point, and divided into three equal parts along the longer axis. The area of protrusions was normalized to the total area of the cell and protrusive areas within the front region were used to generate shape description measurements. Mean values were generated from >25 frames per cell.
Zebrafish xenograft assay
Zebrafish (Danio rerio) were maintained at the University of Manchester Biological Services Unit according to National Home Office regulations under the Animals (Scientific Procedures) Act 1986. Casper strain (roy−/−, nacre−/−) zebrafish were used throughout the study to generate embryos completely lacking pigment, which can otherwise obscure imaging. H1299 cells, A2780 cells, or A2780 cells stably expressing Lifeact-mRFPmars, pLVTHM vector control, or shFHOD3 were stained with carboxyfluorescein succinimidyl ester (CFSE; Life Technologies) 24 h before injection. Suspended cells were injected into the pericardial cavity of 48 h postfertilization zebrafish embryos. cRGDfV was added, where appropriate, 1 d postinjection (dpi). Before imaging, zebrafish embryos were mounted in 1.5% low-melting agarose (LMP; Flowgen Biosciences). Lifeact-mRFPmars–expressing cells were imaged 3 dpi using a spinning disk confocal inverted microscope (Marianas, 3i; with Slidebook 6 software) using a long working distance 63× objective lens (NA 1.15, C Apochromat) at 34°C. Z stacks were rendered using Imaris (version 8.0). Wild-type, vector control, and shFHOD3 4 dpi engrafted tumors were imaged at room temperature using an upright confocal microscope (Leica TCS SP5 AOBS; Leica), with a 20× 0.50 NA Plan Fluotar dipping objective lens and 1.5× confocal zoom and LAS software (Leica). Relative invasion was defined as the mean number of cells located outside the pericardial cavity at 4 dpi normalized to the average number of cells in the control condition.
Inverted invasion assays
Inverted invasion assays were modified from those described previously (Hennigan et al., 1994). In brief, collagen I (final concentration ∼5 mg/ml; BD) supplemented with 25 µg/ml FN was allowed to polymerize in inserts (Transwell; Corning) for 1 h at 37°C. Inserts were then inverted, and cells were seeded directly onto the opposite face of the filter. Transwell inserts were finally placed in 0.1% serum medium, and medium supplemented with 10% FCS and 30 ng/ml EGF was placed on top of the matrix, providing a chemotactic gradient. Where appropriate, cRGDfV was added to the matrix before plug polymerization and also to the medium throughout the system. 48–72 h after seeding, migrating cells were stained with Calcein-AM and visualized by confocal microscopy with serial optical sections being captured at 15-µm intervals using an inverted confocal microscope (TCS SP5 AOBS; Leica) using a 20× objective lens. Invasion was quantified using the area calculator plugin in ImageJ, measuring the fluorescence intensity of cells invading 45 µm or more and expressing this as a percentage of the fluorescence intensity of all cells within the plug.
Integrin recycling assay
Recycling assays were performed as described previously (Caswell et al., 2008). In brief, cells were labeled at 4°C with 0.13 mg/ml NHS-SS-Biotin (Thermo Fisher Scientific), and biotinylated receptors were internalized for 30 min at 37°C in serum-free medium before removal of remaining surface biotin by reduction with sodium 2-mercaptoethane sulfonate. Internalized receptors were chased from the cells by returning the cells to 37°C, surface biotin was removed by a second reduction step, and the fraction of biotinylated integrins was analyzed by ELISA. The antibodies used for capture ELISA detection of α5β1 integrin were obtained from BD. cRGDfV, CK-689, and CK-666 were added to the cells only during the recycling period.
Statistical analysis
Student’s t test (unpaired, two tailed, and unequal variance) or analysis of variance (ANOVA)/Tukey’s multiple comparison test were used to calculate statistical significance as appropriate, using GraphPad Prism version 6 (GraphPad Software). t1/2 (Figs. 1 and S1) was approximated using nonlinear regression (exponential one-phase decay; GraphPad Prism). Bar charts (±SEM) or Tukey box and whisker plots are displayed (whiskers represent 1.5× interquartile range, + indicates mean).
Online supplemental material
Fig. S1 shows retrograde flow in A2780 and H1299 cells in 2D and MDA-MB-231 cells in 3D CDM. Fig. S2 shows the dynamics of Lifeact-GFP in H1299 and MDA-MB-231 cells, and filopodial lifetime of A2780 cells in 3D CDM. Fig. S3 shows the effect of Arp2/3 inhibition/knockdown on migration in 2D and 3D, and the effect of Arp2/3 inhibition on integrin recycling and vesicle localization. Fig. S4 shows formin knockdown by qPCR and the effect of FHOD3 knockdown on 2D migration, and confirms that cancer cells do not express FHOD3 with the muscle-specific exon 26. Fig. S5 shows the localization of RFP-FHOD3wt and an active FHOD3, and knockdown of FHOD3 in H1299-273H cells. Video 1 shows retrograde flow of actin in A2780 cells on CDM. Video 2 shows the dynamics of Lifeact-GFP in A2780 cells on CDM. Video 3 demonstrates that A2780 cells form filopodial actin spikes as they invade in vivo. Video 4 shows the effect of Arp2/3 inhibition on Lifeact-GFP dynamics in A2780 cells on CDM. Videos 5, 6, and 7 show the effect of FHOD3 knockdown and rescue and ROCK inhibition on Lifeact-GFP dynamics in A2780 cells on CDM. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502040/DC1.
Supplementary Material
Acknowledgements
We thank the Bioimaging Facility for their help with microscopy; microscopes used in this study were purchased with grants from BBSRC, the Wellcome Trust, and the University of Manchester Strategic Fund. We thank M. Jackson and G. Howell for assistance with FACS, and David Bradley for technical support. We thank Robert Grosse for providing advice and reagents for formin knockdown, and Roman Gorelik for assistance with DiPer.
The work of our laboratories is supported by the Wellcome Trust (090453/Z/09/Z and 090453/Z/09/A to P.T. Caswell), Worldwide Cancer Research (14-1226 to P.T. Caswell), the European Research Council (ERC-2011-StG-282059 to A. Hurlstone), and The Dowager Countess Eleanor Peel Trust (TH-PRCL.FID2228 to A. Hurlstone). The Wellcome Trust Centre for Cell-Matrix Research is supported by grant 088785/Z/09/Z.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CDM
- cell-derived matrix
- CFSE
- carboxyfluorescein succinimidyl ester
- DAD
- diaphanous autoregulatory domain
- dpi
- day postinjection
- DRF
- Diaphanous-related formin
- FN
- fibronectin
- MSD
- mean square displacement
- paGFP
- photoactivatable GFP
- RCP
- Rab-coupling protein
- ROI
- region of interest
- SIM
- structured illumination microscopy
References
- Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M.B., Guzzardo V., et al. . 2009. A Mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell. 137:87–98. 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- Anderson T.W., Vaughan A.N., and Cramer L.P.. 2008. Retrograde flow and myosin II activity within the leading cell edge deliver F-actin to the lamella to seed the formation of graded polarity actomyosin II filament bundles in migrating fibroblasts. Mol. Biol. Cell. 19:5006–5018. 10.1091/mbc.E08-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjonen A., Kaukonen R., Mattila E., Rouhi P., Högnäs G., Sihto H., Miller B.W., Morton J.P., Bucher E., Taimen P., et al. . 2014. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J. Clin. Invest. 124:1069–1082. 10.1172/JCI67280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J., Breitsprecher D., Kühn S., Winterhoff M., Kage F., Geffers R., Duwe P., Rohn J.L., Baum B., Brakebusch C., et al. . 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22:1005–1012. 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J.J., Hodgson L., and Condeelis J.. 2012. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 24:277–283. 10.1016/j.ceb.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., and Goode B.L.. 2013. Formins at a glance. J. Cell Sci. 126:1–7. 10.1242/jcs.107250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater R.E., Norman J.C., and Caswell P.T.. 2012. Integrin trafficking at a glance. J. Cell Sci. 125:3695–3701. 10.1242/jcs.095810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S.D., Ura S., Hagedorn M., Johnston S.A., May R.C., Soldati T., Machesky L.M., and Insall R.H.. 2011. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 193:831–839. 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., and Norman J.C.. 2006. Integrin trafficking and the control of cell migration. Traffic. 7:14–21. 10.1111/j.1600-0854.2005.00362.x [DOI] [PubMed] [Google Scholar]
- Caswell P., and Norman J.. 2008. Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18:257–263. 10.1016/j.tcb.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Spence H.J., Parsons M., White D.P., Clark K., Cheng K.W., Mills G.B., Humphries M.J., Messent A.J., Anderson K.I., et al. . 2007. Rab25 associates with α5β1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13:496–510. 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Chan M., Lindsay A.J., McCaffrey M.W., Boettiger D., and Norman J.C.. 2008. Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Vadrevu S., and Norman J.C.. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10:843–853. 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Caulin C., Nguyen T., Lang G.A., Goepfert T.M., Brinkley B.R., Cai W.W., Lozano G., and Roop D.R.. 2007. An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J. Clin. Invest. 117:1893–1901. 10.1172/JCI31721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A., Fernandez del Ama L., Ferguson J., Kamarashev J., Wellbrock C., and Hurlstone A.. 2014. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Reports. 8:688–695. 10.1016/j.celrep.2014.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforides C., Rainero E., Brown K.K., Norman J.C., and Toker A.. 2012. PKD controls αvβ3 integrin recycling and tumor cell invasive migration through its substrate Rabaptin-5. Dev. Cell. 23:560–572. 10.1016/j.devcel.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., and Yamada K.M.. 2001. Taking cell-matrix adhesions to the third dimension. Science. 294:1708–1712. 10.1126/science.1064829 [DOI] [PubMed] [Google Scholar]
- Desgrosellier J.S., and Cheresh D.A.. 2010. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 10:9–22. 10.1038/nrc2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B., Morton J.P., Delaney D.W., Ridgway R.A., Wilkins J.A., and Sansom O.J.. 2010. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J. Pathol. 222:129–137. 10.1002/path.2748 [DOI] [PubMed] [Google Scholar]
- Dozynkiewicz M.A., Jamieson N.B., Macpherson I., Grindlay J., van den Berghe P.V.E., von Thun A., Morton J.P., Gourley C., Timpson P., Nixon C., et al. . 2012. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell. 22:131–145. 10.1016/j.devcel.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., and Stuurman N.. 2014. Advanced methods of microscope control using μManager software. J Biol Methods. 1:e10 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Alexander S.. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 147:992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Friedl P., Locker J., Sahai E., and Segall J.E.. 2012. Classifying collective cancer cell invasion. Nat. Cell Biol. 14:777–783. 10.1038/ncb2548 [DOI] [PubMed] [Google Scholar]
- Gardel M.L., Sabass B., Ji L., Danuser G., Schwarz U.S., and Waterman C.M.. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183:999–1005. 10.1083/jcb.200810060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteier J.E., Madrid R., Krautkrämer E., Schröder S., Muranyi W., Benichou S., and Fackler O.T.. 2003. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J. Biol. Chem. 278:38902–38912. 10.1074/jbc.M306229200 [DOI] [PubMed] [Google Scholar]
- Giri A., Bajpai S., Trenton N., Jayatilaka H., Longmore G.D., and Wirtz D.. 2013. The Arp2/3 complex mediates multigeneration dendritic protrusions for efficient 3-dimensional cancer cell migration. FASEB J. 27:4089–4099. 10.1096/fj.12-224352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik R., and Gautreau A.. 2014. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9:1931–1943. 10.1038/nprot.2014.131 [DOI] [PubMed] [Google Scholar]
- Hannemann S., Madrid R., Stastna J., Kitzing T., Gasteier J., Schönichen A., Bouchet J., Jimenez A., Geyer M., Grosse R., et al. . 2008. The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. J. Biol. Chem. 283:27891–27903. 10.1074/jbc.M801800200 [DOI] [PubMed] [Google Scholar]
- Hennigan R.F., Hawker K.L., and Ozanne B.W.. 1994. Fos-transformation activates genes associated with invasion. Oncogene. 9:3591–3600. [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Iskratsch T., Lange S., Dwyer J., Kho A.L., dos Remedios C., and Ehler E.. 2010. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 191:1159–1172. 10.1083/jcb.201005060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Reijntjes S., Dwyer J., Toselli P., Dégano I.R., Dominguez I., and Ehler E.. 2013a Two distinct phosphorylation events govern the function of muscle FHOD3. Cell. Mol. Life Sci. 70:893–908. 10.1007/s00018-012-1154-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskratsch T., Yu C.-H., Mathur A., Liu S., Stévenin V., Dwyer J., Hone J., Ehler E., and Sheetz M.. 2013b FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell. 27:545–559. 10.1016/j.devcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivaska J., and Heino J.. 2011. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu. Rev. Cell Dev. Biol. 27:291–320. 10.1146/annurev-cellbio-092910-154017 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Green D.M., Bridgewater R.E., von Kriegsheim A., Humphries M.J., Norman J.C., and Caswell P.T.. 2013a RCP-driven α5β1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J. Cell Biol. 202:917–935. 10.1083/jcb.201302041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Humphries M.J., and Caswell P.T.. 2013b Role of adhesion receptor trafficking in 3D cell migration. Curr. Opin. Cell Biol. 25:627–632. 10.1016/j.ceb.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya H., Takeya R., Takeuchi K., Watanabe N., Jing N., and Sumimoto H.. 2005. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 10:665–678. 10.1111/j.1365-2443.2005.00867.x [DOI] [PubMed] [Google Scholar]
- Katoh M., and Katoh M.. 2004. Identification and characterization of human FHOD3 gene in silico. Int. J. Mol. Med. 13:615–620. [PubMed] [Google Scholar]
- Kaur S., Kenny H.A., Jagadeeswaran S., Zillhardt M.R., Montag A.G., Kistner E., Yamada S.D., Mitra A.K., and Lengyel E.. 2009. beta3-integrin expression on tumor cells inhibits tumor progression, reduces metastasis, and is associated with a favorable prognosis in patients with ovarian cancer. Am. J. Pathol. 175:2184–2196. 10.2353/ajpath.2009.090028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing T.M., Wang Y., Pertz O., Copeland J.W., and Grosse R.. 2010. Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene. 29:2441–2448. 10.1038/onc.2009.515 [DOI] [PubMed] [Google Scholar]
- Krause M., and Gautreau A.. 2014. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15:577–590. 10.1038/nrm3861 [DOI] [PubMed] [Google Scholar]
- Lai F.P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., Stradal T.E., Dunn G.A., Small J.V., and Rottner K.. 2008. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27:982–992. 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law A.-L., Vehlow A., Kotini M., Dodgson L., Soong D., Theveneau E., Bodo C., Taylor E., Navarro C., Perera U., et al. . 2013. Lamellipodin and the Scar/WAVE complex cooperate to promote cell migration in vivo. J. Cell Biol. 203:673–689. 10.1083/jcb.201304051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., Wickström S.A., and Fässler R.. 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23:397–418. 10.1101/gad.1758709 [DOI] [PubMed] [Google Scholar]
- Lin C.H., Espreafico E.M., Mooseker M.S., and Forscher P.. 1996. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 16:769–782. 10.1016/S0896-6273(00)80097-5 [DOI] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M.T., Barry K.C., Linardopoulou E.V., Osborn G.E., and Parkhurst S.M.. 2009. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 136:2849–2860. 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A., and Mitchison T.. 1999. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146:1097–1106. 10.1083/jcb.146.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malliri A., van der Kammen R.A., Clark K., van der Valk M., Michiels F., and Collard J.G.. 2002. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 417:867–871. 10.1038/nature00848 [DOI] [PubMed] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Maubant S., Cruet-Hennequart S., Dutoit S., Denoux Y., Crouet H., Henry-Amar M., and Gauduchon P.. 2005. Expression of α V-associated integrin β subunits in epithelial ovarian cancer and its relation to prognosis in patients treated with platinum-based regimens. J. Mol. Histol. 36:119–129. 10.1007/s10735-004-4273-0 [DOI] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O., and Smal I.. 2012. Methods for cell and particle tracking. Methods Enzymol. 504:183–200. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Mellor H. 2010. The role of formins in filopodia formation. Biochim. Biophys. Acta. 1803:191–200. 10.1016/j.bbamcr.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Muller P.A.J., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P., et al. . 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell. 139:1327–1341. 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Muller P.A.J., Trinidad A.G., Timpson P., Morton J.P., Zanivan S., van den Berghe P.V.E., Nixon C., Karim S.A., Caswell P.T., Noll J.E., et al. . 2013. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 32:1252–1265. 10.1038/onc.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberg A., Kitzing T., and Grosse R.. 2011. Nucleating actin for invasion. Nat. Rev. Cancer. 11:177–187. 10.1038/nrc3003 [DOI] [PubMed] [Google Scholar]
- Ohta Y., Hartwig J.H., and Stossel T.P.. 2006. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 8:803–814. 10.1038/ncb1437 [DOI] [PubMed] [Google Scholar]
- Okabe S., and Hirokawa N.. 1991. Actin dynamics in growth cones. J. Neurosci. 11:1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S., and Mellor H.. 2005. The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 15:129–133. 10.1016/j.cub.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Pellinen T., and Ivaska J.. 2006. Integrin traffic. J. Cell Sci. 119:3723–3731. 10.1242/jcs.03216 [DOI] [PubMed] [Google Scholar]
- Peng J., Wallar B.J., Flanders A., Swiatek P.J., and Alberts A.S.. 2003. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13:534–545. 10.1016/S0960-9822(03)00170-2 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Gavara N., Chadwick R.S., and Yamada K.M.. 2012. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197:439–455. 10.1083/jcb.201201124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Koo H., and Yamada K.M.. 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 345:1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero E., Caswell P.T., Muller P.A.J., Grindlay J., McCaffrey M.W., Zhang Q., Wakelam M.J.O., Vousden K.H., Graziani A., and Norman J.C.. 2012. Diacylglycerol kinase α controls RCP-dependent integrin trafficking to promote invasive migration. J. Cell Biol. 196:277–295. 10.1083/jcb.201109112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. . 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S., Sahai E., and Marshall C.J.. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 135:510–523. 10.1016/j.cell.2008.09.043 [DOI] [PubMed] [Google Scholar]
- Sawada K., Mitra A.K., Radjabi A.R., Bhaskar V., Kistner E.O., Tretiakova M., Jagadeeswaran S., Montag A., Becker A., Kenny H.A., et al. . 2008. Loss of E-cadherin promotes ovarian cancer metastasis via α5-integrin, which is a therapeutic target. Cancer Res. 68:2329–2339. 10.1158/0008-5472.CAN-07-5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., and Livak K.J.. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shibue T., Brooks M.W., Inan M.F., Reinhardt F., and Weinberg R.A.. 2012. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2:706–721. 10.1158/2159-8290.CD-11-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., Brooks M.W., and Weinberg R.A.. 2013. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell. 24:481–498. 10.1016/j.ccr.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.M., Ezhkova E., Silva J., Heart S., Castillo M., Campos Y., Castro V., Bonilla F., Cordon-Cardo C., Muthuswamy S.K., et al. . 2009. Cyfip1 is a putative invasion suppressor in epithelial cancers. Cell. 137:1047–1061. 10.1016/j.cell.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., and Li R.. 2012. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197:239–251. 10.1083/jcb.201112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Fogelson B., Rubinstein B., Noguera P., Volkmann N., Hanein D., Mogilner A., and Li R.. 2015. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol. Biol. Cell. 26:901–912. 10.1091/mbc.E14-07-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya R., Taniguchi K., Narumiya S., and Sumimoto H.. 2008. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 27:618–628. 10.1038/emboj.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Takeya R., Suetsugu S., Kan-O M., Narusawa M., Shiose A., Tominaga R., and Sumimoto H.. 2009. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 284:29873–29881. 10.1074/jbc.M109.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., and Mayor R.. 2013. Collective cell migration of epithelial and mesenchymal cells. Cell. Mol. Life Sci. 70:3481–3492. 10.1007/s00018-012-1251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I., Thompson P.M., Berlemont S., Plevock K.M., Plotnikov S.V., Zemljic-Harpf A., Ross R.S., Davidson M.W., Danuser G., Campbell S.L., and Waterman C.M.. 2013. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202:163–177. 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoğlu M., Tournier A.L., Jenkins R.P., Hooper S., Bates P.A., and Sahai E.. 2013. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat. Cell Biol. 15:751–762. 10.1038/ncb2775 [DOI] [PubMed] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460. 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan L., Tan C.-T., Chapman A., Nonaka D., Mack N.A., Smith D., Booton R., Hurlstone A.F.L., and Malliri A.. 2015. HUWE1 ubiquitylates and degrades the RAC activator TIAM1 promoting cell-cell adhesion disassembly, migration, and invasion. Cell Reports. 10:88–102. 10.1016/j.celrep.2014.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F.M., Fruhwirth G., Ng T., and Ridley A.J.. 2011. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J. Cell Biol. 193:655–665. 10.1083/jcb.201011038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White D.P., Caswell P.T., and Norman J.C.. 2007. αvβ3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 177:515–525. 10.1083/jcb.200609004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Asokan S.B., Berginski M.E., Haynes E.M., Sharpless N.E., Griffith J.D., Gomez S.M., and Bear J.E.. 2012. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 148:973–987. 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S.D.J., Caswell P., Spence H.J., Carnell M., Insall R.H., Norman J., and Machesky L.M.. 2011. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124:3753–3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.