KLP-7/MCAK regulates kinetochore–microtubule attachment and spindle tension to promote the coalescence of early spindle pole foci, which produces a bipolar structure during the acentrosomal process of oocyte meiotic spindle assembly in C. elegans.
Abstract
During oocyte meiotic cell division in many animals, bipolar spindles assemble in the absence of centrosomes, but the mechanisms that restrict pole assembly to a bipolar state are unknown. We show that KLP-7, the single mitotic centromere–associated kinesin (MCAK)/kinesin-13 in Caenorhabditis elegans, is required for bipolar oocyte meiotic spindle assembly. In klp-7(−) mutants, extra microtubules accumulated, extra functional spindle poles assembled, and chromosomes frequently segregated as three distinct masses during meiosis I anaphase. Moreover, reducing KLP-7 function in monopolar klp-18(−) mutants often restored spindle bipolarity and chromosome segregation. MCAKs act at kinetochores to correct improper kinetochore–microtubule (k–MT) attachments, and depletion of the Ndc-80 kinetochore complex, which binds microtubules to mediate kinetochore attachment, restored bipolarity in klp-7(−) mutant oocytes. We propose a model in which KLP-7/MCAK regulates k–MT attachment and spindle tension to promote the coalescence of early spindle pole foci that produces a bipolar structure during the acentrosomal process of oocyte meiotic spindle assembly.
Introduction
Two sequential meiotic cell divisions produce a haploid oocyte from a diploid precursor. During these two divisions, called meiosis I and II, small bipolar spindles assemble near the cell cortex and ultimately extrude extraneous chromosomes into polar bodies during these highly asymmetric divisions (Fabritius et al., 2011). In many animals, including nematodes, insects, and vertebrates, oocyte meiotic spindles assemble without the centrosomes that dominate mitotic bipolar spindle assembly. During mitosis, at least two distinct pathways for microtubule nucleation function during spindle assembly, one centrosome dependent and the other chromosome dependent (Meunier and Vernos, 2012). During oocyte meiosis, motor proteins and other microtubule-associated proteins, but not centrosomes, orchestrate bipolar spindle assembly (Dumont and Desai, 2012; Meunier and Vernos, 2012; Ohkura, 2015). Our understanding of how spindle poles assemble without centrosomes remains limited, and we are using Caenorhabditis elegans oocyte meiotic cell division as a model system to investigate this process.
With its powerful genetics and transparent anatomy, C. elegans provides a useful model system for investigating oocyte spindle assembly (Yamamoto et al., 2006; Müller-Reichert et al., 2010). Upon ovulation in C. elegans hermaphrodites, the oocyte nuclear envelope breaks down, the oocyte is fertilized as it is engulfed by the spermathecum, and spindle assembly begins. During prometaphase, microtubules accumulate around the chromosomes and rapidly organize into a compact bipolar spindle that lies roughly parallel to the cell membrane. The spindle then shortens and rotates such that its pole-to-pole axis becomes perpendicular to the overlying cell membrane. During anaphase, the homologous chromosomes separate and move toward opposite poles, with one set of sister chromatids extruded into a polar body. The process then repeats itself during meiosis II to segregate the remaining pairs of sister chromatids and establish the haploid oocyte contribution to the zygote (Albertson and Thomson, 1993; McNally et al., 2006).
Thus far, only a few C. elegans genes required for bipolar oocyte meiotic spindle assembly have been identified. Two, mei-1 and mei-2, encode the catalytic and regulatory subunits of a complex called katanin that binds to and severs microtubules (McNally and Vale, 1993; Hartman et al., 1998). Loss of mei-1 or mei-2 results in disorganized and apolar meiotic spindles, suggesting that katanin is required for pole assembly (Clark-Maguire and Mains, 1994b; Srayko et al., 2000, 2006). More recently, we have shown that both the microtubule-severing activity of mei-1 and the microtubule-scaffolding protein ASPM-1 contribute to pole assembly (Connolly et al., 2014). In addition, the kinesin-12 family member klp-18 promotes bipolarity, with loss of klp-18 resulting in monopolar spindles (Segbert et al., 2003; Wignall and Villeneuve, 2009; Connolly et al., 2014; Muscat et al., 2015). Presumably, KLP-18 promotes bipolarity by cross-linking antiparallel microtubules and sliding them in opposite directions, much as vertebrate kinesin-12 family members are thought to do during mitosis (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013). Although such a mechanism can explain how klp-18 promotes bipolarity, why only two poles assemble is not clear. Here, we report our identification of conditional loss-of-function mutations in the C. elegans kinesin-13/mitotic centromere–associated kinesin (MCAK) gene klp-7 and show that this kinesin limits oocyte meiotic spindles to a bipolar state.
Kinesin-13/MCAK family members do not walk along microtubules but rather associate with their minus and plus ends to promote depolymerization (Ems-McClung and Walczak, 2010). During mitosis, these microtubule depolymerases appear to have multiple roles, promoting proper kinetochore–microtubule (k–MT) attachment of paired sister chromatids to opposite poles and the poleward flux of microtubule subunits during anaphase (Wordeman et al., 2007; Ems-McClung and Walczak, 2010). During mitosis in the early C. elegans embryo, KLP-7/MCAK limits the number of microtubules that grow out from centrosomes (Srayko et al., 2005). Less is known about kinesin-13/MCAK during oocyte meiotic spindle assembly. Previous studies have identified roles for MCAKs in promoting chromosome alignment and congression to the metaphase plate in mouse oocytes (Illingworth et al., 2010; Vogt et al., 2010) and in limiting oocyte meiotic spindle length in Drosophila melanogaster and Xenopus laevis (Mitchison et al., 2005; Ohi et al., 2007; Zou et al., 2008). Here, we investigate the meiotic function of KLP-7, the sole kinesin-13/MCAK family member in C. elegans, and present evidence that it functions to limit both microtubule accumulation and pole number during oocyte meiotic spindle assembly.
Results
Two recessive, temperature-sensitive (TS) klp-7 alleles
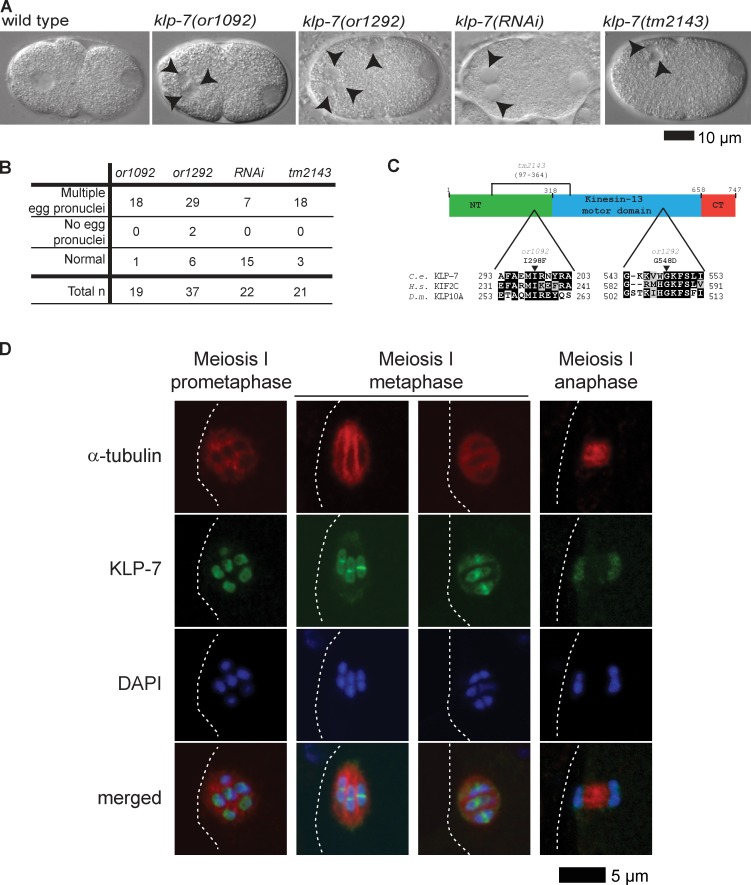
To identify essential genes that mediate oocyte meiotic spindle assembly, we generated a collection of TS, embryonic-lethal mutants and used Nomarski optics to examine live one-cell-stage embryos produced at the restrictive temperature. Wild-type zygotes typically possess a single egg pronucleus that appears after the completion of meiosis I and II, in addition to a single sperm pronucleus (Fig. 1 A; Albertson, 1984; Albertson and Thomson, 1993). We found two recessive mutants called or1092ts and or1292ts that, when L4 larvae raised at the permissive temperature of 15°C were matured to adulthood at the restrictive temperature of 26°C, produced one-cell-stage embryos with multiple egg pronuclei (Fig. 1, A and B), a phenotype associated with defects in oocyte meiotic spindle function (O’Rourke et al., 2011; Connolly et al., 2014).
Figure 1.
Identification of TS klp-7(−) mutants and localization of KLP-7 to meiosis I spindles. (A) Nomarski images of live one-cell-stage wild-type and mutant embryos. Anterior oocyte and posterior sperm pronuclei are to the left and right, respectively. Arrowheads indicate extra oocyte pronuclei in klp-7(−) mutants. (B) Frequency of abnormal number of oocyte pronuclei in one-cell-stage klp-7(−) mutants after the completion of meiosis I and II; wild-type zygotes always have a single oocyte pronucleus. (C) Domain diagram and partial sequence alignment of KLP-7 and orthologues. Arrowheads indicate altered residues in TS alleles; the potentially in-frame tm2143 deletion (see Results section, third paragraph) is marked with a bracket. (D) Immunolocalization of KLPL-7 during meiosis I in fixed wild-type embryos (Materials and methods section KLP-7 immunofluorescence). White dashed lines indicate the oocyte plasma membrane.
To identify the causal mutations in or1092ts and or1292ts mutants, we used both visible marker genetic mapping and whole genome sequencing to identify candidate mutations in klp-7 (see Materials and methods section Positional cloning), which encodes the only kinesin-13/MCAK family member in C. elegans (Siddiqui, 2002; Schlaitz et al., 2007). In or1092ts genomic DNA, we found an isoleucine to phenylalanine change (ATT to TTT) at codon 298 in the neck domain, and in or1292ts, a glycine to aspartic acid change (GGC to GAC) at codon 548 in the motor domain (Fig. 1 C). Consistent with these two mutations being responsible for conditional lethality, or1092ts and or1292ts failed to complement each other, and both failed to complement a deletion allele, klp-7(tm2143) (Tables 1 and 2 and Fig. 1 C). Finally, a GFP fusion to KLP-7 (Schlaitz et al., 2007), which localized to both chromosomes and spindle poles during meiosis I and II (unpublished data; see Fig. 1 D for immunolocalization of KLP-7 in fixed oocyte meiosis I spindles), rescued klp-7(or1092ts) embryonic lethality at the restrictive temperature (Table 1). We conclude that or1092ts and or1292ts are klp-7 alleles.
Table 1. Embryonic viabilityof klp-7(−) mutants.
| Allele | Homozygote embryonic viability (15°C) | Homozygote embryonic viability (26°C) | Heterozygote embryonic viability (26°C) |
|---|---|---|---|
| N2 | 99% (n = 358) | 97% (n = 197) | |
| klp-7(or1092) | 90% (n = 323) | 0.1% (n = 1,530) | 99% (n = 408) |
| klp-7(or1292) | 93% (n = 318) | 0.8% (n = 367) | 97% (n = 604) |
| klp-7(tm2143) | 46% (n = 315) | 0.8% (n = 247) | |
| klp-7(RNAi) | 1% (n = 164) | ||
| Genotype | Embryonic viability (26°C) | ||
| klp-7(or1092)/klp-7(or1292) | 2% (n = 238) | ||
| klp-7(or1092)/klp-7(tm2143)) | 1% (n = 517) | ||
| klp-7(or1292)/klp-7(tm2143)) | 0.5% (n = 220) | ||
| klp-7 Rescue | |||
| klp-7(or1092) unc-68(e587) + gfp::klp-7 | 94% (n = 16) | ||
| klp-7(or1092) unc-68(e587) | 1% (n = 257) |
Embryonic viability (percent hatching) was scored for the wild type and each TS mutant at the permissive temperature (15°C) and after L4 upshifts to the restrictive temperature (26°C). Embryonic viability from heterozygous mutants, after L4 upshifts to the restrictive temperature, was scored to determine whether the mutations are recessive or dominant. Complementation tests also were scored after L4 upshifts of outcross F1s to the restrictive temperature.
Table 2. . Postembryonic phenotypes of klp-7(−) mutants.
| Allele | Sterile hermaphrodites | Males | Number scored |
|---|---|---|---|
| klp-7(or1092) | 99% | 1% | 145 |
| klp-7(or1292) | 100% | 0% | 102 |
| klp-7(tm2143) | 99% | 1% | 166 |
Larval lethality, sterility, and gender were scored after L1 larvae were raised to adulthood at the restrictive temperature.
Genome-wide RNAi screens have reported embryonic lethality and mitotic spindle defects after klp-7(RNAi) knockdown (Gönczy et al., 2000; Kamath et al., 2001; Sönnichsen et al., 2005), but defects in meiotic spindle function have not been reported. Nevertheless, we found that klp-7 RNAi knockdown sometimes resulted in abnormal numbers of oocyte pronuclei (Fig. 1 and Table 1), though with lower penetrance compared with or1092ts and or1292ts. Furthermore, like or1092ts and or1292ts, the deletion allele tm2143 resulted in highly penetrant embryonic lethality, and most of the mutant oocytes had abnormal numbers of egg pronuclei (Fig. 1, A and B; and Table 1). Finally, or1092ts, or1292ts, and tm2143 L1 larvae raised at the restrictive temperature matured into sterile adults (Table 2). We conclude that all three mutant alleles are strong loss-of-function mutations. However, when tm2143 mutants matured to adulthood at 15°C, almost half of their embryos hatched, suggesting that this deletion may not be null even though the deletion, if spliced after transcription to produce an intact reading frame, would remove 268 of 747 predicted amino acids (Materials and methods section Positional cloning), including the phenylalanine at position 298 that is changed to isoleucine in or1092ts. Moreover, the N terminus of vertebrate MCAK has been shown to be required for kinetochore localization (Maney et al., 1998; Walczak et al., 2002).
To further characterize the klp-7 mutant alleles, we used immunolocalization in fixed embryos to examine KLP-7 expression (Fig. 1 D and Fig. S1). In wild-type oocytes during meiosis I, KLP-7 localized to the kinetochore cups described previously for the holocentric bivalents in C. elegans (Howe et al., 2001; Monen et al., 2005; Wignall and Villeneuve, 2009), and also to prominent rings that encircle each bivalent midway along their length. These KLP-7 rings are similar in appearance to those of the kinesin-4/chromokinesin family member KLP-19, which has been shown to be important for the initial capture and alignment of bivalents during oocyte meiosis I (Wignall and Villeneuve, 2009). We detected very little KLP-7 at meiosis I kinetochores in tm2143 and or1292ts mutants and reduced but higher levels in or1092ts mutants. Furthermore, tm2143 mutant spindles were similar in appearance to or1092ts and or1292ts spindles (Fig. S1). These results are consistent with the recessive tm2143, or1092ts, and or1292ts mutations all reducing KLP-7 function. As the tm2143 deletion may remove most of the amino acid residues used to generate the KLP-7 antibodies (Materials and methods section Positional cloning), we cannot conclude whether a mutant KLP-7 protein is expressed in tm2143 mutants. Furthermore, because tm2143 mutants produce many viable embryos when grown at 15°C (Table 1), either this deletion is not null or viability becomes TS in the absence of KLP-7/MCAK function. Because all three mutant alleles were highly penetrant for embryonic lethality and for abnormal numbers of oocyte pronuclei, and because klp-7(tm2143) adults often produced small broods (unpublished data), we focused our analysis on or1092ts and or1292ts.
klp-7 limits microtubule accumulation during oocyte meiotic spindle assembly
To investigate requirements for klp-7, we examined oocyte meiosis I spindle assembly in live klp-7(−) mutants using spinning disk confocal microscopy and transgenic strains that express translational fusions of GFP and mCherry to β-tubulin and Histone H2B to mark microtubules and chromosomes, respectively (see Materials and methods sections C. elegans strains and Microscopy). We found that soon after ovulation and throughout meiosis I in klp-7(−) oocytes, the microtubule signal intensity was greatly increased compared with wild-type oocytes, with the oocyte chromosomes rapidly becoming surrounded by a prominent area of microtubule density (Fig. 2 A and Fig. S2). To quantitatively compare microtubule dynamics in klp-7(−) and wild-type oocytes, we tracked over time the integrated pixel intensity of the GFP::β-tubulin signal during meiosis I (see Materials and methods section Microscopy). In wild-type oocytes, this value was roughly constant until ∼8 min after ovulation and then gradually decreased as meiosis I progressed through anaphase (Fig. 2 B). In contrast, in klp-7(or1292ts) oocytes, the integrated intensity was significantly higher at ovulation compared with the wild type and continued to increase until 8 min after ovulation before decreasing. As other kinesin-13/MCAK family members are known to promote microtubule depolymerization (see Introduction), we conclude that reducing klp-7 function stabilizes microtubules to result in their excessive accumulation during oocyte meiosis I.
Figure 2.
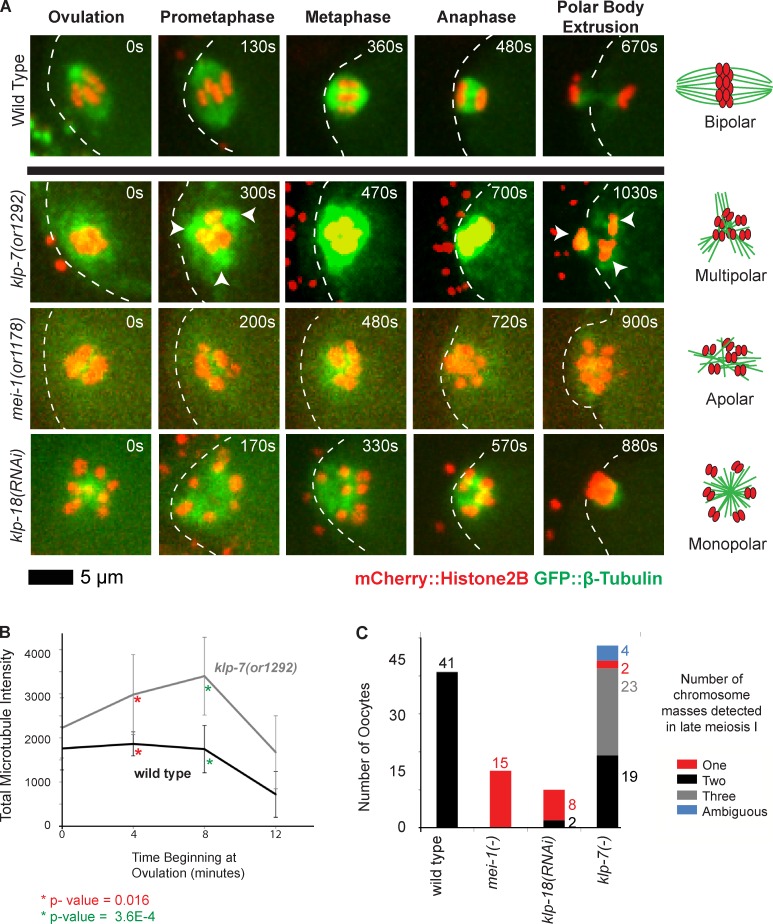
klp-7(−) mutants accumulate an excess of microtubules and exhibit tripolar chromosome segregation during oocyte meiosis I. (A) Time-lapse spinning disk confocal images during meiosis I in live wild-type and mutant embryos expressing mCherry::Histone H2B and GFP::β-tubulin translational fusions to mark chromosomes and microtubules, respectively. White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. Arrowheads indicate possible poles in the klp-7(or1292ts) mutant. Schematics to the right illustrate our interpretations of the pole phenotypes. (B) Integrated GFP::β-tubulin pixel intensity, in arbitrary units, was measured beginning at ovulation and at 4-min intervals (Materials and methods section Microscopy); data are from video recordings of embryos each isolated from individual worms. For wild type, n = 8 and for klp-7(or1292ts), n = 10, at both 4 and 8 min, respectively. Error bars depict the standard deviation among embryos at each time point. P-values, calculated using a Student’s t test, comparing the mean integrated pixel intensities in wild-type and klp-7(−) oocytes for 4 and 8 min are indicated with asterisks. (C) Bar graph shows the number of segregating chromosome masses detected during anaphase for the indicated genotypes, with klp-7(−) referring to the summed results from experiments using RNAi (n = 23), or1092ts (n = 7), and or1292ts (n = 18); each embryo was isolated from a different worm. The numbers of embryos scored as having each phenotype are adjacent to each bar. See Fig. S4 for bar graphs showing the results for each klp-7(−) genotype.
klp-7 limits pole number during oocyte meiotic spindle assembly
We further examined microtubule and chromosome dynamics during meiosis I in live klp-7(−) mutants by comparing them to other mutants with previously characterized oocyte meiotic spindle assembly defects (Fig. 2, A and C; and see Introduction). In wild-type oocytes, the chromosomes and microtubules were initially interspersed but became organized into a bipolar spindle as chromosomes congressed to the metaphase plate, and the chromosomes then moved apart in two discrete masses toward the opposing poles during anaphase. In mei-1(−) oocytes, the meiotic spindle was apolar: chromosomes and microtubules were interspersed throughout meiosis I without any apparent congression or segregation and were often all extruded into the first polar body. In klp-18(−) oocytes, the meiotic spindle was monopolar: the chromosomes were located at the periphery of a central core of microtubules from early in meiosis I, and over time the chromosomes moved into a single, tight aggregate that often was entirely extruded into the first polar body. In contrast, microtubules in klp-7(−) mutant oocytes surrounded the chromosomes throughout most of meiosis I, and the chromosomes often segregated into three distinct masses during anaphase (Fig. 2, A and C; and Figs. S2, S3 A, and S4). Thus, the chromosomes in many klp-7(−) oocytes appeared to move toward extra poles.
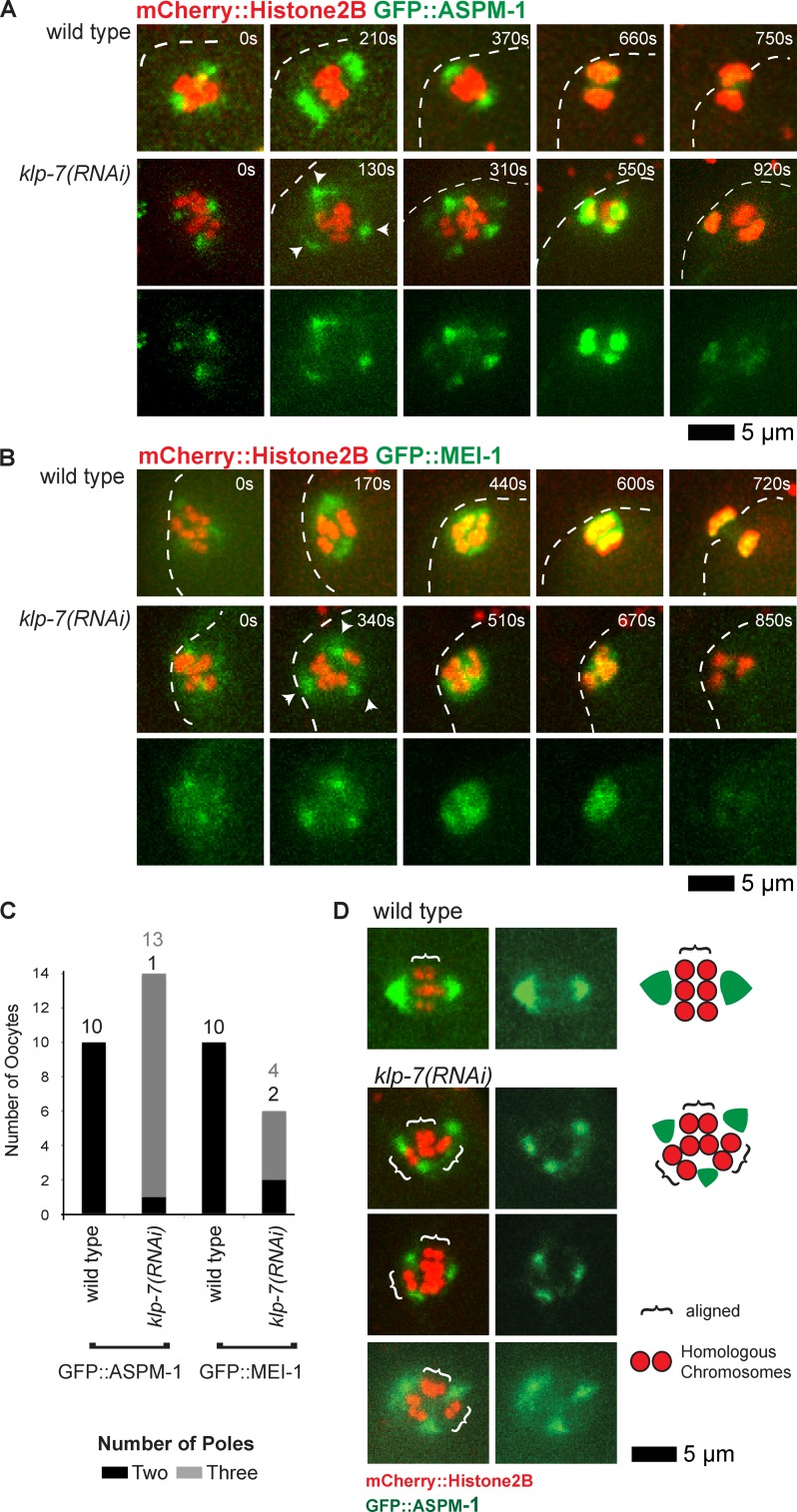
To test the hypothesis that klp-7(−) oocytes assemble extra spindle poles, we next examined GFP::ASPM-1 and GFP::MEI-1, which both mark the meiotic spindle poles early in wild-type oocyte meiosis I (Fig. 3, A and B; Clark-Maguire and Mains, 1994a; van der Voet et al., 2009). As meiosis I proceeded in wild-type oocytes, the two GFP::ASPM-1 foci merged near the metaphase to anaphase transition, encompassing the segregating chromosomes and ultimately residing mostly between the two chromosome masses by late anaphase. Similarly, two GFP::MEI-1 foci in wild-type oocytes merged to surround the chromosomes but then faded to undetectable levels by late anaphase. In all klp-7(−) oocytes, we observed more than two (usually three and sometimes four) foci of both GFP::ASPM-1 and GFP::MEI-1 early in meiosis I (Fig. 3, A–C; and Fig. S4). As meiosis continued, the GFP::ASPM-1 and GFP::MEI-1 foci sometimes coalesced into two loosely organized poles (Fig. 3, B and C) before merging more centrally or fading as chromosomes segregated into two or three distinct masses.
Figure 3.
klp-7(−) mutants assemble extra spindle poles during oocyte meiosis I. (A and B) Time-lapse spinning disk confocal images of meiosis I in live wild-type and mutant embryos expressing mCherry::Histone H2B to mark chromosomes and GFP::ASPM-1 (A) or GFP::MEI-1 (B) translational fusions. White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. Arrowheads indicate possible poles in the klp-7(or1292ts) mutant. (C) Bar graph indicating the number of spindle poles observed during meiosis I in wild-type and klp-7(−) oocytes using both GFP::ASPM-1 and GFP::MEI-1 foci to score pole numbers 400 s after ovulation, or in oocytes that progressed through meiosis more quickly at the last time point before the pole marker coalesced on the chromosomes. The numbers of embryos scored as having each phenotype are adjacent to each bar. See Fig. S4 for bar graphs showing the results for each klp-7(−) genotype. All data are from video micrographs of embryos isolated from different worms in multiple RNAi knockdown experiments. Note that extra poles were almost always observed in klp-7(RNAi) mutants, even though extra oocyte pronuclei were observed less frequently using differential interference contrast optics after the completion of meiosis I and II, compared with TS alleles (Fig. 1 B). (D) Paired homologous chromosomes are aligned along multiple spindle pole axes in klp-7(−) oocytes during meiosis I in oocytes expressing mCherry::Histone H2B to mark chromosomes and GFP::ASPM-1 to mark spindle poles. Brackets indicate homologous chromosome pairs oriented along interpolar axes. Schematics that illustrate representative examples, and a legend, are to the right.
We considered the possibility that extra meiotic spindle poles in klp-7(−) oocytes might result from a failure to eliminate centrosomes during oogenesis (Kim and Roy, 2006). However, as in wild-type oocytes (Leidel et al., 2005), we did not detect any centrosomes in klp-7(−) oocytes using the centriolar markers SAS-6 and SPD-2 (unpublished data). We conclude that klp-7 prevents the assembly of extra acentrosomal oocyte meiotic spindle poles.
To further test our hypothesis that extra oocyte meiotic spindle poles form in klp-7(−) oocytes, we next examined the orientations of paired homologous chromosomes with respect to the wild-type and mutant spindle poles. In bipolar wild-type spindles, homologous chromosomes are oriented with one kinetochore pointing toward each pole and the mid-bivalent region perpendicular to the axis defined by the two poles (Dumont et al., 2010). The mid-bivalent region can be identified by a narrow gap between homologous chromosomes (Fig. 3 D). In klp-7(−) oocytes, homologous chromosome pairs often aligned along an axis between two of three poles and in other cases appeared not to be aligned with respect to any two poles (Fig. 3 D). In most oocytes, one or more homologous pairs of chromosomes oriented properly with respect to one pair of poles, whereas other pairs of homologous chromosomes oriented properly with respect to a different pair of poles. These results suggest that the extra poles detected in klp-7(−) oocytes are functional with respect to paired homologous chromosome capture and alignment, consistent with their frequent segregation into three distinct masses.
Loss of klp-7 partially restores meiotic spindle bipolarity in klp-18(−) mutant oocytes
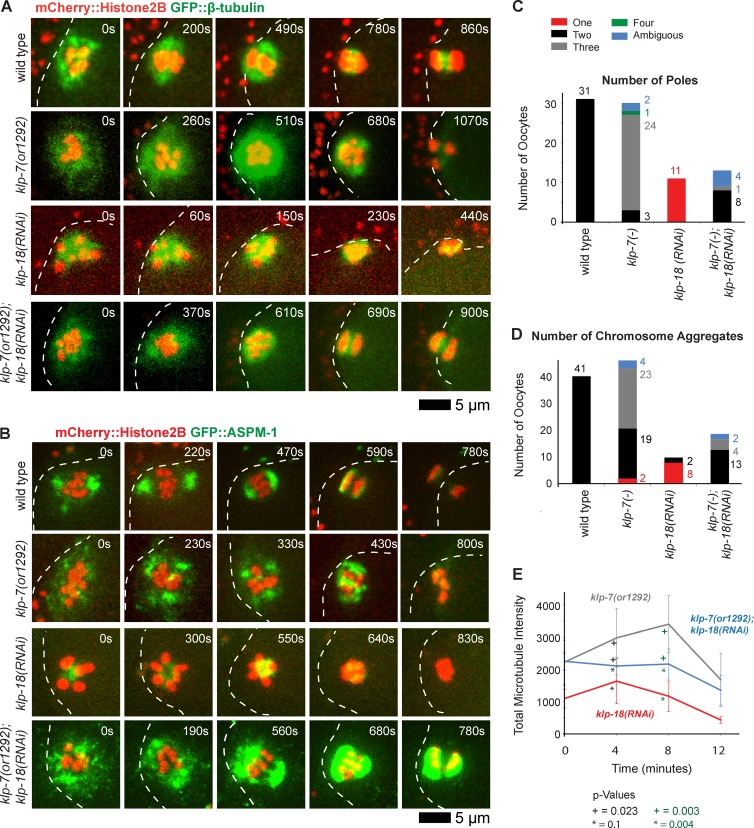
We next used a genetic approach to further test the idea that klp-7 opposes the assembly of extra oocyte meiotic spindle poles. As monopolar oocyte spindles assemble in klp-18(−)/kinesin-12 mutants (see Introduction), we wondered whether reducing klp-7 function in a klp-18(−) background might promote the formation of another pole and restore spindle bipolarity. First, we examined klp-7(−);klp-18(−) double mutant oocytes from a transgenic strain that expressed GFP::β-tubulin and mCherry::Histone H2B to mark microtubules and chromosomes, respectively. Remarkably, we did not observe any monopolar spindles in these double mutants; instead, bipolarity was restored in nearly all cases (4/5 oocytes), with two discrete masses of chromosomes segregating to opposite poles and one mass extruding into a polar body (Fig. 4 A). In 1/5 oocytes examined, three discrete chromosome masses moved apart late in meiosis I, indicating that tripolar oocyte meiotic spindles can assemble in the absence of klp-7, even in a klp-18(−) background.
Figure 4.
Reducing klp-7 function partially restores oocyte meiotic spindle bipolarity in klp-18(−) mutants. (A and B) Time-lapse spinning disk confocal images during meiosis I of live wild-type and mutant embryos expressing either mCherry::Histone H2B and GFP::β-tubulin to mark chromosomes and microtubules, respectively (A), or mCherry::Histone H2B and GFP::ASPM-1 to mark chromosomes and spindle poles, respectively (B). White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. (C and D) As described in Fig. 3, bar graphs indicate the number of GFP::ASPM-1 or GFP::MEI-1 foci in individual oocytes (C) and the number of segregating chromosome masses detected during anaphase from GFP::β-tubulin, GFP::ASPM-1, and GFP::MEI-1 strains expressing mCherry::Histone H2B (D) for the indicated genotypes. The legend (C) provides a color code for pole and aggregate numbers. The numbers from klp-7(RNAi) and klp-7(or1292ts) are merged; see Fig. S4 for the number of embryos analyzed for each genotype to score both pole and aggregate numbers. All data are from video micrographs of individual oocytes, each isolated from different worms in multiple experiments. The numbers of embryos scored as having each phenotype are adjacent to each bar. See Fig. S4 for bar graphs showing the results for each klp-7(−) genotype. (E) Integrated GFP::β-tubulin pixel intensity beginning at ovulation over time during meiosis I from strains of the indicated genotypes. Error bars depict the standard deviation at each time point. P-values comparing the microtubule signals in klp-18(−) and klp-7(−);klp-18(−) oocytes are indicated with asterisks. Note in B and in subsequent figures that GFP::ASPM-1 often appears to accumulate in higher levels in klp-7(−) mutants compared with klp-7(+) genotypes; although we have not quantified this difference, it may result from the increased microtubule accumulation in klp7(−) oocytes or from other influences of KLP-7 on ASPM-1.
To further test our conclusion that loss of klp-7 can restore bipolarity in klp-18(−) oocytes, we also examined klp-7(−);klp-18(−) double mutants from transgenic strains expressing GFP::ASPM-1 and mCherry::Histone H2B to mark oocyte meiotic spindle poles and chromosomes, respectively (Fig. 4, B and C; and Fig. S3 B). As expected, we found that GFP::ASPM-1 localized to a single focus in 11/11 klp-18(−) oocytes and to three or more foci in 20/24 klp-7(−) oocytes. In contrast, GFP::ASPM-1 marked two spindle poles in 8/13 klp-7(−);klp-18(−) double mutant oocytes (Fig. 4 C and Fig. S4); the poles were not as tightly focused and did not move as far apart compared with wild-type oocytes but appeared bipolar (Fig. 4 B). Finally, in klp-7(−);klp-18(−) strains expressing GFP::ASPM-1 (Fig. 4 B) or GFP::β-tubulin (Fig. 4 A), the chromosomes segregated into two distinct masses during anaphase in 13/19 double mutant oocytes, although anaphase bridges were often present (Fig. 4 D and Fig. S4), further documenting the partial rescue of bipolarity. We conclude that reducing klp-7 function can partially restore bipolarity in a monopolar spindle mutant, further supporting our conclusion that klp-7 opposes the assembly of extra oocyte meiotic spindle poles.
We next asked whether loss of klp-7 increased the accumulation of microtubules during meiosis I in a klp-18(−) background (Fig. 4 E), as we observed in klp-7(−) mutants relative to wild-type oocytes (Fig. 2 B). The integrated GFP::β-tubulin pixel intensity in klp-18(−) oocytes increased to a maximum at 4 min after ovulation before decreasing. In klp-7(−);klp-18(−) double mutants, the integrated intensity remained high until 8 min after ovulation before decreasing. These results further support our conclusion that reducing klp-7 function stabilizes microtubules during meiosis I. Moreover, in both klp-7(−) single mutants and klp-7(−);klp-18(−) double mutants, the increased accumulation of microtubules correlated with the assembly of additional oocyte meiotic spindle poles relative to wild-type and klp-18(−) oocytes, respectively.
Disrupting k–MT attachments restores oocyte meiotic spindle bipolarity in klp-7(−) mutants
Interfering with a vertebrate MCAK/kinesin-13 family member has been shown to stabilize incorrect merotelic and syntelic microtubule attachments at kinetochores (Kline-Smith et al., 2004; Ems-McClung and Walczak, 2010), and we have shown that KLP-7/MCAK localized to kinetochores during oocyte meiosis I in C. elegans (Fig. 1 D and Fig. S1). We therefore next asked whether k–MT attachment might influence pole number during oocyte meiotic spindle assembly. More specifically, we hypothesized that if syntelic (both kinetochores in a bivalent attached to a single pole) or merotelic (one kinetochore attached to both poles) k–MT attachments persist in klp-7(−) oocytes, persistent and abnormal k–MT tension might promote the assembly of extra poles. This hypothesis follows from observations in mouse oocytes indicating that early in meiotic spindle assembly, multiple small poles or pole foci (microtubule organizing centers) form and then over time coalesce into two poles (Schuh and Ellenberg, 2007). Moreover, ∼90% of paired homologous chromosomes during oocyte meiosis I experience improper k–MT attachments (Kitajima et al., 2011). Persistent syntelic and merotelic k–MT attachments might then result in abnormal tension that interferes with early pole coalescence. This hypothesis predicts that relieving the abnormal tension by disrupting k–MT attachments should restore spindle bipolarity in klp-7(−) mutants.
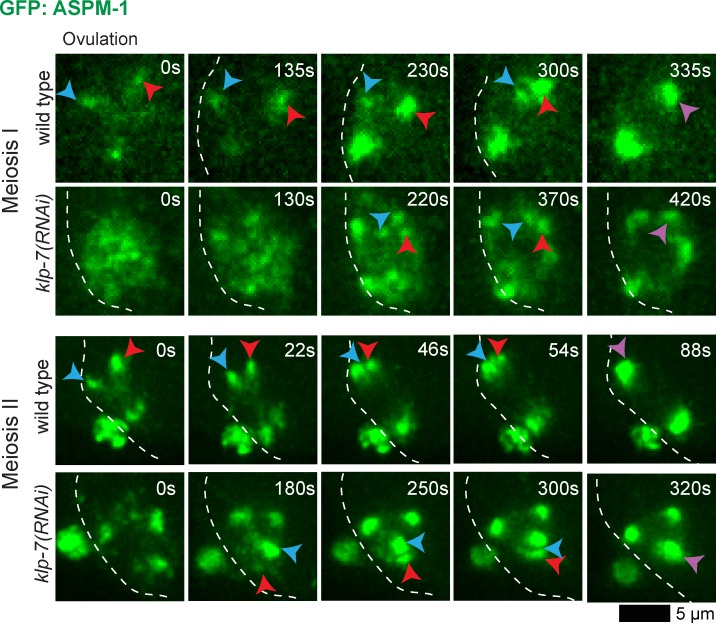
To address a model in which persistent improper k–MT attachments interfere with early pole coalescence in klp-7(−) mutant oocytes, we first examined spindle pole assembly in more detail by using clustered regularly interspaced short palindromic repeats (CRISPRs)/Cas-9 to generate an endogenous GFP::ASPM-1 fusion (Dickinson et al., 2013; Kim et al., 2014). The GFP::ASPM-1 fusion we used in all experiments described thus far (except for Fig. 4 B and Fig. S4) was expressed from an extragenic, extrachromosomal array (Connolly et al., 2014). Like other extrachromosomal arrays in C. elegans, the GFP::ASPM-1 array we generated is unstable, being transmitted to only a small fraction of progeny, and is silenced when the strain is grown at low temperatures (15°C), limiting its usefulness (unpublished data). CRISPR/Cas-9 GFP fusions to endogenous genes are stably transmitted and, in our experience, do not undergo silencing at low temperatures and often reveal previously undetected patterns of protein localization (unpublished data). We therefore made a homozygous viable endogenous GFP::ASPM-1 fusion using CRISPR/Cas-9 (Materials and methods section CRISPR/Cas-9 generation . . .) and found that during both oocyte meiosis I and II, multiple GFP::ASPM-1 foci were present early and then coalesced to form two poles in the wild type (11/11), whereas in 8/10 klp-7(−) mutants, extra spindle poles persisted (Fig. 5 and Fig. S5). These results indicate that in C. elegans, as in mouse oocytes (Schuh and Ellenberg, 2007), multiple small meiotic spindle pole foci coalesce to form two mature spindle poles, and we provide the first evidence for such coalescence using a pole marker rather than microtubules for imaging.
Figure 5.
Meiotic spindle pole coalescence in wild-type and klp-7(−) oocytes. Time-lapse spinning disk microscopy of wild-type and klp-7(RNAi) oocytes using a homozygous viable CRISPR/Cas-9 GFP::ASPM-1 endogenous fusion. White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. Red and blue arrowheads indicate ASPM-1 foci that merged into single poles, marked by purple arrowheads. (Top): Meiosis I images were captured with a 63× objective in utero. (Bottom): Meiosis II images were captured with a 100× objective ex utero.
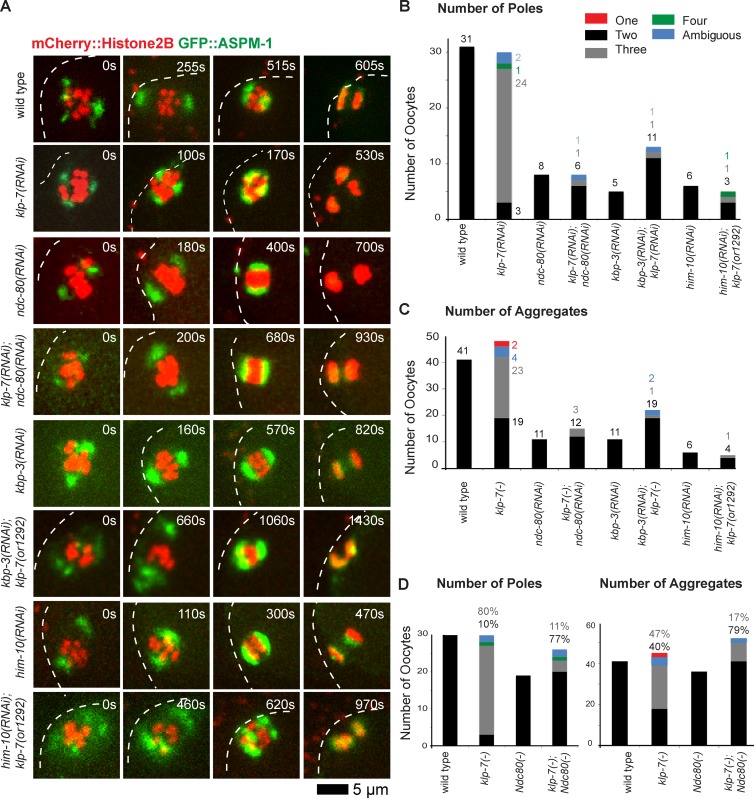
The Ndc80 complex is a part of a large protein network that comprises the kinetochore, with Ndc80 itself directly binding microtubules to provide the primary kinetochore binding site for k–MTs (Cheeseman et al., 2004, 2006; Cheeseman and Desai, 2008; Varma and Salmon, 2012). We therefore examined spindle pole assembly in transgenic strains expressing GFP::ASPM-1 and mCherry::Histone H2B to mark spindle poles and chromosomes, after using RNAi to reduce the function of the Ndc80 complex. We initially used our strain expressing GFP::ASPM-1 from an extrachromosomal array and feeding RNAi to deplete KLP-7 and one of two components of the complex, NDC-80 itself or SPC-25KBP-3 (Fig. 6). As reported previously (Dumont et al., 2010), ndc-80(−) spindles were bipolar but with some disorganized chromosomes that nevertheless eventually segregated toward two poles. Similarly, kbp-3(−) spindles also were bipolar with some defects in chromosome organization. Depleting NDC-80 or KBP-3 restored two ASPM-1 poles in 12/14 klp-7 oocytes, and chromosomes segregated into two aggregates in 31/38 klp-7(−) oocytes, although anaphase bridges were often observed (Fig. 6 D; combining data from strains expressing either mCherry::Histone H2B and GFP::ASPM-1 (Fig. 6) or mCherry::Histone H2B and GFP::β-tubulin (Fig. 7); see Fig. S4 for results from each genotype). We also analyzed Ndc80 knockdowns using our CRISPR-generated GFP::ASPM-1 fusion in the klp-7(or1292ts) strain background so as to target only a single gene for feeding RNAi knockdown (Fig. 6 A). We found that depleting either KPB-3 or a third Ndc80 complex component, HIM-10, restored bipolarity in 8/12 klp-7(or1292ts) oocytes (Fig. S4).
Figure 6.
Disrupting the Ndc80 complex during meiosis I restores bipolarity to klp-7(−) oocytes. (A) Time-lapse spinning disk confocal images during meiosis I in live wild-type, ndc-80(−), kbp-3(−), and klp-7(−) single and double mutant oocytes expressing mCherry::Histone H2B and GFP::ASPM-1 to mark chromosomes and spindle poles, respectively. White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. (B) Bar graph indicates the number of GFP::ASPM-1 foci observed for the indicated genotypes as described in Fig. 3. (C) Bar graph showing the number of segregating chromosome masses detected during anaphase for the indicated genotypes. The legend in B provides a color code for pole and aggregate numbers. (D) Bar graphs showing the same data as in B and C, but as a sum of all Ndc80 complex(−) and klp-7(−); Ndc80 complex(−) mutants. In B–D, klp-7(−) indicates results from both klp-7(or1292ts) and klp-7(RNAi) oocytes, from transgenic strains expressing mCherry::Histone H2B + GFP::ASPM-1 and mCherry::Histone H2B + GFP::MEI-1 (B and D) for pole numbers, and also mCherry::Histone H2B + GFP::β-tubulin (C and D) for aggregate numbers. In D the percentage of embryos, rather than number (B and C), with indicated phenotypes are shown. See Fig. S4 for the numbers of embryos analyzed for each genotype to score both pole and aggregate numbers. All data are from video micrographs of individual oocytes, each isolated from different worms in multiple experiments.
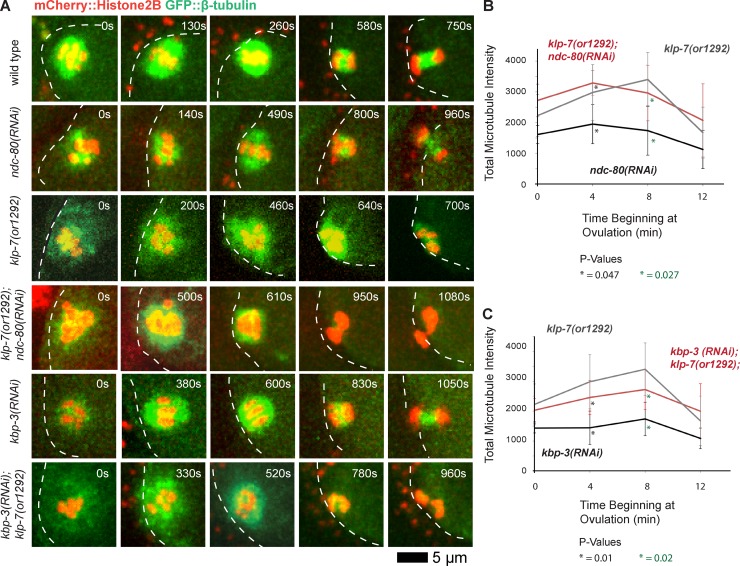
Figure 7.
Disrupting the Ndc80 complex during meiosis I restores bipolarity to klp-7(−) oocytes but does not reduce microtubule accumulation. (A) Time-lapse spinning disk confocal images during meiosis I in live wild-type, ndc-80(−), kbp-3(−), and klp-7(−) single and double mutant oocytes expressing mCherry::Histone H2B and GFP::β-tubulin to mark chromosomes and microtubules, respectively. White dashed lines indicate the oocyte plasma membrane, and times after ovulation are indicated in time-lapse sequence frames. (B and C) Integrated GFP::β-tubulin pixel intensity values in arbitrary units beginning at ovulation during meiosis I (Materials and methods section Microscopy), comparing klp-7(or1292) (n = 10) with ndc-80(RNAi) (n = 3) and klp-7(or1292); ndc-80(RNAi) (n = 5; B) and kbp-3(RNAi) (n = 6) and kbp-3(RNAi);klp-7(or1292) (n = 6; C). All data are from video micrographs of individual oocytes, each isolated from different worms in multiple experiments.
Because feeding RNAi in C. elegans sometimes reduces gene function less effectively than does microinjection of double-stranded RNA into the germline, we also used microinjection to knock down NDC-80 in a klp-7(or1292ts) mutant expressing the CRISPR-generated GFP::ASPM-1 fusion and mCherry::Histone H2B. Consistent with microinjection more effectively reducing NDC-80 function, we observed previously documented mitotic defects—premature and rapid spindle pole separation in one-cell-stage embryos (Oegema et al., 2001)—that we did not observe when using feeding RNAi to deplete Ndc80 components (unpublished data). However, progression through meiosis I was substantially delayed relative to the feeding RNAi experiments, and the spindle morphologies were more severely disrupted and difficult to interpret with respect to pole number and chromosome segregation (unpublished data). Based on the more interpretable results from the weaker feeding RNAi knockdowns of Ndc80 (Figs. 6 and 7), we suggest that extra poles persist in klp-7(−) mutants as a result of abnormal tension caused by persistent inappropriate k–MT attachments.
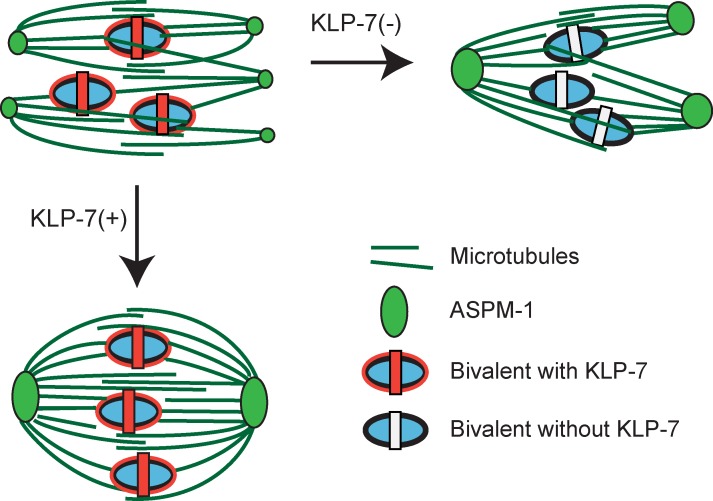
We next considered whether a decrease in microtubule accumulation during oocyte meiotic spindle assembly might provide an alternative explanation for the rescue of bipolarity in klp-7(−) mutant oocytes after Ncd80 knockdown. An excess accumulation of microtubules correlated with the increased spindle pole number both in klp-7(−) mutant oocytes, relative to the wild type, and in klp-7(−);klp-18(−) double mutants, relative to klp-18(−) mutants. We therefore asked whether the rescue of bipolarity in klp-7(−) mutants, after disruption of k–MT attachments by knocking down Ndc80 components, was associated with a decrease in the integrated GFP::β-tubulin intensity during meiosis I. However, we found that the values during meiosis I in ndc-80(−);klp-7(−) and kpb-3(−);klp-7(−) double mutants were higher than the values in ndc-80(−) and kbp-3(−) single mutants and similar to those in klp-7(−) mutants (Fig. 7). Thus, the rescue of bipolarity by Ndc80 knockdown in klp-7(−) mutants does not appear to result from a decreased accumulation of microtubules. Rather, these results support a model in which the persistence of extra oocyte meiotic spindle poles after loss of KLP-7/MCAK is caused by abnormal tension resulting from the persistence of improper k–MT attachments (Fig. 8), and not by changes in microtubule number.
Figure 8.
A model for the role of KLP-7/MCAK in limiting pole number during oocyte meiotic spindle assembly. KLP-7 promotes pole coalescence by relieving abnormal tension caused by inappropriate syntelic and merotelic k–MT attachments. In the absence of KLP-7, incorrect k–MT attachments persist and place abnormal tension on the spindle, interfering with pole coalescence. Other MT attachments to mid-bivalent rings might mediate chromosome positioning through the kinesin-4/chromokinesin family member KLP-19 (Discussion, next to last paragraph), and antiparallel MT–MT interactions in the spindle midzone may contribute to spindle assembly and stability. Note that this schematic does not take into account ultrastructural studies indicating that individual microtubules do not extend from spindle poles to the chromosomes; rather, shorter microtubules appear to be aligned and perhaps are bundled to link poles and chromosomes (Srayko et al., 2006).
Discussion
We identified two recessive and TS alleles of klp-7, the lone kinesin-13/MCAK family member in C. elegans, and thereby discovered a requirement for this widely conserved kinesin in the acentrosomal assembly of C. elegans oocyte meiotic spindles. Consistent with the microtubule depolymerase activity documented for other kinesin-13 family members, we detected a substantial increase in the accumulation of microtubules during oocyte meiosis I spindle assembly. Moreover, we also found that chromosomes in klp-7(−) mutant oocytes often segregated into three discrete masses as a result of the assembly of functional tripolar meiotic spindles. Although the excessive accumulation of microtubules may contribute to the assembly of extra spindle poles, disruption of k–MT attachment rescued meiosis I spindle bipolarity in klp-7(−) mutants without decreasing microtubule accumulation. Thus, the assembly of bipolar oocyte meiotic spindles depends on the proper regulation of microtubule attachment to kinetochores, as discussed in more detail in the third Discussion subsection.
klp-7 limits microtubule accumulation during oocyte meiosis I spindle assembly
The excess accumulation of microtubules during meiosis I in klp-7(−) mutants is consistent with the known function of other kinesin-13/MCAK family members that promote the depolymerization of microtubules from both their minus and plus ends. The Drosophila kinesin-13 family member KLP-10A is thought to act at oocyte meiotic spindle poles to depolymerize microtubules and maintain spindle stability, and in Xenopus extracts MCAK depletion results in abnormally long meiotic spindles (Mitchison et al., 2005; Ohi et al., 2007; Zou et al., 2008), although these studies did not address whether microtubule levels were affected. However, a study of klp-7 during early embryonic mitosis in C. elegans has shown that the loss of klp-7 results in a twofold increase in the number of microtubules that grow out from centrosomes, perhaps reflecting an increase in the stability of microtubules after their nucleation at γ-tubulin ring complexes (Srayko et al., 2005). Our results, using integrated GFP::β-tubulin pixel intensity as a rough proxy for microtubule accumulation, show that a loss of kinesin-13/MCAK function results in the increased accumulation of microtubules during oocyte meiotic spindle assembly.
C. elegans klp-7 limits spindle pole number during oocyte meiotic cell division
The loss of C. elegans klp-7 during oocyte meiosis I resulted not only in an excess accumulation of microtubules but also in the assembly of extra functional spindle poles. Our conclusion that extra oocyte meiotic spindle poles assemble in the absence of klp-7 follows from four observations: (1) in almost half of the klp-7(−) mutants we observed, chromosomes segregated as three discrete masses, consistent with extra functional poles forming; (2) two spindle pole markers, GFP::ASPM-1 and GFP::MEI-1, localized to three or more discrete foci in all klp-7(−) oocytes during prometaphase; (3) homologous pairs of chromosomes often aligned along more than one of the axes defined by the three spindle poles in klp-7(−) oocytes; and (4) reducing klp-7 function partially rescued oocyte meiotic spindle bipolarity in monopolar klp-18(−) mutants.
The partial rescue of spindle bipolarity in klp-7(−);klp-18(−) double mutants is consistent with KLP-7 limiting pole number, but the mechanism underlying bipolar spindle assembly in this double mutant is not clear. The rescue of bipolarity could depend on residual function for either kinesin in the double mutant or on other factors that also promote bipolarity. Presumably, KLP-7 and KLP-18 operate in distinct and parallel processes, with KLP-7 limiting pole number through its role in correcting improper k–MT attachments (see next subsection) and KLP-18 promoting bipolarity by mediating the sliding of antiparallel microtubules in opposite directions (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013).
Recently, another analysis of klp-7(−);klp-18(−) double mutants reported that, as in our analysis, more microtubules accumulated but also that KLP-7/MCAK is not required for chromosome movement during anaphase in larger monopolar spindles (Muscat et al., 2015). This analysis did not address whether bipolarity may have been partially restored in some double mutants or document whether the spindles always appeared monopolar. In our analysis, we scored 4/13 double mutant embryos as having an ambiguous pole number using GFP::ASPM-1 as a marker, and one of these did appear large and monopolar (Fig. S3 B). However, in most cases we observed some rescue of bipolarity after using RNAi to knock down both klp-7 and klp-18, or only to knock down klp-18 in klp-7(or1292ts) mutants (Fig. 4, Fig. S3 B, and Fig. S4).
Although the assembly of extra oocyte meiotic spindle poles has been observed in other organisms (Zenzes and Bielecki, 2004; Li et al., 2006; Eichenlaub-Ritter et al., 2007; Yang et al., 2007; Vogt et al., 2010; Delimitreva et al., 2012; Shen et al., 2015), the results we report here provide, to our knowledge, the first example of highly penetrant extra pole assembly during meiosis I resulting from a loss of gene function. Exposure to chemicals has been reported to induce spindle morphology aberrations in mouse oocytes (Zenzes and Bielecki, 2004; Eichenlaub-Ritter et al., 2007); abnormal spindles in mouse oocytes also have been observed after nuclear transfer of somatic nuclei (Shen et al., 2015) and in the absence of chromosomes (Yang et al., 2007); and abnormal spindles have been observed in arrested primate oocytes after in vitro maturation and fertilization (Delimitreva et al., 2012). However, in all of these examples, extra poles were observed rarely, and the segregation of chromosomes to three poles was not reported.
Although the loss of kinesin-13/MCAK family members has not been reported previously to result in the assembly of oocyte meiotic spindles with extra functional poles, there are suggestions that these kinesins may be involved in the regulation of pole number during meiotic cell divisions in mouse and Drosophila and during mitosis in cultured mammalian cells. Intriguingly, reducing MCAK function in mouse oocytes results in a substantial number of meiotic spindles with disorganized poles, although segregation to three or more poles was not reported (Vogt et al., 2010). Similarly, expression of a dominant-negative allele of the kinesin-13/MCAK family member Klp10A in Drosophila oocytes resulted in shorter meiosis I spindles with loosely organized poles and in one example an apparent small third pole, based on immunofluorescence imaging of tubulin in a single fixed sample (Zou et al., 2008). Finally, in mammalian cells with defects leading to losses of centrosome separation during mitosis, bipolarity was restored by MCAK depletion (Tulu et al., 2006; Zhang et al., 2008; Toso et al., 2009), and MCAK depletion also rescued bipolarity in Xenopus egg extracts, with monopolar oocyte meiotic spindles resulting from compromised Aurora B function (Shao et al., 2012). Although we are not aware of any other studies that have documented tripolar segregation of chromosomes in the absence of MCAK function, an influence on spindle pole organization and number is not unique to C. elegans oocyte meiosis and may therefore reflect a conserved property of this microtubule depolymerase family.
Disrupting k–MT attachment rescues bipolarity in klp-7 mutant oocytes
Our finding that depleting members of the Ndc80 complex rescued bipolarity in klp-7 mutant oocytes indicates that kinetochore function can influence oocyte meiotic spindle pole assembly and number. We found that partially depleting any one of three Ndc80 complex components—NDC-80 itself, KPB-3, or HIM-10—restored bipolarity in many klp-7(−) oocytes. Although full progression through meiosis I was slightly delayed compared with klp-7(−) mutants, the timing of bipolar pole assembly was similar after Ndc80 component depletion, and chromosomes were sometimes misaligned or formed anaphase bridges but ultimately segregated toward two distinct poles (Figs. 6 and 7).
Because MCAK orthologues in other organisms may act to detach improper syntelic and merotelic microtubule attachments during mitosis (Kline-Smith et al., 2004; Ems-McClung and Walczak, 2010), we suggest that KLP-7/MCAK also acts to correct inappropriate k–MT attachments during oocyte meiosis I. In the absence of KLP-7, syntelic and merotelic attachments would then persist because of the function of the Ndc-80 complex, which is thought to provide the primary site of k–MT attachment (Cheeseman et al., 2004, 2006; Cheeseman and Desai, 2008; Varma and Salmon, 2012).
Such persistence of abnormal k–MT attachment in klp-7(−) oocytes might generate abnormal and persistent k–MT tension that prevents poles from coalescing to a bipolar state. Indeed, in mouse oocytes multiple microtubule organizing centers initially form but then coalesce to form a bipolar meiotic spindle (Schuh and Ellenberg, 2007), and ∼90% of the homologous pairs of chromosomes experience inappropriate k–MT attachments (Kitajima et al., 2011). Moreover, based on our live-cell imaging of the pole marker GFP::ASPM-1, multiple small spindle pole foci also appeared early in C. elegans meiosis I and II and then coalesced to form two poles in wild-type oocytes as division progressed. In klp-7(−) oocytes, the early small pole foci failed to fully coalesce and extra poles persisted. We therefore suggest that in the absence of KLP-7/MCAK, persistent and abnormal tension from improper k–MT attachments leads to a diminished coalescence of early premature pole foci and the consequent persistence of extra oocyte meiotic spindle poles (Fig. 7 B).
Although we suggest here that k–MT attachments influence oocyte meiotic spindle assembly, a previous study has shown that kinetochore proteins in C. elegans are largely dispensable for bipolar spindle assembly, chromosome alignment on the metaphase plate, and anaphase chromosome segregation during meiosis I (Dumont et al., 2010). Moreover, the C. elegans kinesin-4/chromokinesin family member KLP-19 localizes to rings that encircle bivalents at their midpoints and is required for proper chromosome positioning and alignment to the metaphase plate during meiosis I (Wignall and Villeneuve, 2009). Thus, it has been proposed that during oocyte meiosis I in C. elegans, microtubules may capture and align chromosomes through attachment to the mid-bivalent rings, with the plus-ended kinesin KLP-19 in part mediating these dynamics (Wignall and Villeneuve, 2009). Furthermore, anaphase chromosome segregation may be largely independent of k–MT attachments (Dumont et al., 2010; Muscat et al., 2015). In these models, k–MT attachments appear largely dispensable. However, defects in chromosome positioning and segregation, albeit mild in nature, are observed in the absence of kinetochore function (Dumont et al., 2010), and we have observed delayed coalescence of pole foci after knocking down the kinetochore component KNL-1 (unpublished data). Moreover, whereas k–MT attachments appear largely dispensable for bipolar spindle assembly in the presence of KLP-7/MCAK, pole coalescence is severely disrupted if KLP-7/MCAK is absent, and this disruption may depend at least in part on the persistence of inappropriate k–MT attachments. We therefore suggest that both k–MT attachments and microtubule attachment to mid-bivalent rings are important for oocyte meiotic spindle assembly (Fig. 8).
Finally, an alternative but not mutually exclusive model for the assembly of extra poles in klp-7 mutant oocytes is that an excessive accumulation of microtubules promotes the assembly of extra poles. Moreover, two other recent studies also have reported an increased accumulation of oocyte spindle microtubules in klp-7(−) mutants (Han et al., 2015; Muscat et al., 2015). Also in support of this alternative hypothesis, in both klp-7(−) single and klp-7(−);klp-18(−) double mutants, the assembly of additional spindle poles correlated with increased microtubule accumulation. We found that KLP-7 was localized to both spindle poles and chromosomes, but we do not know whether the assembly of extra microtubules resulted from loss of klp-7 function at either or both locations. However they originate, the presence of extra microtubules during oocyte meiotic cell division could promote the assembly of extra spindle poles, presumably mediated by nonlimiting pole organizing activities that normally assemble bipolar spindles. However, this model does not explain our finding that depletion of the Ndc80 complex restored bipolarity in klp-7 mutant oocytes without reducing the accumulation of microtubules compared with klp-7(−) single mutants. Although the accumulation of excess microtubules and abnormal k–MT tension might both promote the assembly of extra oocyte meiotic spindle poles, our data suggest that abnormal k–MT tension is primarily responsible for the persistence of extra oocyte meiotic spindle poles in the absence of KLP-7/MCAK function. We anticipate that further investigation of kinetochore function will prove informative with respect to the process of acentrosomal oocyte meiotic spindle pole assembly.
Materials and methods
C. elegans strains
The N2 Bristol strain was used as the wild-type strain with standard nematode protocols used as previously described (Brenner, 1974). The TS alleles were isolated in a screen for TS–embryonic lethal mutant using the methods described previously (Kemphues et al., 1988; O’Connell et al., 1998; Encalada et al., 2000). TS mutations were isolated from a lin-2(e1309) background and outcrossed into the N2 strain as described previously (Encalada et al., 2000). TS strains were maintained at the permissive temperature of 15°C and were shifted to the restrictive temperature 26°C to induce the loss-of-function phenotype. For imaging oocyte meiosis, worms were shifted for 2–5 h to restrictive temperature, bypassing larval lethality and sterility phenotypes that result from earlier upshifts.
All strains used for the manuscript are listed in Table S1. Transgenic strains were crossed into a him-5 background, and males from the him-5 strains were used to cross the transgenes into TS mutant strains. Many of the strains were provided by, or made using strains provided by, the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) which is funded by the National Institutes of Health National Center for Research Resources.
The deletion allele klp-7(tm2143), provided by Shohei Mitani (National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan, Tokyo, Japan), is incompletely penetrant for embryonic lethality at 15°C (Table 1). The 875-bp deletion begins in the intron 3′ of exon 2 and ends within exon 4 (unpublished data; see Fig. S2 A in Ghosh-Roy et al., 2012). If exon 2 is spliced to exon 5 a premature stop codon would occur at codon 185, as predicted in Ghosh-Roy et al. (2012), but this prediction was not tested. If, instead, exon 2 is spliced to the first AG 3′ of the deletion point in exon 4, preceding codon 365, an in-frame 268–amino acid deletion would result. Neither prediction has been verified (e.g., by RT-PCR); we therefore plan, in future work, to use CRISPR/Cas-9 to generate deletion alleles and definitively determine the klp-7 null phenotype.
KLP-7 immunofluorescence
After freeze-cracking, embryos were fixed in −20°C methanol for 15 min and then incubated with 1:1,000 rabbit anti–KLP-7, a gift from K. Oegema at the University of California, San Diego, La Jolla, CA (Oegema et al., 2001), and 1:50,000 mouse anti–α-tubulin (DM1A; Sigma-Aldrich) antibodies at 4°C overnight. As previously described, the KLP-7 antibody was generated by amplifying a fragment of KLP-7 from cDNA using the primers 5′-CGCGCGAGATCTCAGAGAAAACGAGCCGAGAA-3′ and 5′-GCGCGCGAATTCTCAAGGAGCCATACGAACAGGAAC-3′. Fragments were digested with BglII-–EcoRI and cloned into pGEX6P-1 (GE Healthcare) to produce GST-KLP-7 (Oegema et al., 2001). After washing, the slides were incubated with 1:1,000 anti-rabbit–Alexa 488 and 1:1,000 anti-mouse–Alexa 555 (Life Technologies) for 2 h at room temperature. Slides were then incubated with 100 ng/ml DAPI (Roche) for 15 min before mounting. Images were collected with a confocal microscope (LSM 700; Carl Zeiss), and a z projection is shown in Fig. 1 D. A predicted but unverified in-frame splice product of klp-7(tm2143) would remove amino acids 97–364 (Fig. 1 and see the last paragraph of the preceding subsection); the peptide used as the antigen for generating the KLP-7 antisera included amino acids 14–190, and thus, the antisera might not recognize an in-frame deletion allele protein product (Oegema et al., 2001).
CRISPR/Cas-9 generation of GFP::ASPM-1 transgenic strain
For gfp::aspm-1 donor construct, a 2.4-kb fragment (LGI:9,225,478–9,227,634) amplified from fosmid WRM0619aD11 was cloned into the pJET1.2 vector (Thermo Fisher Scientific). The GFP-coding region amplified from pSO26 (O’Rourke et al., 2007) was inserted at the N terminus of aspm-1 by Gibson assembly (Gibson et al., 2009). A point mutation was then introduced to disrupt the protospace adjacent motif site (CGG to CGC) by inverse PCR. eft-3p::Cas-9 plasmid with aspm-1 N terminus targeting small guide RNA (5′-GTACAAATGGCCACCTAAAACGG-3′) was made from pDD162 plasmid (Dickinson et al., 2013). The gfp::aspm-1 donor construct, Cas-9–sgRNA plasmid, and selection marker pRF4 were injected into wild-type N2 young adults. The animals carrying pRF4 were searched for GFP expression as described in Kim et al. (2014).
Positional cloning
We determined that both or1092 and or1292 were located between an ∼10- and 13-Mb region on linkage group III using a genome-wide single nucleotide polymorphism mapping and whole genome sequencing approach (Doitsidou et al., 2010). We next used visible-marker mapping and found that each mutation resides within a 6.31-centimorgan interval between unc-69 and dpy-18, with or1092ts and or1292ts mapping to ∼4.5 centimorgans. Data from genome-wide RNAi knockdown screens indicated klp-7 as a potential candidate because embryos were reported as having large polar bodies, although there was no reference to an abnormal number of maternal pronuclei (Sönnichsen et al., 2005). We identified the mutations using the whole genome sequencing data and discovered or1092 to be an isoleucine to a phenylalanine substitution at position 298 and or1292 to be a glycine to an aspartic acid at position 548.
Phenotypic analysis
Scoring embryonic viability
Viability counts of embryos (percent hatching) were determined by singling out at least seven L4s onto individual plates, growing them at the permissive or restrictive temperature until broods were produced, and then removing the mother and counting the eggs (Table 1). We allowed the embryos to develop for 18–24 h and then counted the number of unhatched embryos that remained. Embryonic viability in the broods of F1s was scored at the restrictive temperature (described in Materials and methods section C. elegans strains). A test for dominance was also performed by examining F1 heterozygous TS mutant (TS mutant/+) as described above in C. elegans strains (Table 1).
Scoring multiple maternal pronuclei
All embryos examined were shifted from 15°C to 26°C for 2–5 h as young adults. RNAi was performed by feeding (as described in Materials and methods section RNAi ), with the exception that L1s were plated at 15°C and shifted to 26°C alongside the mutant strains. One-cell embryos were examined using Nomarski imaging by cutting young adults open in 1 µl M9 and mounting eggs on 4% agar pads.
Scoring larval lethality
Larval lethality was scored by placing L1 larvae, synchronized by hypochlorite hatch-off at 26°C, and raised to adulthood. Sterility was called if the worm exhibited either an empty gonad or a gonad filled with unfertilized oocytes.
RNAi
RNAi to reduce gene function was performed by placing L1 larvae synchronized by hypochlorite hatch-off onto 60-mm nematode growth media agar plates with 100 mg/ml ampicillin and 1-mM isopropyl-β-d-thiogalactopyranoside and seeded with the double-stranded RNA–expressing HT115 Escherichia coli (Fire et al., 1998; Kamath et al., 2001). For co-depletions, we used the same process but seeded plates with an equal mixture of the double-stranded RNA–expressing E. coli strains. We maintained the affected worms at room temperature and examined their phenotypes in zygotes within whole mounted young adults. TS strains were treated the same way, but maintained at 15°C and shifted to 26°C as young adults for 2–5 h. Finally, RNAi by soaking was performed for scoring embryonic lethality for klp-7(RNAi) by purifying double-stranded RNA and diluting in a soaking buffer according to WormBook. L4 N2 worms were soaked for 24 h at 26°C and then recovered. The number of eggs that hatched were scored. All RNAi strains were obtained from the Ahringer RNAi library, and the corresponding primers are listed in Table S2.
Microscopy
Live imaging of fluorescent fusion proteins in utero during meiosis was accomplished by mounting worms on 6% agar pads, with 1 µl each of 1-µm polystyrene beads and M9 on microscope slides covered with a coverslip. Oocytes were analyzed on an inverted microscope (DMI 4000B) fitted with a 63× 1.40–0.60 Plan Apo oil objective lens (HCX; Leica) in a room maintained at 25°C. Time-lapse videos were obtained with an electron multiplying charge coupled device digital camera (Hamamatsu Photonics) using Volocity software (PerkinElmer). Six stacks that were 1.5 µm thick were used to collect images every 10 s where not noted. Ex utero imaging of meiosis II was accomplished by slicing open worms in L15 buffer on microscope slides gently covered by a coverslip dotted with Vaseline. After recording, the videos were cropped and adjusted for contrast, brightness, or color balance in ImageJ (National Institutes of Health). Measurements of microtubule intensity were taken in ImageJ by generating a threshold (set on Otsu) in a cropped area around the spindle in the merged z stack at varying time points throughout meiosis. Using ImageJ, both the area of the threshold (in pixels) and the mean gray value (MGV) were automatically calculated. Additionally, an area was selected around the entire egg (excluding the spindle), and the MGV was calculated for the cytoplasm. These three measurements were placed into the following formula: MGV(spindle)/MGV(cytoplasm) × area(spindle) = total microtubule intensity.
Online supplemental material
Fig. S1 shows KLP-7 immunolocalization in wild-type and klp-7(−) oocytes in the oocyte meiotic spindle. Fig. S2 gives more examples of the multipolar spindle phenotype observed in klp-7(or1292ts), klp-7(or1092ts), and the deletion allele klp-7(tm2143). Fig. S3 provides additional examples of the multipolar spindle phenotype using GFP::ASPM-1 to mark spindle poles. It also provides more examples (including the exceptional cases) of klp-7(−);klp-18(−) double mutants during meiotic spindle assembly, as well as more examples of klp-7(−); Ndc-80(−) double mutants. Fig. S4 provides an expanded quantitation of spindle poles and chromosome aggregates for each genotype scored. Fig. S5 provides more examples of pole coalescence using the CRISPR-generated GFP::ASPM-1 in wild-type and klp-7(−) oocytes. A list of the strains used can be found in Table S1, and the sequences of oligos used to generate the double-stranded RNA can be found in Table S2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412010/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201412010.dv.
Supplementary Material
Acknowledgements
We thank the Caenorhabditis Genetics Center and the Shohei Mitani laboratory for strains, Craig Mello for advice on CRISPR technology, and Eisuke Sumiyoshi for advice on ex utero imaging. We are grateful to Chris Doe and Diana Libuda for comments on the manuscript and to Andrew Chisholm, Iain Cheeseman, and members of the Bowerman laboratory for productive discussions.
This work was supported by a National Institutes of Health grant (GM049869) to A.A. Connolly, C.-H. Chuang, and B. Bowerman and a Human Frontier Science Fellowship to K. Sugioka.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CRISPR
- clustered regularly interspaced short palindromic repeat
- k–MT
- kinetochore–microtubule
- MCAK
- mitotic centromere–associated kinesin
- MGV
- mean gray value
- TS
- temperature sensitive
References
- Albertson D.G. 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101:61–72. 10.1016/0012-1606(84)90117-9 [DOI] [PubMed] [Google Scholar]
- Albertson D.G., and Thomson J.N.. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15–26. 10.1007/BF00710603 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore–microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R. III, Oegema K., and Desai A.. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997. 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Clark-Maguire S., and Mains P.E.. 1994a Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126:199–209. 10.1083/jcb.126.1.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire S., and Mains P.E.. 1994b mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics. 136:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly A.A., Osterberg V., Christensen S., Price M., Lu C., Chicas-Cruz K., Lockery S., Mains P.E., and Bowerman B.. 2014. Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol. Biol. Cell. 25:1298–1311. 10.1091/mbc.E13-11-0687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delimitreva S., Tkachenko O.Y., Berenson A., and Nayudu P.L.. 2012. Variations of chromatin, tubulin and actin structures in primate oocytes arrested during in vitro maturation and fertilization—what is this telling us about the relationships between cytoskeletal and chromatin meiotic defects? Theriogenology. 77:1297–1311. 10.1016/j.theriogenology.2011.10.035 [DOI] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Poole R.J., Sarin S., Bigelow H., and Hobert O.. 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE. 5:e15435 10.1371/journal.pone.0015435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., and Desai A.. 2012. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 22:241–249. 10.1016/j.tcb.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., and Desai A.. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901. 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U., Winterscheidt U., Vogt E., Shen Y., Tinneberg H.R., and Sorensen R.. 2007. 2-methoxyestradiol induces spindle aberrations, chromosome congression failure, and nondisjunction in mouse oocytes. Biol. Reprod. 76:784–793. 10.1095/biolreprod.106.055111 [DOI] [PubMed] [Google Scholar]
- Ems-McClung S.C., and Walczak C.E.. 2010. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 21:276–282. 10.1016/j.semcdb.2010.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada S.E., Martin P.R., Phillips J.B., Lyczak R., Hamill D.R., Swan K.A., and Bowerman B.. 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228:225–238. 10.1006/dbio.2000.9965 [DOI] [PubMed] [Google Scholar]
- Fabritius A.S., Ellefson M.L., and McNally F.J.. 2011. Nuclear and spindle positioning during oocyte meiosis. Curr. Opin. Cell Biol. 23:78–84. 10.1016/j.ceb.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., and Mello C.C.. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Ghosh-Roy A., Goncharov A., Jin Y., and Chisholm A.D.. 2012. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Dev. Cell. 23:716–728. 10.1016/j.devcel.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gönczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 408:331–336. 10.1038/35042526 [DOI] [PubMed] [Google Scholar]
- Han X., Adames K., Sykes E.M.E., and Srayko M.. 2015. The KLP-7 residue S546 is a putative Aurora kinase site required for microtubule regulation at the centrosome in C. elegans. PLoS ONE. 10:e0132593 10.1371/journal.pone.0132593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., and McNally F.J.. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. 10.1016/S0092-8674(00)81578-0 [DOI] [PubMed] [Google Scholar]
- Howe M., McDonald K.L., Albertson D.G., and Meyer B.J.. 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153:1227–1238. 10.1083/jcb.153.6.1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C., Pirmadjid N., Serhal P., Howe K., and Fitzharris G.. 2010. MCAK regulates chromosome alignment but is not necessary for preventing aneuploidy in mouse oocyte meiosis I. Development. 137:2133–2138. 10.1242/dev.048306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., and Ahringer J.. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:H0002 10.1016/S0092-8674(88)80024-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K.J., Priess J.R., Morton D.G., and Cheng N.S.. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 52:311–320. 10.1016/S0092-8674(88)80024-2 [DOI] [PubMed] [Google Scholar]
- Kim D.Y., and Roy R.. 2006. Cell cycle regulators control centrosome elimination during oogenesis in Caenorhabditis elegans. J. Cell Biol. 174:751–757. 10.1083/jcb.200512160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K.S., Seth M., Conte D. Jr., Shirayama M., and Mello C.C.. 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 197:1069–1080. 10.1534/genetics.114.166389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Ohsugi M., and Ellenberg J.. 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 146:568–581. 10.1016/j.cell.2011.07.031 [DOI] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., and Walczak C.E.. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., and Gönczy P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Li G.P., Bunch T.D., White K.L., Rickords L., Liu Y., and Sessions B.R.. 2006. Denuding and centrifugation of maturing bovine oocytes alters oocyte spindle integrity and the ability of cytoplasm to support parthenogenetic and nuclear transfer embryo development. Mol. Reprod. Dev. 73:446–451. 10.1002/mrd.20436 [DOI] [PubMed] [Google Scholar]
- Maney T., Hunter A.W., Wagenbach M., and Wordeman L.. 1998. Mitotic centromere–associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801. 10.1083/jcb.142.3.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., and Vale R.D.. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429. 10.1016/0092-8674(93)90377-3 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., and McNally F.J.. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S., and Vernos I.. 2012. Microtubule assembly during mitosis – from distinct origins to distinct functions? J. Cell Sci. 125:2805–2814. 10.1242/jcs.092429 [DOI] [PubMed] [Google Scholar]
- Mitchison T.J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E.D., and Kapoor T.M.. 2005. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 16:3064–3076. 10.1091/mbc.E05-02-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J., Maddox P.S., Hyndman F., Oegema K., and Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. 10.1038/ncb1331 [DOI] [PubMed] [Google Scholar]
- Müller-Reichert T., Greenan G., O’Toole E., and Srayko M.. 2010. The elegans of spindle assembly. Cell. Mol. Life Sci. 67:2195–2213. 10.1007/s00018-010-0324-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat C.C., Torre-Santiago K.M., Tran M.V., Powers J.A., and Wignall S.M.. 2015. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 4:e06462 10.7554/eLife.06462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K.F., Leys C.M., and White J.G.. 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 149:1303–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Dorfman M.D., Carter J.C., and Bowerman B.. 2007. Dynein modifiers in C. elegans: Light chains suppress conditional heavy chain mutants. PLoS Genet. 3:e128 10.1371/journal.pgen.0030128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Carter C., Carter L., Christensen S.N., Jones M.P., Nash B., Price M.H., Turnbull D.W., Garner A.R., Hamill D.R., et al. 2011. A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS ONE. 6:e16644 10.1371/journal.pone.0016644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Burbank K., Liu Q., and Mitchison T.J.. 2007. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr. Biol. 17:953–959. 10.1016/j.cub.2007.04.057 [DOI] [PubMed] [Google Scholar]
- Ohkura H. 2015. Meiosis: An overview of key differences from mitosis. Cold Spring Harb. Perspect. Biol. 7: a015859 10.1101/cshperspect.a015859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz A.L., Srayko M., Dammermann A., Quintin S., Wielsch N., MacLeod I., de Robillard Q., Zinke A., Yates J.R. III, Müller-Reichert T., et al. 2007. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 128:115–127. 10.1016/j.cell.2006.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., and Ellenberg J.. 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 130:484–498. 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Segbert C., Barkus R., Powers J., Strome S., Saxton W.M., and Bossinger O.. 2003. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol. Biol. Cell. 14:4458–4469. 10.1091/mbc.E03-05-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Ma C., Zhang X., Li R., Miller A.L., Bement W.M., and Liu X.J.. 2012. Aurora B regulates spindle bipolarity in meiosis in vertebrate oocytes. Cell Cycle. 11:2672–2680. 10.4161/cc.21016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Wang Z., Shen X., Zheng Z., Zhang Q., Feng X., Hu L., and Lei L.. 2015. Abnormal dynamic changes in β-tubulin in somatic nuclear transfer cloned mouse embryos. Zygote. 23:76–82. [DOI] [PubMed] [Google Scholar]
- Siddiqui S.S. 2002. Metazoan motor models: Kinesin superfamily in C. elegans. Traffic. 3:20–28. 10.1034/j.1600-0854.2002.30104.x [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L.B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.M., Artelt J., Bettencourt P., Cassin E., et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Srayko M., Buster D.W., Bazirgan O.A., McNally F.J., and Mains P.E.. 2000. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., and Hyman A.A.. 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell. 9:223–236. 10.1016/j.devcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Srayko M., O’toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Sturgill E.G., and Ohi R.. 2013. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol. 23:1280–1290. 10.1016/j.cub.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Janssen A., Geers E.F., Alvarez-Fernández M., and Medema R.H.. 2009. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19:1703–1711. 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., and McAinsh A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372. 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., and Wadsworth P.. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541. 10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M., Berends C.W., Perreault A., Nguyen-Ngoc T., Gönczy P., Vidal M., Boxem M., and van den Heuvel S.. 2009. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Gα. Nat. Cell Biol. 11:269–277. 10.1038/ncb1834 [DOI] [PubMed] [Google Scholar]
- Vanneste D., Takagi M., Imamoto N., and Vernos I.. 2009. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 19:1712–1717. 10.1016/j.cub.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Varma D., and Salmon E.D.. 2012. The KMN protein network – chief conductors of the kinetochore orchestra. J. Cell Sci. 125:5927–5936. 10.1242/jcs.093724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt E., Sanhaji M., Klein W., Seidel T., Wordeman L., and Eichenlaub-Ritter U.. 2010. MCAK is present at centromeres, midspindle and chiasmata and involved in silencing of the spindle assembly checkpoint in mammalian oocytes. Mol. Hum. Reprod. 16:665–684. 10.1093/molehr/gaq025 [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Gan E.C., Desai A., Mitchison T.J., and Kline-Smith S.L.. 2002. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr. Biol. 12:1885–1889. 10.1016/S0960-9822(02)01227-7 [DOI] [PubMed] [Google Scholar]
- Wignall S.M., and Villeneuve A.M.. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11:839–844. 10.1038/ncb1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., and von Dassow G.. 2007. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J. Cell Biol. 179:869–879. 10.1083/jcb.200707120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto I., Kosinski M.E., and Greenstein D.. 2006. Start me up: Cell signaling and the journey from oocyte to embryo in C. elegans. Dev. Dyn. 235:571–585. 10.1002/dvdy.20662 [DOI] [PubMed] [Google Scholar]
- Yang J.W., Lei Z.L., Miao Y.L., Huang J.C., Shi L.H., OuYang Y.C., Sun Q.Y., and Chen D.Y.. 2007. Spindle assembly in the absence of chromosomes in mouse oocytes. Reproduction. 134:731–738. 10.1530/REP-07-0149 [DOI] [PubMed] [Google Scholar]
- Zenzes M.T., and Bielecki R.. 2004. Nicotine-induced disturbances of meiotic maturation in cultured mouse oocytes: Alterations of spindle integrity and chromosome alignment. Tob Induc Dis. 2:151–161. 10.1186/1617-9625-2-3-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., and Walczak C.E.. 2008. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 19:2752–2765. 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Hallen M.A., Yankel C.D., and Endow S.A.. 2008. A microtubule-destabilizing kinesin motor regulates spindle length and anchoring in oocytes. J. Cell Biol. 180:459–466. 10.1083/jcb.200711031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.