A subset of inner and outer kinetochore proteins responsible for lateral attachment and spindle checkpoint signaling in Xenopus egg extracts collaboratively coalesce into micrometer-scale filamentous protrusions in the absence of microtubules.
Abstract
It is widely accepted that the kinetochore is built on CENP-A–marked centromeric chromatin in a hierarchical order from inner to outer kinetochore. Recruitment of many kinetochore proteins depends on microtubule attachment status, but it remains unclear how their assembly/disassembly is orchestrated. Applying 3D structured illumination microscopy to Xenopus laevis egg extracts, here we reveal that in the absence of microtubule attachment, proteins responsible for lateral attachment and spindle checkpoint signaling expand to form micrometer-scale fibrous structures over CENP-A–free chromatin, whereas a core module responsible for end-on attachment (CENP-A, CENP-T, and Ndc80) does not. Both outer kinetochore proteins (Bub1, BubR1, Mad1, and CENP-E) and the inner kinetochore component CENP-C are integral components of the expandable module, whose assembly depends on multiple mitotic kinases (Aurora B, Mps1, and Plx1) and is suppressed by protein phosphatase 1. We propose that phospho-dependent coexpansion of CENP-C and outer kinetochore proteins promotes checkpoint signal amplification and lateral attachment, whereas their selective disassembly enables the transition to end-on attachment.
Introduction
Accurate chromosome segregation during mitosis depends on the kinetochore, which supports chromosome movements by attaching to microtubules and also acts as a signaling hub to control the spindle assembly checkpoint (SAC; Foley and Kapoor, 2013). The kinetochore is thought to assemble on centromeric chromatin according to a temporal and spatial hierarchy of functionally distinct complexes. In addition, the kinetochore changes its size and composition in response to microtubule attachment status (Rieder, 1982; Thrower et al., 1996; McEwen et al., 1998; Hoffman et al., 2001). The molecular basis of this adaptive response is not clear.
Based on classic electron microscopy images of a trilaminar structure, the kinetochore has been divided into inner and outer regions (Cleveland et al., 2003). Recent studies using super-resolution fluorescence microscopy techniques and immunoelectron microscopy, combined with epistasis analyses has led to a hierarchical view of kinetochore assembly upon centromeric chromatin marked by the centromere-specific histone H3 variant CENP-A (see Fig. 8 A, left; Wan et al., 2009; Suzuki et al., 2011). The inner kinetochore includes the constitutive centromere–associated network (CCAN) proteins, which recognize CENP-A and are present on centromeres throughout the cell cycle (Perpelescu and Fukagawa, 2011; Westhorpe and Straight, 2013). During mitosis, two CCAN proteins, CENP-C and CENP-T, recruit outer kinetochore protein complexes via distinct mechanisms. CENP-T directly recruits the Ndc80 complex (Ndc80C; Schleiffer et al., 2012; Nishino et al., 2013), whereas CENP-C recruits the KMN (KNL1, Mis12, and Ndc80) network, composed of the KNL1 complex (KNL1C), the Mis12 complex (Mis12C), and the Ndc80C through direct binding to the Mis12C (Przewloka et al., 2011; Screpanti et al., 2011). Thus, both CENP-C and CENP-T contribute to the recruitment of the Ndc80C, which generates the stable attachments to microtubule ends (end-on attachments) essential for anaphase chromosome movement. In contrast, KNL1 and its binding partner Zwint primarily act as a scaffold for the transient SAC proteins (including Bub1, BubR1, Bub3, Mad1, Mad2, the Rod/ZW10/Zwilch (RZZ) complex, and Spindly; Varma and Salmon, 2012). KNL1 promotes microtubule attachment indirectly by recruiting the motor proteins dynein and CENP-E, which interact with the lateral sides of microtubules (Rieder and Alexander, 1990; Wood et al., 1997; Kapoor et al., 2006), whereas KNL1’s direct microtubule binding activity functions primarily in silencing the SAC (Espeut et al., 2008). These motor proteins and checkpoint components have been localized to an outermost “fibrous corona” region that can extend over 100 nm into the cytoplasm from the outer kinetochore plate (McEwen et al., 1998; Hoffman et al., 2001).
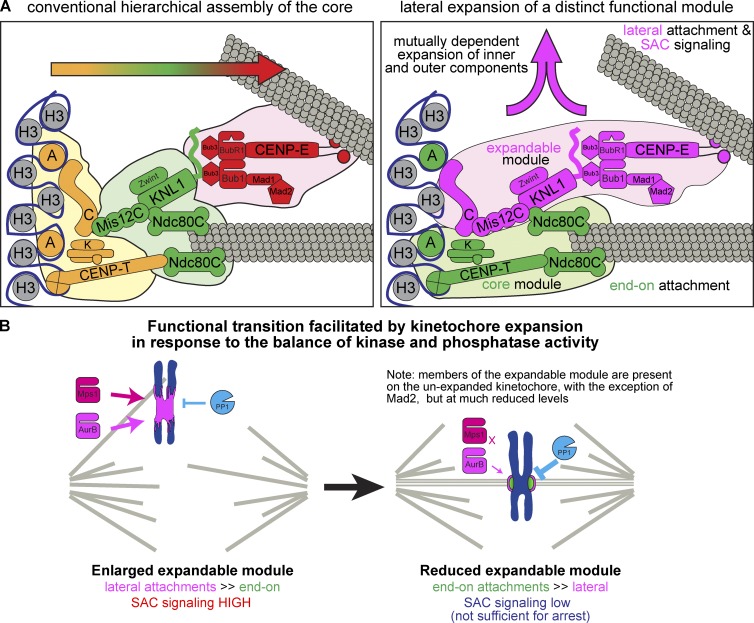
Figure 8.
Assembly of distinct functional modules within the kinetochore. (A) Comparison of the conventional view of kinetochore structure and assembly with our model of the kinetochore as distinct functional domains: an expandable and core module, each comprised of both inner and outer kinetochore components. Arrows indicate the direction of kinetochore assembly. (B) Schematic of the functional transition promoted by the dynamics of the expandable module.
Each kinetochore has the capacity to recruit multiple microtubule-binding proteins, which confer specific mechanisms of chromosome movement at distinct times during mitosis (Magidson et al., 2011). During prometaphase, dynein captures the lateral sides of microtubules and rapidly moves chromosomes toward the poles (Rieder and Alexander, 1990; Skibbens et al., 1993), whereas CENP-E helps congress chromosomes to the metaphase plate by sliding along preestablished kinetochore microtubules attached to other chromosomes (Kapoor et al., 2006). Throughout prometaphase, metaphase, and anaphase, the Ndc80C enables kinetochores to track dynamic microtubule ends to support oscillatory movement of chromosomes and chromosome segregation (Cheeseman et al., 2006; DeLuca et al., 2006). The transitions from unattached to lateral attachment to end-on attachment coincide with the removal of SAC proteins, which preferentially (Bub1 and BubR1) or exclusively (Mad1 and Mad2) associate with kinetochores that are not attached to microtubules (Chen et al., 1996; Howell et al., 2004; Shah et al., 2004). If mitotic cells are exposed to nocodazole for an extended period, some outer kinetochore proteins (BubR1 and Mad2; CENP-E and dynein) form large C-shaped “collars” or rings that encircle both sister kinetochores (Thrower et al., 1996; Hoffman et al., 2001), but the functional importance of these structures is not known.
When a kinetochore establishes stable end-on attachment, SAC proteins, CENP-E and dynein are actively removed by a dynein-mediated “stripping” mechanism and other dynein-independent mechanisms (Howell et al., 2001; Wojcik et al., 2001; Gassmann et al., 2010). In contrast, the Ndc80C stably associates with kinetochores and has been proposed to represent a “core” kinetochore (DeLuca et al., 2005). In addition, although the amount of CENP-E at the kinetochore is reduced in metaphase, a fraction of CENP-E remains on the kinetochore to support end-on attachment (Gudimchuk et al., 2013). Similarly, although Mad1 and Mad2 are completely removed from the attached kinetochores, a reduced but substantial amount of Bub1, BubR1, and Bub3 is retained (Chen et al., 1996; Howell et al., 2004; Shah et al., 2004). Although antagonism between microtubule binding by the Ndc80C and the RZZ proteins that bridge KNL1 and dynein has been shown (Gassmann et al., 2008; Cheerambathur et al., 2013), it remains to be established how only a subset of kinetochore proteins are selectively removed upon end-on attachment. Applying 3D structured illumination microscopy (SIM) to kinetochores assembled in Xenopus laevis egg extracts (Gustafsson et al., 2008; Schermelleh et al., 2008), we here present novel insights into architectural organization of the kinetochore.
Results
Outer kinetochore proteins undergo dramatic expansion in the absence of microtubules
To investigate the assembly of vertebrate kinetochores and their response to microtubule attachment status, we monitored kinetochore architecture using 3D SIM in Xenopus egg extracts, where kinetochore formation occurs on replicated sperm chromosomes in a synchronous manner. Although BubR1 forms ∼0.5-µm “collars” extending around sister centromeres in the absence of microtubules in human telomerase-immortalized retinal pigmented epithelial (RPE-hTERT) cells as has been previously reported using somatic tissue culture cells (Fig. 1, A and B; Hoffman et al., 2001), in Xenopus egg extracts BubR1 signals protruded in long slender stretches that appear continuous at this level of resolution and can extend over a micrometer from the center of the closest CENP-A focus (Fig. 1, C and D; and Videos 1 and 2). Quantification of the intensity or volume of the BubR1 signal shows a similar level of increase, suggesting that these fibers are formed by loading new protein at a relatively constant density rather than redistribution of a fixed pool of protein (Figs. 1 E and S1, A and B). Although the absolute degree of expansion is sensitive to extract quality and is thus somewhat affected by immunodepletion procedures, this dramatic expansion provides an excellent system to characterize microtubule-dependent changes in kinetochore structure.
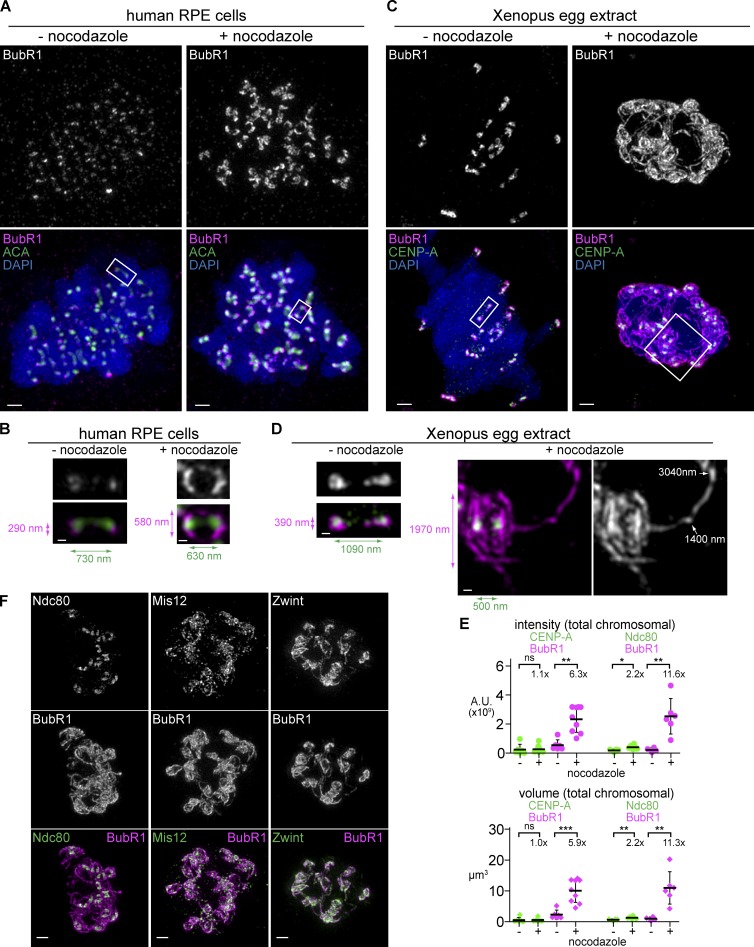
Figure 1.
Kinetochores expand by forming micrometer-scale fibrous structures that extend throughout the chromosome mass in the absence of microtubules. (A–D) Comparison of mitotic chromosomes and kinetochores in human RPE-hTERT cells (A and B) and Xenopus egg extracts (C and D) in the presence (right) or absence (left) of 33 µM nocodazole to completely depolymerize microtubules (not depicted). Each image represents a set of chromosomes derived from a single nucleus. Centromeres are stained (green) with CREST antisera (A and B) or CENP-A (C and D) and costained for the outer kinetochore protein BubR1 (magenta) and DAPI (blue). All chromosomes (A and C; bars, 1 µm) and blowups of sister centromeres (B and D; bars, 0.2 µm) are shown at the same magnification to allow size comparison. (B and D) Colored arrows indicate lengths of maximum BubR1 signal perpendicular to the centromere–centromere axis (magenta) and the distance between centroids of centromere signals (green). White arrows indicate length measurements of traces along the filamentous structure beginning from the centroid of the nearest CENP-A focus. (E) Quantification of the total kinetochore staining on the chromosome cluster. Mean and standard deviation plotted in black. Mean values and fold change indicated above. Mann–Whitney test: ns, P > 0.5; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (n = 4–10 nuclei). (F) Staining of KMN network components (green) in nocodazole-treated extracts costained with BubR1 (magenta, chromosomes not depicted). Bars, 1 µm. All images are maximum intensity projections of 3D-SIM datasets. A.U., arbitrary unit.
The expandable module is composed of kinetochore components that promote lateral attachment and the SAC
We next systematically examined which kinetochore submodules show expansion in the absence of microtubule attachment. Outermost kinetochore components, such as CENP-E, Xorbit, and dynein, were present throughout the expanded kinetochores (Fig. S1 C). In addition, Mad1 and Mad2 were both stained heavily on the expanded kinetochore, despite being weakly present (Mad1) or undetectable (Mad2) on untreated kinetochores (Fig. S1, D and E).
In contrast, Ndc80 showed a slight increase in volume and intensity in nocodazole-treated Xenopus egg extracts, but it remained mostly at the core of each kinetochore and failed to robustly localize along the extended fibers (Fig. 1, E and F). However, although Ndc80 is a member of the KMN network, other KMN components did expand: Mis12 and Zwint, which binds tightly to the C terminus of KNL1, were both found throughout the expanded region, and thus, substantial amounts of each protein are spatially segregated from the Ndc80C (Fig. 1 F).
CENP-C is a component of the expandable kinetochore
The different behaviors among the KMN network raised the question of how the internal CCAN components would respond to the lack of microtubules. Consistent with its role in directly recruiting the Ndc80C, CENP-T remained in small foci (like CENP-A staining) in nocodazole-treated extracts (Fig. 2 A). CENP-K, a component of the CENP-H/I/K complex contributing to Ndc80C recruitment (Cheeseman et al., 2008; Basilico et al., 2014; Kim and Yu, 2015), also remained in small foci (Fig. 2 A).
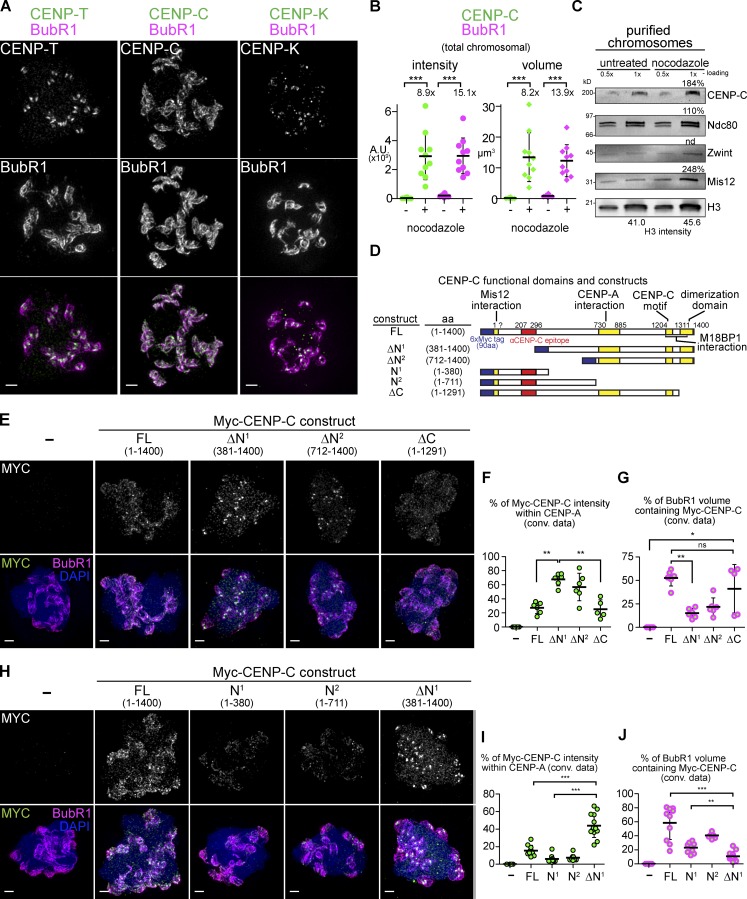
Figure 2.
CENP-C is a component of the expanded kinetochore. (A) Staining of CCAN proteins (green) in nocodazole-treated extracts costained with BubR1 (magenta, chromosomes not depicted). (B) Quantification of the total kinetochore staining on the chromosome cluster. Mean, standard deviation, and fold change indicated in black (n = 8–10 nuclei). (C) Western blots of chromosomes purified from untreated or nocodazole-treated extracts. Percent values calculated by correcting raw values with the ratio of H3 signal. (D) Schematic of Xenopus CENP-C protein and constructs used in D–I. (E) Mitotic chromosomes assembled in extracts in which the indicated 6×Myc–CENP-C construct was expressed. (F and G) Quantification of data from D, comparing the Myc signal to CENP-A or BubR1 staining. (H–J) Sample preparation and quantification as in E–G. Mann–Whitney test: ns, P > 0.5; *, P < 0.05; **, P < 0.01; ***, P < 0.001. All images are maximum intensity projections of 3D-SIM datasets; bars, 1 µm. A.U., arbitrary unit.
Surprisingly, CENP-C is present at high levels throughout the expanded kinetochore and, like BubR1, increases in both intensity and volume (Fig. 2, A and B). We also observed an increase in CENP-C recovered on chromosomes purified from nocodazole-treated extracts (Fig. 2 C). CENP-C harbors multiple well-characterized functional domains (Fig. 2 D) of which the central CENP-A interaction domain, CENP-C motif, and dimerization domain, but not the Mis12C-interacting domain, are required for CENP-C to localize to the kinetochore (Milks et al., 2009). To define the domains that target CENP-C to the expanded kinetochore, we expressed full-length (FL) and truncated versions of CENP-C containing six N-terminal Myc tags in egg extracts (Milks et al., 2009). Although FL Myc–CENP-C was able to recapitulate the localization of endogenous CENP-C, a truncation mutant lacking either 380 amino acids (CENP-C-ΔN1) or 711 amino acids (CENP-C-ΔN2) of the N terminus was found almost exclusively at the CENP-A–marked core and not on the expanded region (Fig. 2, E–G; and Fig. S2 F). In contrast, Myc-tagged N-terminal fragments lacking the C-terminal 109 amino acids responsible for dimerization (CENP-C-ΔC) were targeted to the expanded region containing BubR1 (Fig. 2, E–G). Shorter N-terminal fragments (CENP-C-N1 and CENP-C-N2) lacking the CENP-A interaction domain were also targeted to the expanded region but with much reduced efficiency (Fig. 2, H–J; and Fig. S2 G). These results indicate that the N-terminal 380 amino acids of CENP-C, which interacts with the Mis12C, drives preferential targeting of CENP-C to the expandable module rather than to the core module. However, this N terminal region itself is insufficient for robust localization to the expandable module, suggesting that CENP-C expansion requires both Mis12C interaction and additional functionality, perhaps the nucleosome interaction.
Kinetochore expansion is rapid and can be observed in prometaphase extracts in the presence of microtubules
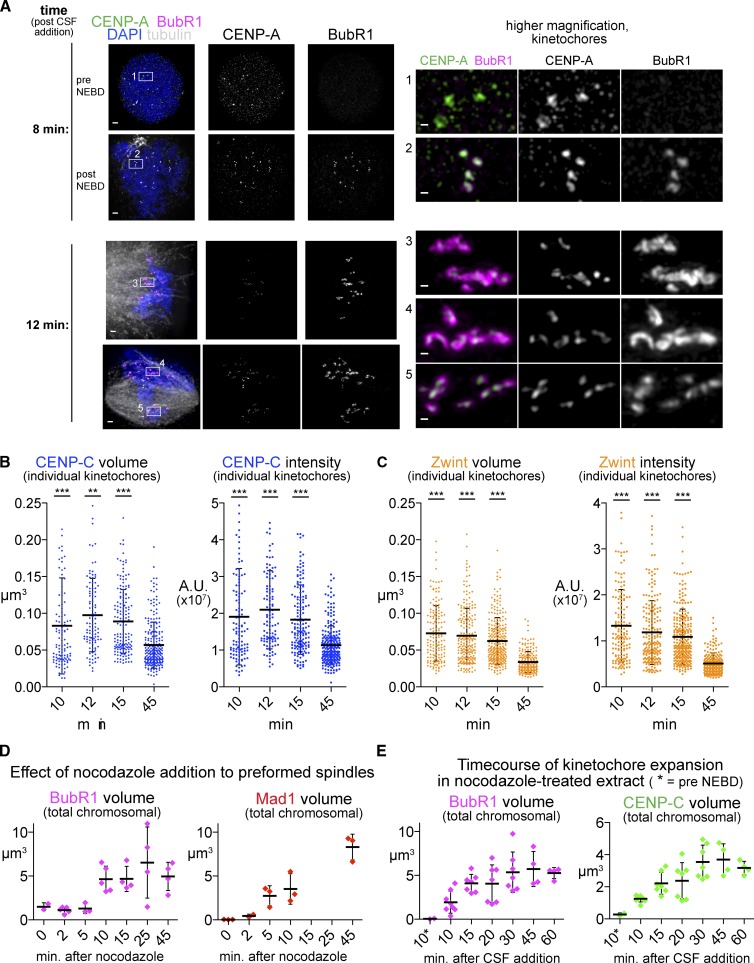
We next tested whether similar kinetochore expansion occurs during unperturbed spindle formation at the entry into M phase. Like in tissue culture systems, prometaphase kinetochores in Xenopus egg extracts accumulate outer kinetochore and checkpoint proteins such as BubR1, which can be seen soon after nuclear envelope breakdown enlarged on the early, disordered spindles (Fig. 3 A). Other expanded kinetochore proteins such as CENP-C and Zwint also showed increased staining in prometaphase extracts before completion of metaphase spindle formation. Quantification of signal intensity and volume suggested that, like expansion in nocodazole-treated extracts, this was caused by an increase in the total amount of protein, not a restructuring of a fixed amount of protein (Fig. 3, B and C). Time course experiments revealed expansion within 10 min after treatment of fully formed spindles with nocodazole (Fig. 3 D) or after nuclear envelope breakdown in the presence of nocodazole (Fig. 3 E), well within the timescale of unperturbed prometaphase.
Figure 3.
The kinetics of expansion. (A) Examples of kinetochore staining at time points shortly after nuclear envelope breakdown (∼10 min after addition of fresh CSF extract to induce M phase entry). (left) Images of two nuclei at each time point to illustrate the types of figures seen at that time point. Bars, 1 µm. (right) Higher magnification images of a group of kinetochores from each nucleus. Note that after 8 min, small amounts of BubR1 have loaded only in nuclei that have lost their circularity and have presumably completed nuclear envelope breakdown (NEBD). In contrast, after 12 min, much more BubR1 staining can be seen, particularly in kinetochores in locations where the spindle is more disorganized (inset 3 and 4; bars, 0.2 µm). All images are maximum intensity projections of 3D-SIM datasets. (B and C) Quantification of individual kinetochore signals for the indicated protein over a time course during spindle assembly. Significance relative to the 45-min value. Mann–Whitney test: ***, P < 0.001; **, P < 0.01. n > 100 kinetochores from 5–8 nuclei. (D and E) Quantification of total signal from chromosome clusters for the indicated kinetochore component over a time course to indicate the rate of kinetochore expansion. Mean and standard deviation are plotted in black. A.U., arbitrary unit.
Assembly of the expandable kinetochore does not follow the same hierarchical mechanism as the core
The data presented here show that the architectural composition of the kinetochore can be divided in two distinct, spatially separated modules made of proteins known to have different functions (Fig. 4 A): (1) a core module containing CENP-A, CENP-T, CENP-K, and the Ndc80C, which is responsible for end-on attachment, and (2) an expandable module containing CENP-C, Mis12C, KNL1C, Bub1, BubR1, Mad1, Mad2, CENP-E, and dynein, which are responsible for SAC signaling and lateral attachment. Although the core module exhibits little or no change in response to microtubule attachment status, the expandable module expands over CENP-A–free chromatin in the absence of microtubule attachment. On the metaphase plate within a bipolar spindle, a majority of the components of the expandable module are reduced to the vicinity of the core, with the exception of Mad2, which dissociates to below the detection limit. The dynamic nature of the expandable module relative to the core suggests that its assembly mechanisms are distinct. Thus, we next tested which components are required for the kinetochore expansion.
Figure 4.
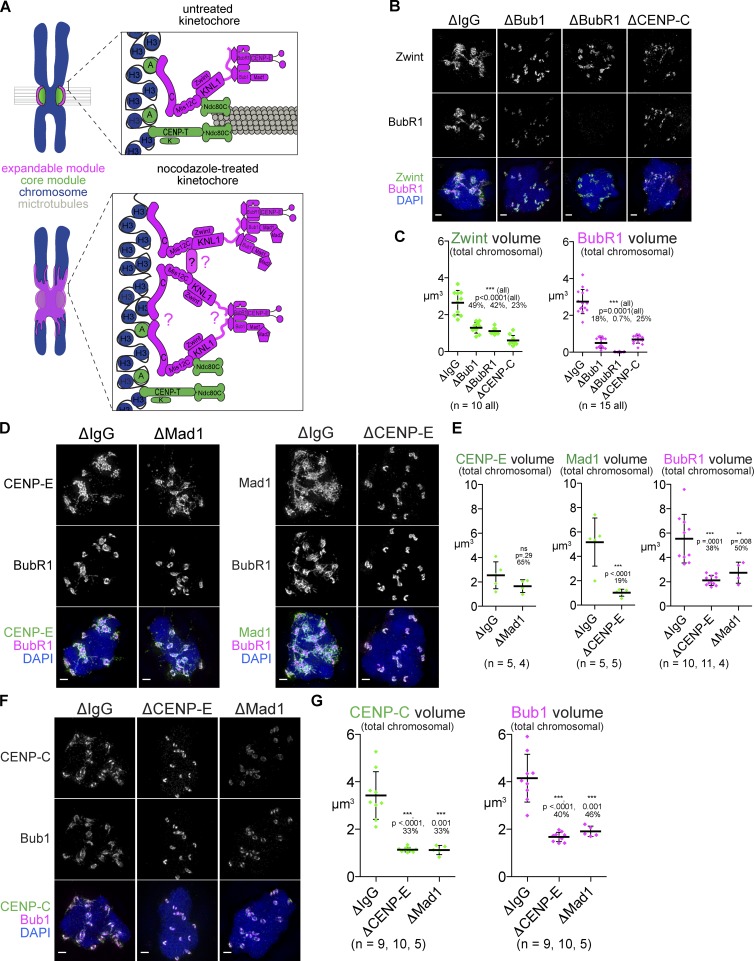
Formation of the expanded kinetochore requires outer kinetochore proteins and CENP-C. (A) Model of kinetochore expansion. Proteins present on the expanded kinetochore are shown in magenta, whereas the core kinetochore proteins are in green. Question marks indicate sites of potential oligomerization. (B–G) Mitotic chromosomes assembled in Xenopus egg extracts after depletion indicated, treated with nocodazole, and stained for the kinetochore proteins indicated. Bars, 1 µm. (C, E, and G) Quantification of total signal from chromosome clusters in extracts from B, D, and F. P-values from Mann–Whitney test and percentage of mean value relative to ΔIgG are indicated above each condition. Mean and standard deviation are plotted in black. All images are maximum intensity projections of 3D-SIM datasets.
Previous studies in Xenopus extracts demonstrated that loading of the outer kinetochore proteins Bub1, BubR1, and CENP-E are mutually dependent (Chen, 2002; Mao et al., 2003; Vigneron et al., 2004; Wong and Fang, 2006). We tested whether these outer kinetochore components of the expandable module contribute to loading of inner kinetochore proteins in kinetochore expansion. It has been suggested that Zwint loads together with KNL1 independently of the presence of Bub1 and BubR1 (Emanuele et al., 2005; Kiyomitsu et al., 2011; Petrovic et al., 2014). However, Zwint was significantly reduced on chromosomes in ΔBub1 or ΔBubR1 extracts (Fig. 4, B and C; and Fig. S2, A and B). This result suggests a reversal of the conventional loading hierarchy.
Expansion of Zwint and BubR1 was also inhibited in ΔCENP-C extracts, but unexpectedly, their kinetochore recruitment was not completely eliminated (Fig. 4, B and C; and Fig. S2 B). Reduced but significant levels of CENP-T and Ndc80 at kinetochores (∼30% of CENP-T remaining), but not Mis12, were detected in ΔCENP-C extract (Fig. S2, C and D), suggesting either that there is a Mis12C-independent recruitment of KNL1, perhaps through CENP-H/I/K, or that the remaining Mis12C is below our detection limit. In contrast, CENP-T depletion did not affect kinetochore expansion (Fig. S2, E and F) despite a clear loss of Ndc80 at the core, as expected for the important role for CENP-T in Ndc80 recruitment (Fig. S2 F). Thus, two CCAN proteins, CENP-C and CENP-T, play distinct functions in the formation of the expandable module and core module, respectively.
Next, we tested the importance of two outer kinetochore proteins CENP-E and Mad1, which both play roles in the SAC but are not thought to affect recruitment of inner kinetochore proteins. Depletion of either protein (Fig. S2 G) caused a significant loss of BubR1 or Bub1 (Fig. 4, D–G; and Fig. S2, H and I). Although CENP-E was not significantly reduced in Mad1-depleted extracts (ΔMad1), Mad1 was dramatically reduced in CENP-E–depleted extract (ΔCENP-E; Fig. 4, D and E; and Fig. S2 H). Strikingly, like Bub1, BubR1, and Mad1, CENP-C was also lost in either ΔMad1 or ΔCENP-E extract (Fig. 3, F and G; and Fig. S2 I). The loss of a centromeric protein after depletion of outer kinetochore components confirmed that the linear, hierarchical assembly established for the core kinetochore may not be shared by the expandable module.
During the course of our experiments into the requirement for the N terminus of CENP-C in expansion, we noticed that overexpression of FL Myc-CENP-C or CENP-C-ΔC caused a decrease in the amount of kinetochore expansion, whereas constructs lacking the N terminus (CENP-C-ΔN1 or CENP-C-ΔN2) did not have this effect on expansion (Fig. S3, A–C). Thus, the N terminus of CENP-C exerts a dominant-negative effect on expansion when overexpressed.
The dominant-negative effect of CENP-C overexpression on kinetochore expansion prompted us to test whether the effect of CENP-C depletion was caused by codepletion of a critical interacting protein, such as the Mis12C. However, the levels of Mis12 and other kinetochore proteins (Bub1, Ndc80, Zwint, and Plx1) were not affected in ΔCENP-C extracts (Fig. S4 A). Unfortunately, we were not able to rescue the loss of kinetochore expansion in ΔCENP-C extracts with either added-back Myc-tagged or untagged CENP-C, despite the fact that the untagged protein expanded beyond the central core and colocalized well throughout the residual BubR1-staining region (Fig. S4, B and C). We suspect that the failure to rescue expansion is a result of the dominant-negative effects of the C-terminally truncated products that are abortively produced from exogenously added mRNA and can be detected by anti–CENP-C antibody whose epitopes reside in the N terminus (Fig. S4 D), though the possibility of codepleting an unknown interacting protein cannot be ruled out. Whatever the mechanism of CENP-C’s dominant-negative/depletion effect, these data suggest that CENP-C is an integral component of the expandable module.
CENP-C–dependent kinetochore expansion promotes the SAC signal
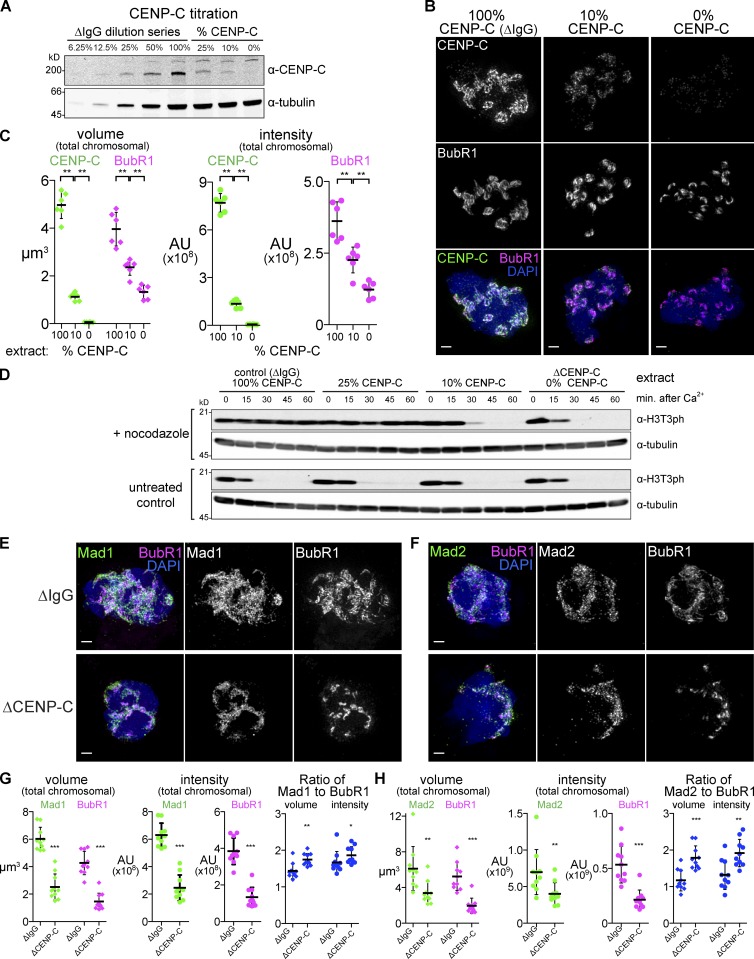
Accumulation of proteins involved in the SAC in the expanded kinetochore suggests that the strength of the checkpoint signaling may correlate with expansion. To test this, we wished to selectively compromise kinetochore expansion without affecting the core or depleting the integral components of SAC signaling. Since we saw that CENP-C depletion dramatically reduced expansion but residual KMN protein and BubR1 were still recruited (Fig. 4, B and C), we wondered whether the degree of expansion and SAC strength can be sensitized by the dose of CENP-C. To address this point, we immunodepleted CENP-C from extracts to levels below the detection limit (<6% of the endogenous level, ΔCENP-C extracts) by Western blotting and near background levels by immunofluorescence (Fig. 5, A–C) and then used combinations of CENP-C–depleted and mock-depleted (ΔIgG) extracts to titrate the level of CENP-C. We found that reducing the level of CENP-C reduced BubR1 staining in a dose-dependent manner; reduced, but substantial expansion of BubR1 and CENP-C can be seen in the presence of 10% CENP-C (Fig. 5, B and C), indicating that this level of CENP-C is sufficient to promote expansion beyond the core formation.
Figure 5.
CENP-C–dependent kinetochore expansion supports the SAC signal. (A) Extracts were immunodepleted of CENP-C and mixed with control extract to produce intermediate levels of CENP-C. (B) Mitotic chromosomes assembled in CENP-C titration extracts shown in A and treated with nocodazole. Bars, 1 µm. (C) Quantification of total signal for samples as in B. (D) Extracts described in A were assayed for checkpoint activity by challenging M-phase extract to cycle into interphase upon addition of calcium, which does not occur over 60 min in control extracts but occurred after 30 min in extracts with 10% and more rapidly in extracts with 0% of endogenous CENP-C. Cell cycle state was monitored by the presence of phosphorylated Threonine 3 of Histone H3 (H3T3ph). (E and F) Mitotic chromosomes assembled in nocodazole-treated control (ΔIgG) or CENP-C–depleted (ΔCENP-C) extracts and stained for the indicated checkpoint protein. (G and H) Quantification of total signal from E and F. Mean and standard deviation in black. Mann–Whitney test: ***, P < 0.0005; **, P < 0.01; *, P < 0.05. All images are maximum intensity projections of 3D-SIM datasets. AU, arbitrary unit.
We next tested whether inhibition of kinetochore expansion by CENP-C depletion has an impact on SAC activity. Metaphase Xenopus egg extracts arrested with the cytostatic factor (CSF; CSF extracts) can normally be released into interphase by adding calcium, but extracts containing nocodazole and a high concentration of nuclei activate the SAC and delay the cell cycle transition (Minshull et al., 1994). Cell cycle progression can be monitored by the loss of M-phase modifications such as Cdk1-dependent phosphorylation (Fig. S4 E) or H3T3ph, which is undetectable by 30 min after calcium addition (Fig. 5 D, bottom). When extracts were treated with nocodazole, these M-phase modifications remained in mock-depleted extracts or extracts with 25% of normal CENP-C even 60 min after calcium addition. However, in extracts with 0% or 10% of CENP-C levels, mitotic markers disappeared within 30 min after calcium addition (Fig. 5 D, top). Notably, the kinetics of mitotic exit in 10% CENP-C extract is slower than that in 0% CENP-C implying that the amplitude of the SAC depends on the dose of CENP-C. Furthermore, CENP-C depletion reduced but did not eliminate the SAC components Mad1 and Mad2, a similar reduction to that of BubR1 (Fig. 5, E–H). These results suggest that CENP-C’s ability to sustain the checkpoint is directly correlated with the amount of kinetochore expansion present and that when CENP-C protein is limiting, the SAC fails despite recruitment of outer kinetochore proteins to the residual smaller kinetochore.
Characterizing the distinct roles of Aurora B and Bub1 in kinetochore expansion
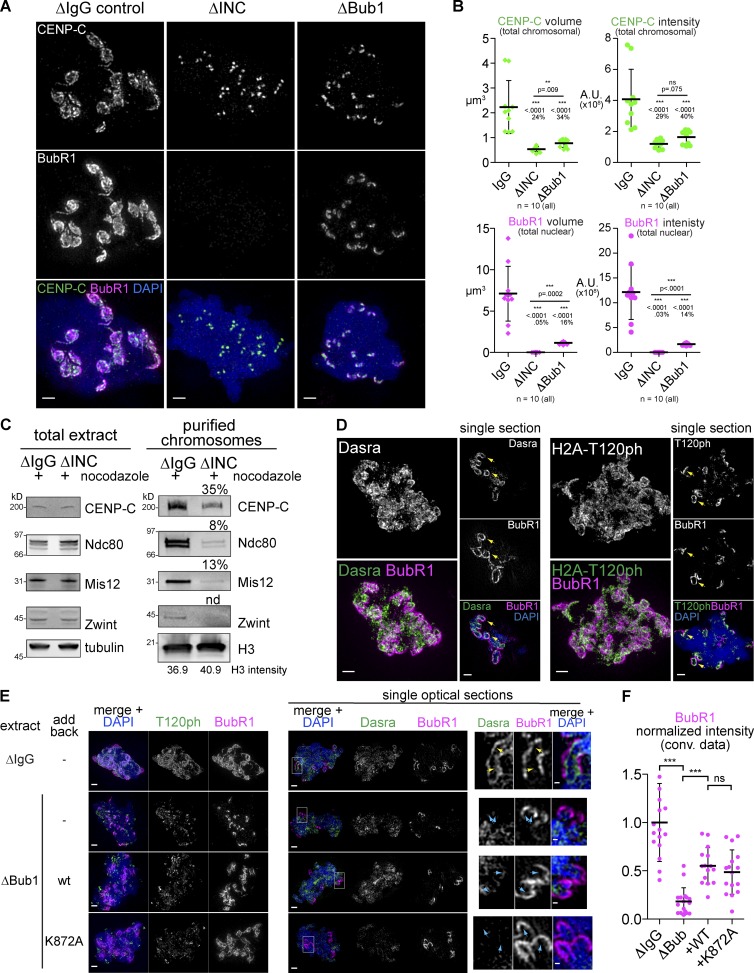
What is the mechanism that induces kinetochore expansion? In Xenopus egg extracts, one of the most upstream kinases important for recruitment of outer kinetochore proteins, including SAC regulators, is Aurora B, the kinase subunit of the chromosomal passenger complex (CPC; Kallio et al., 2002; Vigneron et al., 2004; Emanuele et al., 2008). As expected, there was a dramatic loss of BubR1 localization in CPC-depleted (ΔINC) extract (Fig. 6, A and B; and Fig. S5 A). CPC depletion did not affect CENP-C recruitment to the core module but inhibited expansion of CENP-C (Fig. 6, A and B). Western-based detection of CENP-C on purified chromosomes also revealed the CPC-dependent enrichment of CENP-C on chromosomes, like other proteins known to be dependent on Aurora B in Xenopus such as Ndc80, Mis12, and Bub1 (Fig. 6 C and Fig. S5 B).
Figure 6.
Aurora B is required for the core outer kinetochore, whereas Bub1 is specifically required for the expandable module. (A) Mitotic chromosomes assembled in Xenopus egg extracts immunodepleted of Bub1 kinase (ΔBub1) or the CPC using antibodies recognizing INCENP (ΔINC) and treated with nocodazole. (B) Quantification of the total signal, P values from Mann–Whitney test, and percentage of control mean value are shown. A.U., arbitrary unit. (C) Western blots of total extract (left) and chromosomes purified from nocodazole-treated control (ΔIgG) or CPC-depleted (ΔINC) extracts (right) show a reduction in CENP-C and a more severe loss of Ndc80, Mis12, and Zwint signal. (D) Mitotic chromosomes assembled in nocodazole-treated extracts. (left) Maximum intensity projections of whole nuclei. (right) Single optical section from the same dataset to highlight the inner centromeric staining underlying the expanded kinetochores (yellow arrows). (E) Mitotic chromosomes assembled in indicated extracts. Higher magnification images of a single optical section within the chromosome mass of Dasra-stained samples (area of inset shown with white boxes) are shown in the right-most panels to highlight the mislocalization of the CPC in all Bub1-depleted nuclei (cyan arrows) relative to controls (yellow arrows). wt, wild type. (F) Quantification normalized to the intensity of the BubR1 staining in control depletion (ΔIgG). Mean and standard deviation plotted in black. Mann–Whitney test: ***, P < 0.0001; ns, P = 0.28. All images are maximum intensity projections, except when “single section” is indicated, of 3D-SIM datasets. Bars: (A, D, and E, left) 1 µm; (E, right) 0.2 µm.
One of the mechanisms by which Aurora B becomes active at the kinetochore is through a Bub1-dependent positive feedback loop in which Bub1, which is recruited to the kinetochore in an Aurora B–dependent manner, phosphorylates H2A on T120, which recruits Shugoshin to the inner centromere, where it recruits the CPC (Boyarchuk et al., 2007; Yamagishi et al., 2010). Because Bub1 was required for kinetochore expansion (Fig. 4 B), we wondered whether this feedback pathway was involved in mediating expansion. The CPC itself (monitored with its subunit Dasra A) and the H2A T120ph mark both enriched on the chromatin underlying the expanded kinetochores visualized with BubR1 (Fig. 6, D and E, yellow arrows; and Video 3). To test the importance of Bub1’s kinase activity, wild-type (wt) or a kinase-dead version of Bub1 (K872A) were added back to extracts depleted of Bub1 (ΔBub1 extracts; Fig. 6, E and F; and Fig. S5, C and D). For unclear reasons, not only the K872A mutant but also wild-type Bub1 failed to restore H2A T120ph (Fig. 6 E), but they were able to partially rescue expansion of Bub1, BubR1, and CENP-C (Fig. 6, E and F; and Fig. S5, C and D). Both depletion of Bub1 and loss of Bub1 kinase activity caused the CPC to be mislocalized, no longer directly underlying the expanded BubR1 signals (Fig. 6 E, cyan arrows) even though it could still be detected on other chromatin regions. Consistent with this, codepletion of Sgo1 and Sgo2 did not affect kinetochore expansion (Fig. S5, E and F). Together, although Bub1 kinase activity may partially contribute to kinetochore expansion, these data suggest that Bub1 does so independently of its kinase activity. This is consistent with the notion that Bub1 kinase activity may help but is not essential for SAC activation (Sharp-Baker and Chen, 2001; Funabiki and Wynne, 2013). In addition, although kinetochore expansion leads to coexpansion of the CPC enrichment on underlying chromatin, this CPC localization does not seem to be essential for the kinetochore expansion, arguing against a model in which the kinetochore expansion is driven by a positive feedback involving Bub1 and Aurora B deposition and activation.
A balance of kinase and phosphatase activities tune the extent of kinetochore expansion
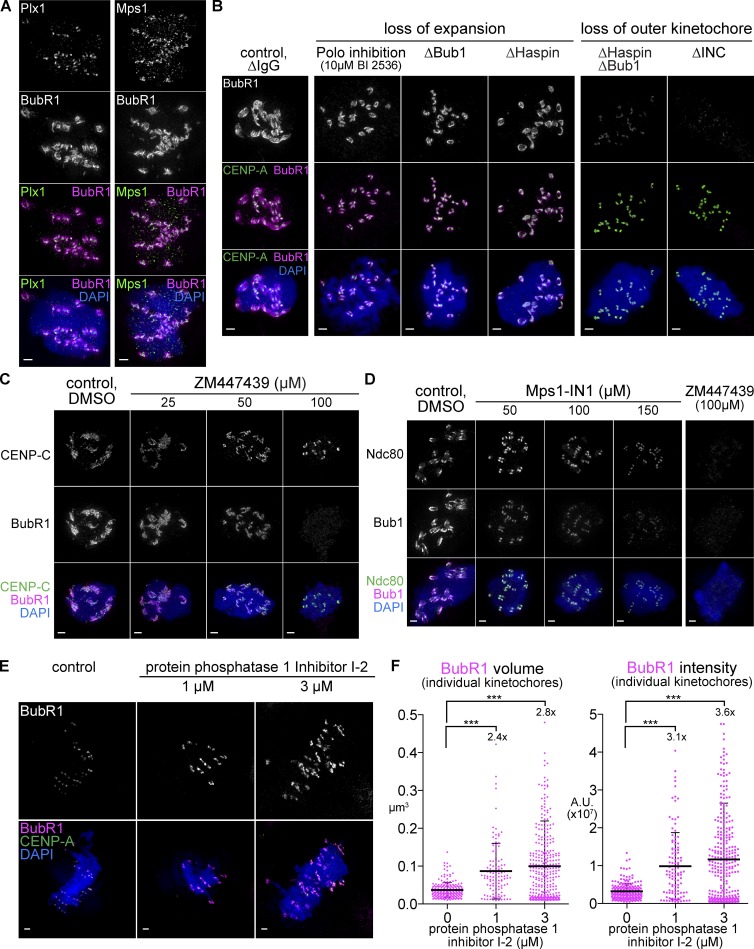
In addition to Aurora B and Bub1, two other kinases play roles in kinetochore assembly and function, the checkpoint regulator Mps1 and the Xenopus homologue of Polo like kinase 1 (Plx1). In contrast to Aurora B and Bub1, both Mps1 and Plx1 remained restricted to the core kinetochore and did not localize well to the expanded region despite the fact that each has well-characterized targets at the outer kinetochore (KNL1 is a critical target for Mps1; London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012), and BubR1 is a target of polo (Fig. 7 A and Fig. S5 G; Elowe et al., 2007; Matsumura et al., 2007; Suijkerbuijk et al., 2012). The lack of Plx1 and Mps1 expansion may reflect the role of CCAN proteins and the Ndc80C in kinetochore targeting of Polo-like kinase and Mps1, respectively (Martin-Lluesma et al., 2002; Kang et al., 2006; Hori et al., 2008).
Figure 7.
A balance of kinase and phosphatase activity tunes the extent of expansion. (A–D) Mitotic chromosomes assembled in the indicated nocodazole-treated extract. (E) Mitotic spindles assembled in extracts treated with 1–3 µM PP1 inhibitor I-2 and processed for immunofluorescence against BubR1 (top, white; bottom, magenta). (F) Quantification of BubR1 signal for individual kinetochores, mean, standard deviation, and fold change in black. Mann–Whitney test: ***, P < 0.0001. All images are maximum intensity projections of 3D-SIM datasets; bars, 1 µm. A.U., arbitrary unit.
To test the role of Plx1 in expansion, we treated extracts with the inhibitor BI 2536 and saw a similar effect to that of Bub1 depletion, with a loss of expansion but some BubR1 remaining near the core region (Fig. 7 B and Fig. S5 H). Consistent with the fact that Plx1 activates Haspin kinase (Ghenoiu et al., 2013; Zhou et al., 2014), which promotes activation of Aurora B through phosphorylating histone H3 Thr3 and enriching the CPC (Kelly et al., 2010; Zierhut et al., 2014), we found that Haspin depletion also inhibits kinetochore expansion (Fig. 7 B and Fig. S5 H). Furthermore, codepletion of Bub1 and Haspin from extract, like CPC depletion, almost completely inhibited localization of BubR1 (Fig. 7 B and Fig. S5 H). Although Aurora B may not have to be enriched directly beneath the kinetochore to support kinetochore expansion (Fig. 6, E and F), these results suggest that chromatin-dependent activation of Aurora B through Haspin and Bub1 pathways is critical for recruitment of outer kinetochore components even at the core.
Based on these results, we hypothesized that the expanded pool of kinetochore protein is more sensitive to kinase activity than the core. Indeed, inhibiting Aurora B using ZM447439 or Mps1 using Mps1-IN1 showed dose-dependent effects with the highest level of ZM447439 sufficient to completely remove BubR1 staining, like CPC depletion, whereas components of the core module, CENP-C and Ndc80, are more resistant to these treatments (Fig. 7, C and D; and Fig. S5 I). Together, these data show that the expandable module requires the full activity of multiple mitotic kinases, whereas a lower level of kinase activity is sufficient for localization to the core.
Because kinetochore expansion depends on kinases, we asked whether disassembly of this structure upon microtubule attachment requires a phosphatase activity, particularly protein phosphatase 1 (PP1), which is critical for SAC silencing (Pinsky et al., 2009; Vanoosthuyse and Hardwick, 2009; Rosenberg et al., 2011). Indeed, significantly more outer kinetochore material was observed despite formation of bipolar spindles when extracts were treated with inhibitor 2 (I-2), a specific inhibitor of PP1 (Fig. 7, E and F; Emanuele et al., 2008). The levels of kinetochore expansion seen after I-2 treatment were not as extreme as those in nocodazole-treated extracts, but instead were similar to kinetochores in early time points after nuclear envelope breakdown (Fig. 3, A–C). The simplest explanation for this intermediate level of expansion is that the extent of kinetochore expansion is correlated with phosphorylation level, not merely by the presence or absence of microtubule attachment. Altogether, the aforementioned results suggest that modulation of phosphorylation by multiple protein kinases and dephosphorylation by PP1 controls assembly and disassembly of the expandable module.
Discussion
Using Xenopus egg extracts, here we demonstrate that kinetochore assembly is not unidirectional. Instead, kinetochores can expand laterally in response to the lack of microtubule attachment in a manner dependent on both inner kinetochore CENP-C and outer kinetochore components (Fig. 8 A). This expansion generates platforms to recruit proteins responsible for SAC activation (Mad1, Mad2, Bub1, and BubR1) and motors supporting lateral attachment (dynein and CENP-E). However, CENP-A, CENP-T, CENP-K, and Ndc80, which is responsible for end-on attachment, do not show apparent coexpansion. We propose that expansion allows a spatial separation of distinct functional complexes, which supports the transition from lateral attachment and SAC activation to end-on attachment and SAC silencing by selectively disassembling most of the expandable module upon microtubule attachment (Fig. 8 B).
Adaptive kinetochore assembly in Xenopus egg extracts
Expansion of outer kinetochore proteins in the absence of microtubules has been seen in Caenorhabditis elegans and mammalian tissue culture cells (Thrower et al., 1996; Hoffman et al., 2001; Stear and Roth, 2004). In Xenopus egg extracts, this expansion is not limited to the outermost kinetochore proteins. The KMN network proteins Zwint and Mis12 as well as the CCAN member CENP-C expand. The expansion we observe in Xenopus has not to our knowledge been previously commented on, likely because the kinetochores assembled in Xenopus extract without passage through the cell cycle have smaller kinetochores and do not show expansion (unpublished data; Maresca and Heald, 2006).
Compared with mammalian somatic tissue culture systems, the level of expansion in Xenopus egg extracts is extreme, forming long, fibrous protrusions. We suspect that this dramatic expansion is a result of the large stockpile of kinetochore proteins in Xenopus egg extracts, but it may also reflect features of kinetochore assembly that are specific to oocytes. Extraordinarily large kinetochore expansion may ensure rapid and efficient kinetochore attachment in the fast, synchronized cell cycles characteristic of the early Xenopus embryo (Hara et al., 1980). They may also promote the onset of competency for SAC-mediated arrest that accompanies the midblastula transition, which is limited by the ratio of chromatin to cytoplasm (Kimelman et al., 1987; Minshull et al., 1994). Interestingly, a recent study of mouse embryos documented increased levels of CENP-C on maternal chromosomes relative to the paternal chromosomes, which persisted through the first few mitotic divisions (De La Fuente et al., 2015). Despite the difference in the degree of expansion among these systems, we hypothesize that similar principles and mechanisms control adaptive kinetochore assembly in response to microtubule attachment status.
Inner and outer kinetochore proteins are reciprocally required for kinetochore expansion
Our time course analyses suggest that the expanded kinetochores nucleate from the core regions marked by CENP-A and then extend over CENP-A–free chromatin. The appearance of fibers rather than even growth in all directions leads us to hypothesize that expansion is mediated by formation of copolymers, in which multiple different inner and outer kinetochore proteins (here including, but not limited to, CENP-C, Bub1, BubR1, CENP-E, and Mad1) coassemble in a mutually dependent manner.
Previous work in chicken cells showed that artificial kinetochores formed by chromosomal tethering of CENP-T recruited most outer kinetochore components and Aurora B in the absence of CENP-C (Hori et al., 2013). It would be interesting to see whether these kinetochores are able to expand in the absence of microtubules or whether, as in Xenopus, they require CENP-C to do so. Furthermore, a recent study showed that when CENP-T is tethered to chromosomes, it can recruit KNL1 and the Mis12C, but this recruitment requires the Ndc80C, an inversion of the conventional recruitment hierarchy (Rago et al., 2015).
Our finding that CENP-C is able to localize to regions far away from CENP-A nucleosomes and that this depends on its N-terminal Mis12C-interacting domain suggests that its role in adaptive kinetochore assembly is separable from its other roles in CENP-A nucleosome recognition. However, the N-terminal Mis12C-interacting module alone is inefficiently targeted to the expanded module, suggesting that the DNA or chromatin interaction also supports CENP-C’s role in expansion, perhaps through its capacity to interact with H3-containing nucleosomes (Ribeiro et al., 2010).
We currently do not know which molecules confer the fibrous property of the expandable module. KNL1 is a primary candidate because it contains a long disordered region that recruits Bub1 and BubR1 upon phosphorylation by Mps1 (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Petrovic et al., 2014), can form an oligomer (Petrovic et al., 2014; Kern et al., 2015), and has been observed in long fibers away from chromatin during meiosis I in C. elegans (Monen et al., 2005). Mps1-mediated recruitment of Bub1 and BubR1 may facilitate further oligomerization, as it has been recently shown that BubR1 heterodimerizes with Bub1 (Overlack et al., 2015). In addition, several components of the expandable module have the capacity to homodimerize, including Mad1, CENP-E, and CENP-C, which may also contribute to the expansion (Sugimoto et al., 1997; Kim et al., 2008, 2012).
Phospho-dependent regulation of the assembly and disassembly of the adaptive kinetochore explains the functional transitions kinetochores undergo
Assembly of the expandable module depends on phosphorylation by multiple kinases including Aurora B, Haspin, Bub1, Plx1, and Mps1. The essential targets of Mps1 are likely to be on KNL1, whereas the requirement for Plx1 and Haspin may reflect their activation of Aurora B (Ghenoiu et al., 2013; Zhou et al., 2014). Critical substrates of Aurora B in kinetochore formation are not established in Xenopus egg extracts, but Mis12C is a likely target as Aurora B–dependent phosphorylation of Dsn1(Mis13), a subunit of the Mis12C, facilitates CENP-C interaction in human and budding yeast (Yang et al., 2008; Akiyoshi et al., 2013; Kim and Yu, 2015).
Because the size of the expandable module can be experimentally controlled by differential doses of Aurora B inhibitor, Aurora B appears to be required not only for the initial establishment of the outer kinetochore but also for the expansion process. Similarly, an Mps1 inhibitor causes dose-dependent inhibition of kinetochore expansion. Interestingly, in both cases although the activity of the kinase is crucial to expansion, the kinase localization does not have to be predominantly on the expanded region, as we found that Aurora B accumulation underlying the expanded region is not critical (Fig. 6 E) and that Mps1 and Plx1 are localized mainly to the core (Fig. 7 A). Thus, like the models for Aurora B, there must be substantial diffusion of these pools of kinase to their substrates, which is consistent with the importance of Mps1 turnover at the kinetochore (Howell et al., 2004; Nijenhuis et al., 2013). Conversely, inhibition of PP1 by inhibitor 2 permitted kinetochore expansion even in the presence of microtubules. These results demonstrate that the size of the expandable module is tunable by phosphorylation level, determined by the balance of kinase and phosphatase activities.
Dynamic, phospho-dependent kinetochore expansion is consistent with the observation that the SAC exhibits a graded response to the number of unattached kinetochores (Minshull et al., 1994; Chen, 2004; Collin et al., 2013). It may also explain why CENP-E is not essential for SAC activation induced by a loss of microtubule assembly but contributes to SAC maintenance in response to a single unattached kinetochore in mouse tissue-culture cells (Weaver et al., 2003); in the presence of multiple unattached kinetochores, the large CENP-E–dependent kinetochore expansion may not be required for SAC activation in somatic cells. Along the same lines, CENP-E was recently shown to contribute to stable end-on attachment in human tissue culture cells in addition to its role in chromosome congression via supporting lateral attachment (Kapoor et al., 2006; Gudimchuk et al., 2013). CENP-E can support these distinct functions through an altered level of kinetochore recruitment relative to the Ndc80C as MT attachments are made.
Our study suggests that microtubule attachment status not only controls recruitment of a subset of outer kinetochore components to a static platform but instead causes a fundamental reorganization of kinetochore structure. This dynamic kinetochore expansion may represent a novel example of phospho-dependent multiprotein copolymerization.
Materials and methods
Kinetochore assembly and SAC assays in Xenopus egg extracts
To assemble kinetochores, 1,600/µl demembranated sperm were added to CSF-arrested meiotic metaphase II Xenopus egg extracts (Murray, 1991) and cycled through interphase using 0.3 mM CaCl2 for 90 min followed by addition of 1 vol of fresh CSF-arrested extract for 45 min at 20°C. For nocodazole treatments, 33 µM nocodazole was added immediately after addition of fresh CSF extract. To test SAC function, high sperm concentrations (5,000–10,000/µl) were added to mitotic extracts, cycled, and treated with nocodazole and then were challenged to release into interphase by adding 0.6 mM CaCl2 and monitored by Western blotting for 60 min.
Chromosome purification
The method previously described was followed (Funabiki and Murray, 2000). In brief, extracts containing 2,500/µl sperm were cycled in the presence of 5 µM biotin-16-dUTP, diluted in dilution buffer (DB; 10 mM K-Hepes, 50 mM β-glycerophosphate, 50 mM NaF, 20 mM EGTA, 2 mM EDTA, 0.5 mM spermine, 1 mM PMSF, and 200 mM sucrose), layered over 60% sucrose in DB, and spun at 11,000 g. The chromosome pellet was then incubated with M-280 streptavidin Dynabeads (Invitrogen) for 30 min at 4°C with constant rotation and washed with DB containing 30% sucrose.
Immunodepletion, mRNA reconstitution, and kinase or phosphatase inhibition
Antibodies or control IgG were coupled to protein A Dynabeads (Invitrogen) according to the manufacturer’s directions. For Haspin depletion, 0.1 mg/ml cycloheximide was added to inhibit translation, which was not necessary for other depletions. For reconstitution experiments, in vitro transcribed mRNA using SP6 polymerase was added to CSF extract before cycling to a concentration range of 40–300 ng/µl, depending on the desired level of protein expression. For expression of Myc–CENP-C, 40 ng/µl mRNA was added only to the fresh CSF used for cycling to metaphase, as expression of high concentrations of the N terminus of CENP-C was found to inhibit kinetochore expansion. Constructs used for mRNA expression contained FL wild-type or K872A Bub1 or the indicated CENP-C truncations (Fig. 2 D) coding sequence following an SP6 promotor and six Myc tags in a pCS2 vector. Depletion conditions and inhibitor concentrations are provided in Table 1 and Table 2. Kinase and phosphatase inhibitors were added after addition of fresh CSF used for cycling interphase extracts back to metaphase at the following concentrations: 10 µM BI-2536 (JS Research Chemicals Trading); 50–150 µM Mps1-IN1 (a gift from N.S. Gray, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA; Kwiatkowski et al., 2010); 1–3 µM Protein Phosphatase Inhibitor 2 (New England Biolabs, Inc.); and 25–100 µM ZM447439 (ChemieTek).
Table 1. Extract depletion conditions.
| Antibody | Antibody/100-µl beads | Antibody beads to deplete 50 µl extract |
|---|---|---|
| µg | µl | |
| Bub1 | 10 | 100 |
| BubR1 | 10 | 100 |
| CENP-C | 1.8 | 16.7 |
| CENP-E | 10 | 100 (2 rounds of 50) |
| Haspin | 7 | 100 (2 rounds of 50) |
| INCENP | 10 | 100 |
| Mad1 | 10 | 100 (2 rounds of 50) |
| Sgo1&2 | 16 each | 200 (2 rounds of 100) |
Table 2. Kinase inhibition treatment conditions.
| Inhibitor | Concentration |
|---|---|
| µM | |
| BI 2536 | 10 |
| Mps1-IN1 | 50–150 |
| ZM447439 | 25–100 |
| I-2 | 1–3 |
Immunofluorescence, immunoblots, and antibodies
For immunofluorescence, mitotic spindles or chromosomes in the absence of microtubules were fixed in 2% formaldehyde, spun down onto glass coverslips, treated with methanol for 3–5 min, rehydrated in TBS-TX (10 mM Tris, 150 mM NaCl, and 0.1% Triton X-100), and blocked overnight in AbDil (TBS-TX and 2% BSA) before antibody incubations (Desai et al., 1999; Funabiki and Murray, 2000). Bovine tubulin was conjugated to Alexa Fluor 488, 594, or 647 fluorophores (Life Technologies) and added to extracts at 0.2 µM before cycling. Primary and secondary antibodies for immunofluorescence were diluted in AbDil. Samples stained with fluorescently labeled rabbit primary antibodies (done for both BubR1 and Bub1) after staining with a different rabbit primary antibody, which was detected using secondary anti–rabbit antibodies, were blocked with 100 µg/ml rabbit IgG for 30 min before addition of the direct-labeled primary antibody without intervening wash steps, and control single-stained samples were prepared to ensure specificity of the fluorescent signal. For immunoblots, nitrocellulose membranes were blocked with autoclaved 4% nonfat milk in PBS at room temperature, and primary antibodies were diluted in Odyssey Blocking Buffer (LI-COR Biosciences) with 0.1% Tween 20 and detected with IRDye 800CW and 680LT secondary antibodies using the Odyssey Infrared Imaging System (LI-COR Biosciences). All antibody epitopes and species are listed in Table 3. For antibodies against Xenopus proteins, Bub1 and BubR1 antibodies were generated according to Chen (2002) and Sharp-Baker and Chen (2001), and CENP-A antibodies were generated according to Maddox et al. (2003). Dasra antibodies were reported previously (Sampath et al., 2004). CENP-C, -E, -K, -T, and dynein antibodies were provided by A.F. Straight. Mis12, Ndc80, and Zwint antibodies were provided by P.T. Stukenberg (University of Virginia Medical Center, Charlottesville, VA). Mad1 antibodies were provided by R.H. Chen (Academia Sinica, Taipei, Taiwan) and T. Lorca (Centre de Recherche de Biochimie Macromoléculaire, Montpellier, France), who also provided CENP-E antibodies. Mad2 and Mps1 antibodies were provided by Y. Mao (Columbia University, New York, NY). Xorbit antibodies were provided by R. Heald (University of California Berkeley, Berkeley, CA). Sgo1 and Sgo2 antibodies were provided by A. Losada (Spanish National Cancer Research Center, Madrid, Spain). For antibodies against human proteins, BubR1 (MAB3612; EMD Millipore) and ACA (#15-235; Antibodies Inc.) antibodies were purchased. Antibody concentrations used for immunofluorescence and immunoblots are listed in Table 3.
Table 3. Antibodies used in this study.
| Reactivity | Epitope | IF condition | WB condition | Species | Reference or source |
|---|---|---|---|---|---|
| aa | |||||
| Xenopus | |||||
| Bub1 | 274–467 | 5 µg/ml | 1.5 µg/ml | Rabbit | This study; Sharp-Baker and Chen, 2001 |
| BubR1 | 189–369 | 5 µg/ml | 1.5 µg/ml | Rabbit | This study; Chen, 2002 |
| CENP-A | 1–50 | 1.5 µg/ml | ND | Rabbit | This study; Maddox et al., 2003 |
| CENP-C | 207–296 | 1 µg/ml | 1 µg/ml | Rabbit | Milks et al., 2009 |
| CENP-E | 826–1,106 | 2 µg/ml | ND | Rabbit | Milks et al., 2009 |
| CENP-E | 2,397–2,954 | Rabbit | T. Lorca; Wood et al., 1997; Vigneron et al., 2004 | ||
| CENP-K | 276–287 | 2 µg/ml | ND | Rabbit | Milks et al., 2009 |
| CENP-T | 1–603 | 0.3 µg/ml | 0.3 µg/ml | Rabbit | A.F. Straight |
| Dasra A | 284–296 | 1.4 µg/ml | 7 µg/ml | Rabbit | Sampath et al., 2004) |
| dynein | Undiluted hybridoma supernatant | ND | Mouse | A.F. Straight | |
| H2A_T120ph | 1:100 | ND | Rabbit | Abcam (ab111492) | |
| H3_T3ph | ND | 1:5,000 | Rabbit | EMD Millipore (07-424) | |
| Mad1 | FL | 1:200 | ND | Rabbit | R.H. Chen; Chen et al., 1998 |
| Mad1 | 1–12 | Rabbit | T. Lorca; Vigneron et al., 2004 | ||
| Mad2 | FL | 1:500 | ND | Rabbit | Y. Mao; Chen et al., 1996; Mao et al., 2005 |
| Mps1 | 1–14 | 1:200 | ND | Rabbit | Y. Mao; Abrieu et al., 2001 |
| Mis12 | FL | 1:500 | 1:500 | Rabbit | Emanuele et al., 2005 |
| Ndc80 | FL | 1:500 | 1:1,000 | Rabbit | McCleland et al., 2003 |
| α-Tubulin | ND | 1:20,000 | Mouse | Sigma-Aldrich (T9026) | |
| Xorbit | 799–1,457 | 1:4,000 | ND | Rabbit | R. Heald; Hannak and Heald, 2006 |
| Zwint | FL | 1:500 | 1:1,000 | Rabbit | Emanuele et al., 2005 |
| Human | |||||
| ACA | CENP-A,B,C | 1:800 | ND | Human | Antibodies, Inc. (15-235) |
| BubR1 | 1:1,000 | ND | Rabbit | EMD Millipore (MAB3612) |
IF, immunofluorescence; WB, Western blotting.
Human cell culture and immunofluorescence
RPE-hTERT cells were grown in DMEM/F-12 media (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals) and Penicillin-Streptomycin (Life Technologies). For immunofluorescence, cells grown on coverslips were treated for 4 h with 10 nM MG132 and DMSO or 3.3 µM nocodazole with or without 2 µM ZM447439, fixed in 2% paraformaldehyde in PBS for 10 min, and permeabilized with PBS containing 0.5% triton X-100 for 10 min. Incubations with antibody and DAPI were done as for extract.
Microscopy and image analysis
All microscopy data were acquired using a DeltaVision optical microscope experimental (OMX) V4/Blaze 3D-SIM super-resolution microscope system (GE Healthcare) maintained by the Rockefeller University Bio-Imaging Resource Center. This OMX system is fitted with a 100×/1.40 NA UPLSAPO oil objective (Olympus) and three electron-multiplying charge-coupled device cameras (Evolve; Photometrics) that were used in EM gain mode fixed at 170 electrons per count. Immersion oils ranging in refractive index from 1.512–1.518 were used depending on the ambient temperature (kept between 20 and 23°C) and fluorochromes used (including Alexa Fluor 488, 594, 555, and 647). 405-, 488-, and 568-nm laser lines were used for excitation, and the corresponding emission filter sets were 436/31, 528/48, and 609/37 nm, respectively. Two identical stacks of optical sections with 125-nm spacing were collected for each dataset, first using conventional wide-field illumination of all channels and then using structured illumination of selected channels. The system produces an effective xy pixel size of 40 nm in the 3D-SIM mode. After acquisition, all datasets were processed using softWoRx v. 6.1 software (GE Healthcare) using Optical Transfer Functions (OTFs) generated from point spread functions acquired with 100 nm (green and red) or 170 nm (blue) FluoSpheres (Invitrogen/Life Technologies). Wide-field datasets were deconvolved using constrained iterative deconvolution using softWoRx default settings, and structured illumination datasets were reconstructed using channel-specific k0 values, custom OTFs, and Wiener filters of 0.005, 0.002, and 0.002 for the blue, green, and red channels, respectively, using softWoRx. Image registration was performed with parameters refined using 100 nm TetraSpeck beads (Invitrogen/Life Technologies). Image registration parameters and OTFs were frequently refined by the resource center staff. Imaris software (Bitplane) and ImageJ (National Institutes of Health) were used for 3D visualization and generating projection images for visualization. Image segmentation and quantification of intensity, volume, and extent were performed using MATLAB (MathWorks). Because the expanded outer kinetochores resulting from nocodazole treatment were usually continuous around sister centromeres and often continuous among clusters of multiple centromeres, we found quantification of whole nuclear signal to be more robust than attempts to assign outer kinetochore signal to specific centromeres (performed using watershed segmentation, only for Xenopus data in Fig. S1 [A and B] for comparison to total nuclear measurements).
Online supplemental material
Fig. S1 shows further characterization of kinetochore expansion and Myc–CENP-C expression. Fig. S2 shows further characterization of the requirement of both outer kinetochore components and CENP-C for expansion. Fig. S3 shows a dominant-negative effect of CENP-C overexpression. Fig. S4 shows further characterization of the effects of CENP-C immunodepletion and addback on expansion and the SAC. Fig. S5 shows further characterization of the roles of kinases on expansion including depletion and chemical inhibition. Video 1 shows a 3D rendering of a chromosome cluster with extensive kinetochore expansion. Video 2 shows a 3D rendering focusing on a single pair of sister kinetochores from the same chromosome mass shown in Video 1. Video 3 shows a 3D rendering of an inner centromeric histone modification underlying expanded kinetochores. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506020/DC1.
Supplementary Material
Acknowledgments
We thank R.H. Chen, R. Heald, T. Lorca, A. Losada, Y. Mao, N. Gray, A. Straight, and P.T. Stukenberg for reagents, A. North and K. Thomas in the Rockefeller University Bio-Imaging Resource Center (BIRC) for help with data acquisition and processing, and C. Jenness, A. Straight, J. Xue, and C. Zierhut for critical reading of the manuscript.
D.J. Wynne was supported by a Ruth L. Kirschstein Institutional National Research Service Award postdoctoral fellowship (F32GM103147). This work was supported by a grant from National Institutes of Health to H. Funabiki (R01GM075249) and to the BIRC (S10RR031855) from the National Center For Research Resources.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health. The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere–associated network
- CPC
- chromosomal passenger complex
- CSF
- cytostatic factor
- DB
- dilution buffer
- FL
- full length
- OMX
- optical microscope experimental
- OTF
- optical transfer function
- PP1
- protein phosphatase 1
- RPE-hTERT
- human telomerase-immortalized retinal pigmented epithelial
- SAC
- spindle assembly checkpoint
- SIM
- structured illumination microscopy
References
- Abrieu A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., and Labbé J.C.. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93. 10.1016/S0092-8674(01)00410-X [DOI] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C.R., and Biggins S.. 2013. The aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics. 194:785–789. 10.1534/genetics.113.150839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y., Salic A., Dasso M., and Arnaoutov A.. 2007. Bub1 is essential for assembly of the functional inner centromere. J. Cell Biol. 176:919–928. 10.1083/jcb.200609044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur D.K., Gassmann R., Cook B., Oegema K., and Desai A.. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 342:1239–1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., and Desai A.. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594. 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.-H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158:487–496. 10.1083/jcb.200204048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H. 2004. Phosphorylation and activation of Bub1 on unattached chromosomes facilitate the spindle checkpoint. EMBO J. 23:3113–3121. 10.1038/sj.emboj.7600308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.-H., Waters J.C., Salmon E.D., and Murray A.W.. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. 10.1126/science.274.5285.242 [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Shevchenko A., Mann M., and Murray A.W.. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., and Sullivan K.F.. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Collin P., Nashchekina O., Walker R., and Pines J.. 2013. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 15:1378–1385. 10.1038/ncb2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R., Baumann C., and Viveiros M.M.. 2015. ATRX contributes to epigenetic asymmetry and silencing of major satellite transcripts in the maternal genome of the mouse embryo. Development. 142:1806–1817. 10.1242/dev.118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., and McEwen B.F.. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. 10.1091/mbc.E04-09-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Elowe S., Hümmer S., Uldschmid A., Li X., and Nigg E.A.. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21:2205–2219. 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., McCleland M.L., Satinover D.L., and Stukenberg P.T.. 2005. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol. Biol. Cell. 16:4882–4892. 10.1091/mbc.E05-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S.M., and Stukenberg P.T.. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254. 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Gaussen A., Bieling P., Morin V., Prieto S., Fesquet D., Surrey T., and Abrieu A.. 2008. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol. Cell. 29:637–643. 10.1016/j.molcel.2008.01.004 [DOI] [PubMed] [Google Scholar]
- Foley E.A., and Kapoor T.M.. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., and Murray A.W.. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. 10.1016/S0092-8674(00)00047-7 [DOI] [PubMed] [Google Scholar]
- Funabiki H., and Wynne D.J.. 2013. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma. 122:135–158. 10.1007/s00412-013-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399. 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971. 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenoiu C., Wheelock M.S., and Funabiki H.. 2013. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol. Cell. 52:734–745. 10.1016/j.molcel.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudimchuk N., Vitre B., Kim Y., Kiyatkin A., Cleveland D.W., Ataullakhanov F.I., and Grishchuk E.L.. 2013. Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15:1079–1088. 10.1038/ncb2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M.G., Shao L., Carlton P.M., Wang C.J., Golubovskaya I.N., Cande W.Z., Agard D.A., and Sedat J.W.. 2008. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94:4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., and Heald R.. 2006. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat. Protoc. 1:2305–2314. 10.1038/nprot.2006.396 [DOI] [PubMed] [Google Scholar]
- Hara K., Tydeman P., and Kirschner M.. 1980. A cytoplasmic clock with the same period as the division cycle in Xenopus eggs. Proc. Natl. Acad. Sci. USA. 77:462–466. 10.1073/pnas.77.1.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., and Fukagawa T.. 2008. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 19:843–854. 10.1091/mbc.E07-06-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60. 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., and Salmon E.D.. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964. 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Kallio M.J., McCleland M.L., Stukenberg P.T., and Gorbsky G.J.. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900–905. 10.1016/S0960-9822(02)00887-4 [DOI] [PubMed] [Google Scholar]
- Kang Y.H., Park J.E., Yu L.R., Soung N.K., Yun S.M., Bang J.K., Seong Y.S., Yu H., Garfield S., Veenstra T.D., and Lee K.S.. 2006. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol. Cell. 24:409–422. 10.1016/j.molcel.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., and Khodjakov A.. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science. 311:388–391. 10.1126/science.1122142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., and Funabiki H.. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D.M., Kim T., Rigney M., Hattersley N., Desai A., and Cheeseman I.M.. 2015. The outer kinetochore protein KNL-1 contains a defined oligomerization domain in nematodes. Mol. Biol. Cell. 26:229–237. 10.1091/mbc.E14-06-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Yu H.. 2015. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208:181–196. 10.1083/jcb.201407074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Heuser J.E., Waterman C.M., and Cleveland D.W.. 2008. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J. Cell Biol. 181:411–419. 10.1083/jcb.200802189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sun H., Tomchick D.R., Yu H., and Luo X.. 2012. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. USA. 109:6549–6554. 10.1073/pnas.1118210109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D., Kirschner M., and Scherson T.. 1987. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 48:399–407. 10.1016/0092-8674(87)90191-7 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., and Yanagida M.. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011. 10.1128/MCB.00815-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N., Jelluma N., Filippakopoulos P., Soundararajan M., Manak M.S., Kwon M., Choi H.G., Sim T., Deveraux Q.L., Rottmann S., et al. 2010. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat. Chem. Biol. 6:359–368. 10.1038/nchembio.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906. 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P., Straight A., Coughlin P., Mitchison T.J., and Salmon E.D.. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382. 10.1083/jcb.200301088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., and Khodjakov A.. 2011. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 146:555–567. 10.1016/j.cell.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Abrieu A., and Cleveland D.W.. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 114:87–98. 10.1016/S0092-8674(03)00475-6 [DOI] [PubMed] [Google Scholar]
- Mao Y., Desai A., and Cleveland D.W.. 2005. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J. Cell Biol. 170:873–880. 10.1083/jcb.200505040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., and Heald R.. 2006. The long and the short of it: linker histone H1 is required for metaphase chromosome compaction. Cell Cycle. 5:589–591. 10.4161/cc.5.6.2581 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., and Nigg E.A.. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- Matsumura S., Toyoshima F., and Nishida E.. 2007. Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J. Biol. Chem. 282:15217–15227. 10.1074/jbc.M611053200 [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Hsieh C.E., Mattheyses A.L., and Rieder C.L.. 1998. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 107:366–375. 10.1007/s004120050320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks K.J., Moree B., and Straight A.F.. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell. 20:4246–4255. 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Sun H., Tonks N.K., and Murray A.W.. 1994. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 79:475–486. 10.1016/0092-8674(94)90256-9 [DOI] [PubMed] [Google Scholar]
- Monen J., Maddox P.S., Hyndman F., Oegema K., and Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. 10.1038/ncb1331 [DOI] [PubMed] [Google Scholar]
- Murray A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. 10.1016/S0091-679X(08)60298-8 [DOI] [PubMed] [Google Scholar]
- Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M.H.J., Snel B., Perrakis A., and Kops G.J.P.L.. 2013. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201:217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436. 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack K., Primorac I., Vleugel M., Krenn V., Maffini S., Hoffmann I., Kops G.J., and Musacchio A.. 2015. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife. 4:e05269 10.7554/eLife.05269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M., and Fukagawa T.. 2011. The ABCs of CENPs. Chromosoma. 120:425–446. 10.1007/s00412-011-0330-0 [DOI] [PubMed] [Google Scholar]
- Petrovic A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S., et al. 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 53:591–605. 10.1016/j.molcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Nelson C.R., and Biggins S.. 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19:1182–1187. 10.1016/j.cub.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Venkei Z., Bolanos-Garcia V.M., Debski J., Dadlez M., and Glover D.M.. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21:399–405. 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Rago F., Gascoigne K.E., and Cheeseman I.M.. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25:671–677. 10.1016/j.cub.2015.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Vagnarelli P., Dong Y., Hori T., McEwen B.F., Fukagawa T., Flors C., and Earnshaw W.C.. 2010. A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. USA. 107:10484–10489. 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 1982. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79:1–58. 10.1016/S0074-7696(08)61672-1 [DOI] [PubMed] [Google Scholar]
- Rieder C.L., and Alexander S.P.. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81–95. 10.1083/jcb.110.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg J.S., Cross F.R., and Funabiki H.. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947. 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S.C., Ohi R., Leismann O., Salic A., Pozniakovski A., and Funabiki H.. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202. 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P.M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M.C., Agard D.A., Gustafsson M.G.L., et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 320:1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613. 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398. 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J.V., Botvinick E., Bonday Z., Furnari F., Berns M., and Cleveland D.W.. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H., and Chen R.-H.. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250. 10.1083/jcb.153.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899. 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., and Salmon E.D.. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875. 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear J.H., and Roth M.B.. 2004. The Caenorhabditis elegans kinetochore reorganizes at prometaphase and in response to checkpoint stimuli. Mol. Biol. Cell. 15:5187–5196. 10.1091/mbc.E04-06-0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., and Himeno M.. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132–141. 10.1023/A:1018422325569 [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk S.J.E., Vleugel M., Teixeira A., and Kops G.J.P.L.. 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 23:745–755. 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Hori T., Nishino T., Usukura J., Miyagi A., Morikawa K., and Fukagawa T.. 2011. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J. Cell Biol. 193:125–140. 10.1083/jcb.201012050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower D.A., Jordan M.A., and Wilson L.. 1996. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskeleton. 35:121–133. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., and Hardwick K.G.. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181. 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., and Salmon E.D.. 2012. The KMN protein network—chief conductors of the kinetochore orchestra. J. Cell Sci. 125:5927–5936. 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S., Prieto S., Bernis C., Labbé J.C., Castro A., and Lorca T.. 2004. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 15:4584–4596. 10.1091/mbc.E04-01-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684. 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Bonday Z.Q., Putkey F.R., Kops G.J., Silk A.D., and Cleveland D.W.. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551–563. 10.1083/jcb.200303167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe F.G., and Straight A.F.. 2013. Functions of the centromere and kinetochore in chromosome segregation. Curr. Opin. Cell Biol. 25:334–340. 10.1016/j.ceb.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E., Basto R., Serr M., Scaërou F., Karess R., and Hays T.. 2001. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3:1001–1007. 10.1038/ncb1101-1001 [DOI] [PubMed] [Google Scholar]
- Wong O.K., and Fang G.. 2006. Loading of the 3F3/2 antigen onto kinetochores is dependent on the ordered assembly of the spindle checkpoint proteins. Mol. Biol. Cell. 17:4390–4399. 10.1091/mbc.E06-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S., and Cleveland D.W.. 1997. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 91:357–366. 10.1016/S0092-8674(00)80419-5 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., and Watanabe Y.. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., et al. 2008. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283:26726–26736. 10.1074/jbc.M804207200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Tian X., Zhu C., Wang F., and Higgins J.M.. 2014. Polo-like kinase-1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep. 15:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Jenness C., Kimura H., and Funabiki H.. 2014. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 21:617–625. 10.1038/nsmb.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.