TRIM20 and TRIM21 are mediators of IFN-γ–induced autophagy, which act as autophagic receptor regulators that target specific inflammasome components and type I interferon response regulators for degradation by precision autophagy.
Abstract
The present paradigms of selective autophagy in mammalian cells cannot fully explain the specificity and selectivity of autophagic degradation. In this paper, we report that a subset of tripartite motif (TRIM) proteins act as specialized receptors for highly specific autophagy (precision autophagy) of key components of the inflammasome and type I interferon response systems. TRIM20 targets the inflammasome components, including NLRP3, NLRP1, and pro–caspase 1, for autophagic degradation, whereas TRIM21 targets IRF3. TRIM20 and TRIM21 directly bind their respective cargo and recruit autophagic machinery to execute degradation. The autophagic function of TRIM20 is affected by mutations associated with familial Mediterranean fever. These findings broaden the concept of TRIMs acting as autophagic receptor regulators executing precision autophagy of specific cytoplasmic targets. In the case of TRIM20 and TRIM21, precision autophagy controls the hub signaling machineries and key factors, inflammasome and type I interferon, directing cardinal innate immunity response systems in humans.
Introduction
Autophagy is a eukaryotic intracellular pathway that carries out key aspects of cytoplasmic homeostasis (Mizushima et al., 2011; Frake et al., 2015; Kroemer, 2015) in addition to its independent nutritional and metabolic roles (Rabinowitz and White, 2010; Kenific and Debnath, 2015). The principal aspects of how the core autophagy machinery operates have been defined and are conserved form yeast to humans (Mizushima et al., 2011). In mammalian cells, the key factors (Simonsen and Tooze, 2009) activating autophagy include Beclin 1 (Liang et al., 1999) and ULK1/2 (mammalian Atg1 paralogues; Chan and Tooze, 2009), in a complex phosphorylation cascade downstream of AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (Egan et al., 2011; Kim et al., 2011, 2013; Russell et al., 2013). In addition to these upstream regulators, the autophagic machineries include mammalian Atg8s (mAtg8s: LC3A, LC3B, LC3C, GABARAP, GAPARAPL1, and GABARAPL2) and their recruiter ATG12–ATG5–ATG16L1 complex, the emblematic autophagy factors believed to support the elongation and completion of autophagosomal membranes (Mizushima et al., 2011; Weidberg et al., 2011) as they envelop and sequester the intended autophagic targets. The breath of targets for autophagy in mammalian cells (Randow and Youle, 2014; Stolz et al., 2014) illustrates the versatility of autophagy as a process but also poses the question regarding how autophagy recognizes the cargo. Much progress has been made, including realization that many of the targets such as faulty mitochondria (Youle and Narendra, 2011), protein aggregates (Rubinsztein et al., 2012), damaged endosomes/lysosomes (Maejima et al., 2013), and invading microorganisms (Deretic et al., 2015) are earmarked for autophagy by molecular tags such as ubiquitin (Stolz et al., 2014) and galectins (Randow and Youle, 2014). Furthermore, only five autophagic receptors recognizing such tags have been described (Birgisdottir et al., 2013; Rogov et al., 2014). Nevertheless, the number of proteinaceous, membranous, and many complex targets for selective autophagy keeps growing, and yet the number of autophagic receptors has not followed this expansion. This poses the question of whether the thus far discovered receptors suffice to carry out all of selective autophagy in the cell or whether there are additional receptors to be uncovered. A recent study suggests that the tripartite motif (TRIM; RING domain, B-Box, and coiled-coil domain [CCD]) class of proteins, a large family of proteins with broad range of functions for inflammation and cell growth (Reymond et al., 2001; Kawai and Akira, 2011), may represent a new class of autophagic receptors (Mandell et al., 2014).
Autophagy has many biological effects that include immunological processes and inflammation (Ma et al., 2013; Mathew et al., 2014; Deretic et al., 2015). One aspect of this role of autophagy is regulation of inflammasome activity (Schroder and Tschopp, 2010), whereby autophagy antagonizes inflammasome activation, through several proposed mechanisms (Saitoh et al., 2008; Nakahira et al., 2011; Zhou et al., 2011; Shi et al., 2012) that are, however, not fully understood. Another distinct manifestation is autophagic suppression of type I IFN responses through STING and RIG-I (retinoic acid-inducible gene 1)–like receptors (Jounai et al., 2007; Saitoh et al., 2009; Tal et al., 2009; Konno et al., 2013; Liang et al., 2014; Deretic et al., 2015). In this work, we show that a subset of TRIMs is involved in selective autophagy of the components of the inflammasome and type I IFN regulatory systems. These TRIMs form a platform for the type of autophagy termed precision autophagy (Deretic et al., 2015) and directly bridge their degradative targets with the core autophagic machinery, including autophagy initiation (ULK1 and Beclin 1), elongation (ATG16L1), and execution (mAtg8s) factors.
Results
TRIMs participate in IFN-γ–induced autophagy
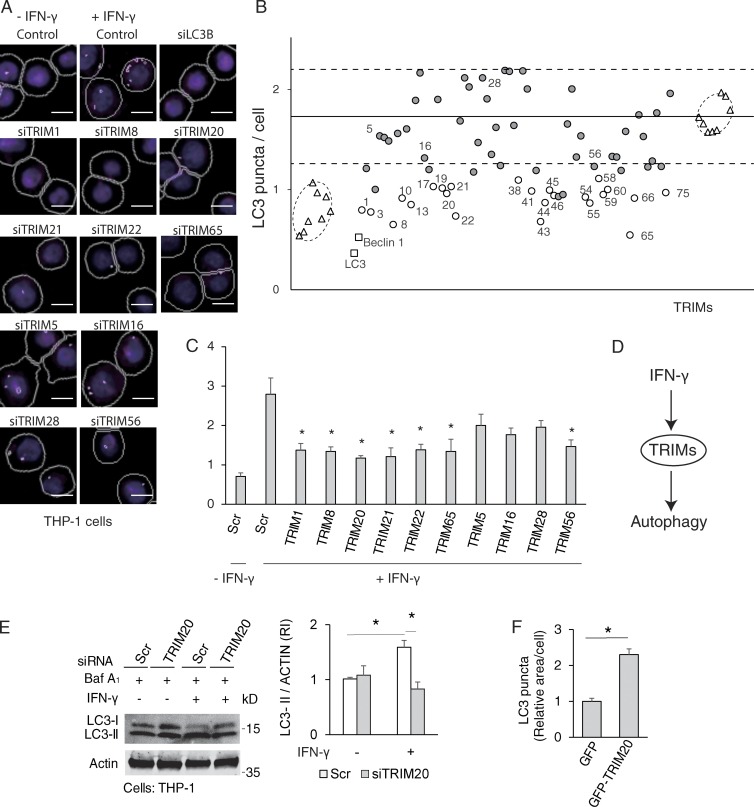
IFN-γ induces autophagy (Inbal et al., 2002; Gutierrez et al., 2004; Fabri et al., 2011) and influences cytokine networks and polarization of immune systems (Ghezzi and Dinarello, 1988; Schroder and Tschopp, 2010; Mishra et al., 2013), whereas TRIMs are involved in immune responses (Kawai and Akira, 2011) and, through an assortment of proposed mechanisms affect autophagy (Niida et al., 2010; Tomar et al., 2012; Barde et al., 2013; Pizon et al., 2013; Yang et al., 2013; Khan et al., 2014; Mandell et al., 2014; Pineda et al., 2015). IFN-γ can induce expression of a subset of TRIMs (Carthagena et al., 2009). We wondered whether TRIMs might be contributing mediators to autophagy induction by IFN-γ. We used an image-based high content (HC) analysis of LC3 puncta (Fig. 1 A) to screen for effects of TRIM knockdowns on IFN-γ–induced autophagy in human myelomonocytic cells. IFN-γ–induced autophagy in THP-1 cells (Fig. 1 A), showing dose dependence (Fig. S1 A) and in primary human macrophages (Fig. S1 B). For standardization, we used THP-1 cells for the screen (Fig. 1 B and Fig. S1 C). Out of the 70 human TRIMs tested, knockdowns of 24 different TRIMs reduced endogenous LC3 puncta per cell under IFN-γ treatment (Fig. 1 B, open circles; Fig. S1 C shows average ± range values from two independent screens). We followed this up by individual knockdowns of a subset of six positive and four neutral TRIMs from the screen (Fig. 1 C). All six TRIMs that were positive hits from the screen, TRIM1, TRIM8, TRIM20, TRIM21, TRIM22, and TRIM65 (knockdowns were evaluated in Fig. S1 D), were required for optimal induction of autophagy by IFN-γ (Fig. 1 C). Of the neutral TRIMs, TRIM56 that was marginally positive in the screen, showed a borderline but statistically significant effect (Fig. 1 C). Thus, TRIMs contribute to autophagy induction in response to IFN-γ (Fig. 1 D).
Figure 1.
TRIMs regulate IFN-γ–induced autophagy. (A) THP-1 cells were subjected to TRIM knockdown and treated with 1,000 U/ml IFN-γ for 4 h, and high content (HC) analysis was performed using a Cellomics HCS scanner (epifluorescence) and iDEV software. HC (magenta, endogenous LC3B immunofluorescence; blue, nuclei stained with Hoechst). Mask overlay, software-defined objects (primary objects, cell outlines; internal secondary objects, LC3 puncta). (B) Average count of LC3 puncta per cell from cells treated as in A (data from two 96-well plates with identical siRNA arrangements; the corresponding data are shown in Fig. S1 C). Encircled are IFN-γ–treated wells (right) and wells with vehicle controls (bottom left). TRIM knockdowns that reduced LC3 puncta readout in both of the two experiments by 3 SDs (horizontal dot lines) from the average of IFN-γ–treated controls (horizontal solid line) are indicated by corresponding TRIM numbers (open circle). TRIMs that were chosen in follow up experiments in Fig. 1 C are also indicated with number. (C) Similar to B, except that THP-1 cells were subjected to specific TRIM or scrambled (Scr; control) knockdowns and were analyzed in quadruplicates or more repeats. (D) Model of TRIMs-mediated IFN-γ–induced autophagy based on the results obtained in Fig. 1 and Fig. S1 thus far. (E) THP-1 cells were treated with TRIM20 or scrambled siRNAs, incubated with or without IFN-γ for 4 h in the presence of bafilomycin A1 (Baf A1), and LC3-II conversion was determined by immunoblots. RI, relative intensity. (F) HeLa cells were transfected with GFP or GFP-TRIM20, and HC analysis was performed. Data, means ± SE; n ≥ 3. *, P < 0.05 (ANOVA, C and E, or t test in F). Bars, 5 µm.
TRIM20 induces autophagy
We focused on TRIM20 as a TRIM strongly induced by IFN-γ (Carthagena et al., 2009; Chae et al., 2011). We confirmed that TRIM20 expression was responsive to IFN-γ in our system and tested its kinetics and dose–response (Fig. S1, E and F). We next used HC analysis to establish in a dose–response setting that TRIM20 was required for IFN-γ–induced autophagy (Fig. S1, G and H). This was confirmed in immunoblot assays of LC3 lipidation in the presence of bafilomycin A1, an inhibitor of autophagic flux (LC3-II conversion; Fig. 1 E). Mirroring these findings, overexpression of GFP-TRIM20 increased LC3 puncta (Fig. 1 F) and enhanced LC3-II conversion in immunoblots (Fig. S1 I); as expected, the LC3-II band was revealed only in bafilomycin A1–treated cells, which protects it from degradation through autophagic flux. These results indicate that activation of autophagy by IFN-γ depends on TRIM20 and that elevated expression of TRIM20, a TRIM whose transcription is known to be strongly activated by IFN-γ (Carthagena et al., 2009; Chae et al., 2011), induces autophagy.
TRIM20 interacts with ULK1, Beclin 1, and ATL16L1
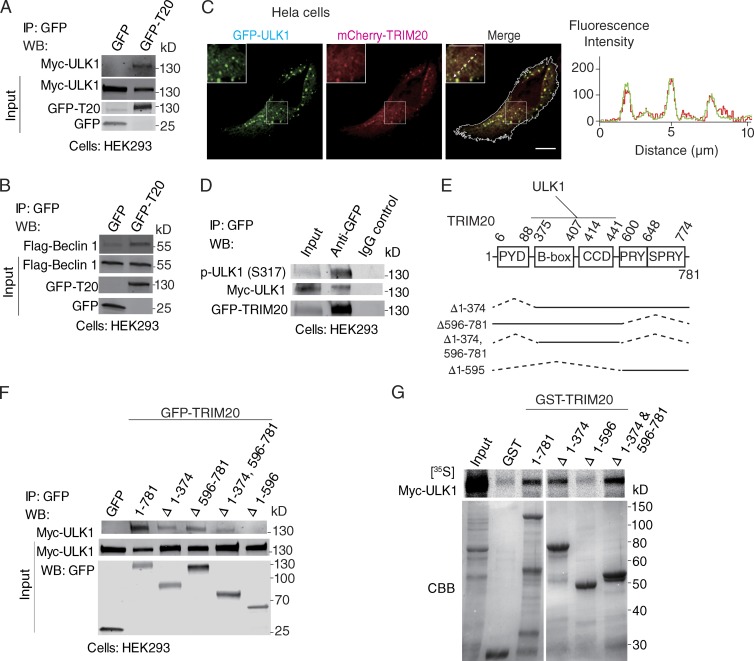
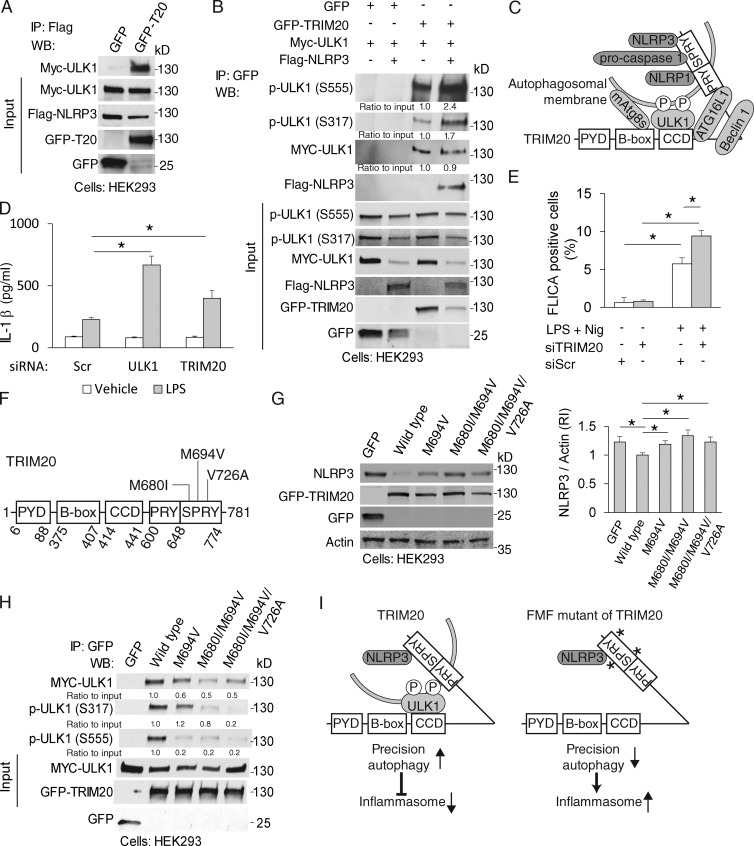
We next examined how TRIM20 induced autophagy. Autophagy requires ULK1 and Beclin 1, both of which play pivotal roles in autophagy initiation in mammalian cells (He and Levine, 2010; Mizushima et al., 2011). We detected GFP-TRIM20 in immunoprecipitates with coexpressed Myc-ULK1 and Flag-Beclin 1 (Fig. 2, A and B) and with endogenous ULK1 and Beclin 1 (Fig. S2, A and B). TRIM20 puncta colocalized with ULK1 in the cytoplasm (Fig. 2 C). Induction of autophagy depends on a phosphorylation cascade, which includes activation of ULK1 by phosphorylation at Ser-317 (Kim et al., 2011). Active p-ULK1 (Ser-317) coimmunoprecipitated with TRIM20 (Fig. 2 D).
Figure 2.
TRIM20 interacts with ULK1 and Beclin 1. (A and B) Coimmunoprecipitation analysis of GFP-TRIM20 (T20) with Myc-ULK1 (A) or Flag-Beclin 1 (B) in HEK293 cells extracts. IP, immunoprecipitation; WB, Western blot. (C) Confocal microscopy of HeLa cells coexpressing mCherry-TRIM20 with GFP-ULK1. Line tracing corresponds to arrow. White outline, cell boundary defined by background fluorescence. Insets, enlarged areas as indicated by white outlined squares. Bars, 10 µm. (D) Coimmunoprecipitation analysis of TRIM20 complexes with p-ULK1 (Ser-317) in HEK293 cells. (E) TRIM20 domains and deletion constructs used. Dotted lines, deleted regions. (F) Coimmunoprecipitation analysis of interactions between deletion variants of TRIM20 (as GFP fusions) with Myc-ULK1 in HEK293 cells. (G) GST pull-down analysis of radiolabeled Myc-ULK1 with GST-tagged deletion variants of TRIM20. (top) Autoradiogram of pull-down products. (bottom) Coomassie brilliant blue (CBB)–stained SDS-polyacrylamide gel with GST deletion variants of TRIM20. Data are representative of three or more experiments.
We next mapped ULK1-binding regions within TRIM20 (Fig. 2 E). Like the majority of TRIMs (Reymond et al., 2001; Kawai and Akira, 2011), TRIM20 has B box, CCD, and PRY/SPRY domains, but lacks an E3 ligase RING domain, and is uniquely endowed with a pyrin domain (PYD). TRIM20 constructs lacking PYD and PRY/SPRY domains still bound ULK1 in immunoprecipitation assays (Fig. 2 F). Direct biding between TRIM20 and ULK1 was established in GST pull-down experiments (Fig. 2 G). Both in vivo and in vitro experiments pointed to the middle portion (including B-box and CCD) of TRIM20 as being critical for association with ULK1, whereas the N-terminal PYD and the C-terminal SPRY domains were dispensable (Fig. 2 E).
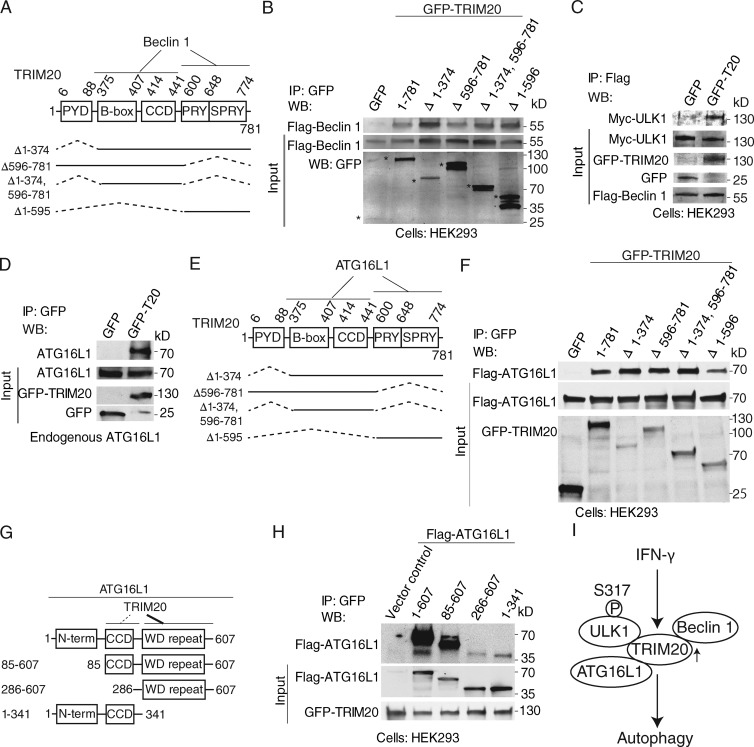
Beclin 1 showed a more complex domain requirement for inclusion in TRIM20 complexes, with either the middle portion (including B-box and CCD) or the C-terminal region (PRY/SPRY) displaying an independent capacity to bring down Beclin 1 in immunoprecipitates (Fig. 3, A and B). We also examined Beclin 1 for regions required for the ability to coimmunoprecipitate with TRIM20 (Fig. S2, C and D). Two Beclin 1 regions appeared to be required: the first one between BH3 (Bcl-2-homology-3) and CCD and the second one overlapping with the evolutionary conserved domain of Beclin 1 (Fig. S2, C and D). Furthermore, in the presence of TRIM20, the immunoprecipitated Beclin 1 complexes were enriched for ULK1 (Fig. 3 C). Thus, TRIM20 can interact simultaneously with multiple autophagy factors and serves as a platform for coassembly of ULK1 and Beclin1.
Figure 3.
TRIM20 assembles ULK1 and Beclin 1 in a complex and interacts with ATG16L1. (A) TRIM20 domains and deletion constructs used. (B) Coimmunoprecipitation analysis of interaction between deletion variants of TRIM20 (as GFP fusions; asterisks denote fusion products on the bottom blot) with Flag-Beclin 1 in HEK293 cells. (C) Coimmunoprecipitation analysis of ULK1 in Beclin 1 complexes in the presence and absence of TRIM20 in HEK293T cell lysates. (D) Coimmunoprecipitation analysis of GFP-TRIM20 with endogenous ATG16L1. (E) TRIM20 domains and deletion constructs used. (F) Coimmunoprecipitation analysis of interaction between deletion variants of TRIM20 with Flag-ATG16L1 in HEK293 cells. (G) ATG16L1 domains and deletion constructs used. (H) Coimmunoprecipitation analysis of interactions between deletion variants of Flag-ATG16L1 and GFP-TRIM20 in HEK293 cells. (I) Model of TRIM20-dependent autophagy induction based on Fig. 2, Fig. 3, and Fig. S2. Data are representative of three or more experiments.
We also found that TRIM20 coimmunoprecipitated with ATG16L1 (Fig. 3 D). TRIM20 displayed a complex domain requirement for inclusion in ATG16L1 complexes, with either the middle portion (including B-box and CCD) or the C-terminal region (PRY/SPRY) showing an independent capacity to bring down ATG16L1 in immunoprecipitates (Fig. 3, E and F). TRIM20 primarily interacted with the WD repeat of ATG16L1 (Fig. 3, G and H). Thus, the TRIM20 platform (Fig. 3 I) contains other autophagy regulators, such as ATG16L1, a component of the autophagy E3-like complex that regulates LC3 conjugation and autophagosome formation (Mizushima et al., 2003).
TRIM20 interacts with a subset of mammalian Atg8 paralogues
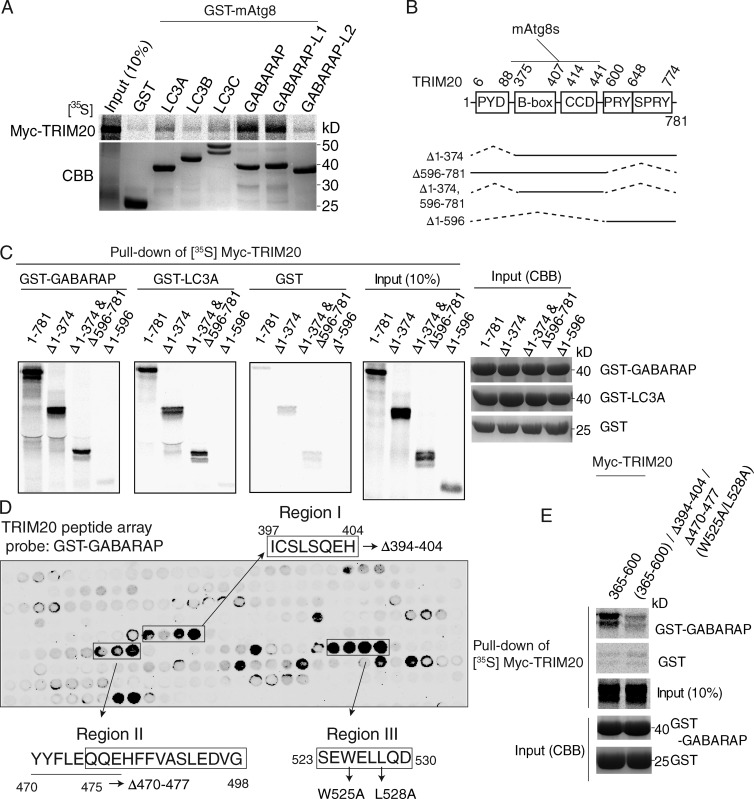
We examined whether TRIM20 possessed the ability to interact with mammalian Atg8 paralogues (mAtg8s), factors required for autophagosomal membrane formation (Mizushima et al., 2011). Although no binding was detected with LC3B, the commonly used marker for autophagic membrane (Kabeya et al., 2000), GST pull-down experiments revealed interactions of TRIM20 with GABARAP and GABARAPL1, and to a lesser extent with LC3A, LC3C, and GABARAPL2 (Fig. 4 A). GABARAP colocalized with TRIM20 (Fig. S2 E). Albeit TRIM20 did not directly interact with LC3B, mCherry-TRIM20 profiles were closely juxtaposed to conventional LC3-positive puncta (Fig. S2 F). The region of TRIM20 (Fig. 4 B) responsible for the interaction with mAtg8s, GABARAP and LC3A, was mapped. A TRIM20 deletion construct spanning residues 375–595 retained capacity to bind GABARAP or LC3A (Fig. 4, B and C). To delimit further the TRIM20 sequence required for mAtg8s binding, we used GST-GABARAP as bait in a binding assay with an array of TRIM20 peptides (Fig. 4 D). Three series of TRIM20 peptides (regions of primary sequence staggered by three amino acid residues), with either three or four positive consecutive binding signals, were identified (Fig. 4 D). The most upstream region (397-ICSLSHQEH-404; region I) did not contain a recognizable LC3-interacting region motif, whereas region II (470-YYFLEQQEHFFVSLEDVG-498) and region III (523-SEWELLQD-530) contained potential LC3-interacting region motifs (Birgisdottir et al., 2013). In follow up mutational analyses, no single or double alterations of the regions I–III abrogated GABARAP binding (Fig. S2 G). Only when all three regions (I, II, and III) were mutated, did this cause loss of GABARAP binding (Fig. 4 E and Fig. S2 G). Thus, all three regions contribute to the binding of TRIM20 to mAtg8s. Collectively, the aforementioned findings and experiments described in previous sections demonstrate that TRIM20 assembles both the key regulators of autophagy (ULK1, Beclin 1, and ATG16L1) and a subset of effector factors (mAtg8s).
Figure 4.
TRIM20 interacts with mammalian Atg8 paralogues (mAtg8s). (A) GST pull-down analysis of interactions between radiolabeled Myc-TRIM20 and GST-tagged mAtg8s. (top) Autoradiogram of pull-down products. (bottom) CBB-stained SDS-polyacrylamide gel with GST-mAtg8s. (B) TRIM20 domains and deletion constructs used. (C) GST pull-down analysis of binding between radiolabeled Myc-TRIM20 deletion variants and GST-GABARAP and GST-LC3A. (D) Identification of GABARAP-interacting regions on TRIM20 by peptide array. Three series of TRIM20 peptides (regions of primary sequence staggered by three amino acid residues), with either three or four positive consecutive binding signals, were identified. The peptide sequences corresponding to the positive binding signals (encompassed spots; defined as regions I, II, and III) were mutated as indicated and were subjected to the GST pull-down experiments in E and Fig. S2 F. (E) GST pull-down analysis of interaction between radiolabeled Myc-TRIM20 triple mutants and GST-GABARAP. Data are representative of three or more experiments.
TRIM20 is a receptor for selective autophagy of inflammasome components
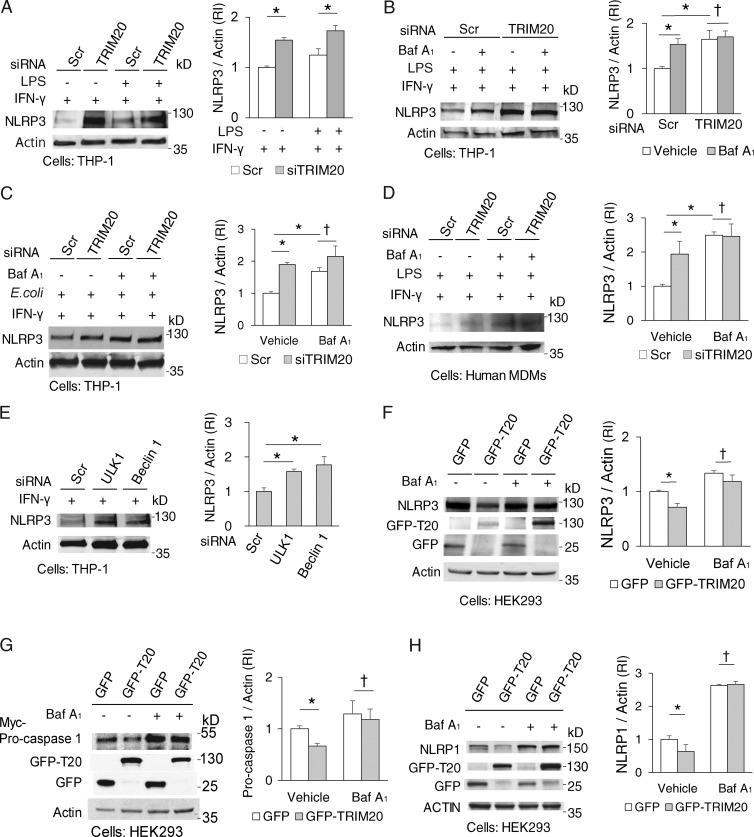
TRIM20, encoded by the MEFV gene, is a risk locus for familial Mediterranean fever (FMF; French FMF Consortium, 1997; The International FMF Consortium, 1997). TRIM20 has 305 FMF-associated variants (http://fmf.igh.cnrs.fr/ISSAID/infevers/), with frequent mutations in its PRY/SPRY domain (Masters et al., 2009). The PYD domain of TRIM20 has been the primary focus of interest in inflammasome regulation as a result of its potential to bind the cognate PYD domain of ASC (Schroder and Tschopp, 2010). However, it has been reported that the PRY/SPRY domain, located at the other end of TRIM20, recognizes, and binds to NLRP3 (Papin et al., 2007). The latter relationship has remained obscure despite the frequency of mutations in the PRY/SPRY domain (Masters et al., 2009). We explored the significance of the interactions between the TRIM20 PRY/SPRY domain and NLRP3 in the context of the aforementioned recognized function of TRIM20 in autophagy. The full-length TRIM20 and a TRIM20 construct containing only the PRY/SPRY domain both interacted with NLRP3 (Fig. S3 A). A knockdown of TRIM20 spared NLRP3 from degradation in cells treated with IFN-γ and lipopolysaccharide (LPS; Fig. 5 A and Fig. S3, B and C). When cells were treated with LPS alone, a knockdown of TRIM20 had no effect on NLRP3 levels (Fig. S3 D), albeit LPS alone increased NLRP3 levels (Fig. S3 E) as expected (Bauernfeind et al., 2009), in keeping with TRIM20 acting to transduce the effects of IFN-γ. Bafilomycin A1 (an inhibitor of autophagic degradation) protected NLRP3, whereas TRIM20 knockdown increased amounts of NLRP3 and eliminated the protective effects of bafilomycin A1 (Fig. 5 B). Similar effects were observed with THP-1 cells exposed to pathogens (adherent-invasive Escherichia coli LF82; Lapaquette et al., 2010) and with primary human peripheral blood monocyte-derived macrophages (MDMs) treated with LPS (Fig. 5, C and D).
Figure 5.
TRIM20 degrades inflammasome components through autophagy. (A) Levels of NLRP3 were determine in lysates from THP-1 cells subjected to TRIM20 or scrambled (Scr) siRNA were activated with 1,000 U/ml IFN-γ for 3 h, and 2.5 µg/ml LPS for 2 h (for optimal TRIM20 expression; Fig. S3 B). RI, relative intensity. (B) Levels of NLRP3 were determined in THP-1 subjected to TRIM20 or control knockdowns and treated or not with bafilomycin A1 (Baf A1). (C) The abundance of NLRP3 protein was determined in THP-1 cells subjected to TRIM20 or control knockdowns and exposed to Escherichia coli strain LF82 and IFN-γ in the presence or absence of bafilomycin A1. (D) The abundance of NLRP3 protein was determined in primary human MDMs subjected to TRIM20 or control knockdowns and exposed to LPS and IFN-γ in the presence or absence of bafilomycin A1. (E) Levels of NLRP3 were determined in THP-1 cells subjected to ULK1, Beclin 1, or scrambled (Scr) siRNA and treated with IFN-γ and LPS. (F–H) Levels of NLRP3 (F), NLRP1 (G), or pro–caspase 1 (H) were determined in cells expressing GFP or GFP-TRIM20 after autophagy induction (Earle's balanced salt solution , 3 h) in the presence or absence of bafilomycin A1. Data, means ± SE; n ≥ 3. *, P < 0.05; †, P ≥ 0.05 (ANOVA).
Degradation of NLRP3 depended on ULK1 and Beclin 1, establishing that disposal of NLRP3 was through autophagy (Fig. 5 E; and Fig. S3 F). Conversely, expression of TRIM20 decreased levels of coexpressed NLRP3 (Fig. 5 F). The destabilizing effect of TRIM20 overexpression on NLRP3 levels was suppressed by bafilomycin A1 (Fig. 5 F). Additionally, TRIM20 was protected by bafilomycin A1 from degradation in the presence of NLRP3 (Fig. S3, G and H), indicating that TRIM20 is degraded along with the delivery of its substrate to autolysosomal compartments.
In addition to NLRP3, other inflammasome components, pro–caspase 1 (Chae et al., 2006; Papin et al., 2007) and NLRP1 (Papin et al., 2007), have been previously shown to interact with the PRY/SPRY domain of TRIM20. When pro–caspase 1 and NLRP1 were coexpressed with TRIM20, they too were subject to degradation inhibitable by bafilomycin A1 (Fig. 5, G and H). These data show that TRIM20 acts as an autophagy receptor for degradation of inflammasome components and that TRIM20 is responsible for delivery of NLRP3 and other tested inflammasome components for autophagic degradation.
Presence of target substrate potentiates assembly of activated autophagic components on the TRIM20 platform
We tested whether the availability of substrate, NLRP3, influenced TRIM20 assembly with ULK1. Although ULK1 was enriched in NLRP3 immunoprecipitates when cells expressed TRIM20 (Fig. 6 A), this was reduced when cells were subjected to TRIM20 knockdown (Fig. S3 I). The presence of NLRP3 did not affect levels of total ULK1 in TRIM20 immunoprecipitates (Fig. 6 B). However, the presence of NLRP3 increased the amount of active p-ULK1 (Ser-317 and Ser-555; Egan et al., 2011; Kim et al., 2011) associated with TRIM20 (Fig. 6 B). Because these two sites of ULK1 are phosphorylated by AMPK (Egan et al., 2011; Kim et al., 2011), we tested whether AMPK is recruited to the TRIM20 complex. AMPK was found in TRIM20 complexes with or without NLRP3 (Fig. S3 J). These data indicate that modulation of TRIM20 action, in the presence of its cognate autophagic target, is reflected in ULK1 phosphorylation state and not in ULK1 or AMPK levels. These and aforementioned data suggest a model in which not only does TRIM20 organize autophagic machinery by serving as a platform for the assembly of ULK1, Beclin 1, ATG16L1, and mAtg8s, but it also recognizes autophagic substrates via its PRY/SPRY domain, and that this substrate recognition enriches ULK1 in its activated state on the TRIM20 platform (Fig. 6 C).
Figure 6.
ULK1 is recruited to NLRP3 complexes by wild-type TRIM20 but not by FMF disease-associate TRIM20 mutants. (A) Coimmunoprecipitation analysis of ULK1 in NLRP3 complexes in HEK293T cells expressing GFP-TRIM20 or GFP alone. IP, immunoprecipitation; WB, Western blot. (B) The effect of NLRP3 expression on the presence of phospho-ULK1 in TRIM20 complexes. Lysates from HEK293 cells transiently expressing Myc-ULK1, GFP-TRIM20 (or GFP alone), and Flag-NLRP3 (or not) were immunoprecipitated with anti-GFP and immunoblots were probed as indicated. (C) Model of TRIM20’s function in autophagy as a regulator-receptor: TRIM20 assembles autophagy machinery (ULK1, Beclin 1, ATG16L1, and mAtg8s) and recognizes substrates (NLRP3, pro–caspase 1, and NLRP1) delivering them for autophagic degradation. The recognition of substrate enriches active p-ULK1 on the TRIM20 platform. (D) The levels of IL-1β were determined in supernatants of THP-1 cells that had been subjected to knockdown of ULK1 or TRIM20, treated with IFN-γ and LPS, and stimulated with nigericin for 30 min. (E) FLICA-positive cells were quantified using THP-1 cells that had been subjected to knockdown of TRIM20, treated with IFN-γ, and then treated with or without LPS (2 h) and nigericin (10 min), and stained for active caspase-1 (with FLICA); >150 cells per experiment were analyzed for quantification. Scr, scrambled. (F) Predominant FMF-associated point mutations of TRIM20 reside in the PRY/SPRY domain. (G) Levels of NLRP3 were determined in lysates of HEK293 cell expressing GFP-TRIM20 (wild-type or FMF-associated variants) or GFP and induced for autophagy by starvation in EBSS for 3 h. (H) Effects of FMF-associated variants on ULK1 presence in TRIM20 complexes. HEK293 cells were transiently transfected with Myc-ULK1, and either GFP-TRIM20 (wild-type or FMF-associated variants) or GFP alone. Lysates were immunoprecipitated with anti-GFP, and immunoblots were probed as indicated. Numbers indicate relative intensity of the band above. (I) Role of FMF-associated mutation in NLRP3 degradation. The presence of NLRP3 promotes phosphorylation of ULK1 in TRIM20 complex, leading to autophagic degradation of NLRP3. TRIM20 mutants recruited less ULK1 and phospho-ULK1, which results in lower autophagic activity and diminishes degradation of inflammasome components. Asterisks denote common FMF-associated point mutations in TRIM20. Data, means ± SE; n ≥ 3. *, P < 0.05 (ANOVA).
Disease-associated mutations in TRIM20 diminish its autophagic potency
A physiologically relevant consequence of TRIM20 mutations in FMF is excessive IL-1β production (Chae et al., 2011; Meinzer et al., 2011; Omenetti et al., 2014). In patients (Omenetti et al., 2014), albeit not in murine systems (Chae et al., 2011), this is dependent on NLRP3 in the context of TRIM20 mutations. A knockdown of ULK1 or TRIM20 elevated IL-1β responses (specifically for IL-1β [Fig. 6 D] because lactate dehydrogenase [LDH] release was unaffected Fig. S4 A). When the cells knocked down for either ULK1 or TRIM20 were also subjected to knockdowns of NLRP3, this normalized IL-1β expression (Fig. S4, B–D). When cells were subjected to inflammasome activation with LPS and nigericin, fluorochrome-labeled inhibitor of caspases (FLICA) staining (based on a fluorogenic probe FAM-YVAD-FMK for detection of in situ caspase 1 activity) revealed active caspase-1 puncta, as reported previously (Broz et al., 2010). The number of FLICA-positive cells increased when cells were subjected to a TRIM20 knockdown (Fig. 6 E and Fig. S4 E). Thus, TRIM20 suppresses caspase-1 activation and IL-1β production.
We then tested whether the disease-causing variants of TRIM20 affected autophagy and clearance of inflammasome components. We chose the three most frequent variants found in FMF patients (Masters et al., 2009), M680I, M694V, and V726A (Fig. 6 F). Compound (double or triple) mutant variants of TRIM20 formed fewer TRIM20 puncta (Fig. S4, F and G). Whereas expression of wild-type TRIM20 resulted in degradation of NLRP3, overexpression of TRIM20 single (M694V), double (M680I and M694V), and triple (M680I, M694V, and V726A) mutants showed diminished degradation of NLRP3 (Fig. 6 G). Furthermore, protein complexes with the M694V, double (M680I+M694V), and triple (M680I+M694V+V726A) TRIM20 mutants harbored less ULK1, a trend that was paralleled by phospho-ULK1 levels (Fig. 6 H). Consistent with this, there were fewer LC3 puncta per cell induced through expression of the triple mutant TRIM20 (M680I+M694V+V726A) than by the wild-type TRIM20 (Fig. S4 H). Thus, the disease-associated mutations in the PRY/SPRY domain of TRIM20 perturb ULK1 recruitment and autophagic degradation of NLRP3 and hence may contribute to the inflammatory phenotype associated with FMF mutations (Fig. 6 I).
TRIM21 interacts with autophagy factors
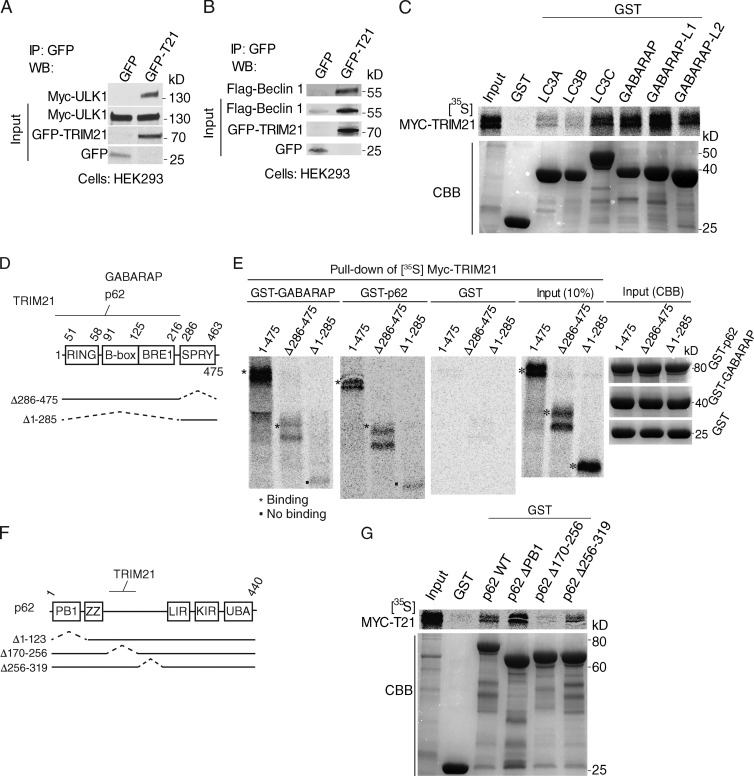
Our IFN-γ screen with TRIM family of proteins yielded additional hits beside TRIM20 (Fig. 1 B), several of which were validated in follow up analyses (Fig. 1 C). Among these was TRIM21 (also known as Ro52/SSA associated with Sjögren syndrome), which is transiently induced by IFN-γ (Fig. S5 A). Incidentally, TRIM20 and 21 could be coimmunoprecipitated (Fig. S5 B). The IFN-γ induction of TRIM21 expression was in agreement with previous studies (Carthagena et al., 2009; Espinosa et al., 2009). TRIM21 has an acknowledged role in regulating type I IFN responses (Higgs et al., 2008; Espinosa et al., 2009; Yoshimi et al., 2009; McEwan et al., 2013; Zhang et al., 2013). In one mechanism, TRIM21 has been reported to cause IKKβ degradation most likely through autophagy, based on its 3-methyladenine protection and LC3 localization (Niida et al., 2010). Based on our detailed studies with TRIM20, we wondered whether TRIM21 might also act as a platform for assembly of autophagic regulatory factors. Indeed, TRIM21 bound both regulators, ULK1 and Beclin 1 (Fig. 7, A and B), and a subset of mAtg8s, most prominently GABARAP (Fig. 7 C). GABARAP binding to TRIM21 did not require the SPRY domain of TRIM21 (Fig. 7, D and E). Unlike TRIM20, which does not bind Sequestosome1/p62 (p62; Mandell et al., 2014), a well-known autophagic receptor (Birgisdottir et al., 2013), TRIM21 did bind p62 (Fig. 7, F and G). The TRIM21-binding region within p62 was delimited to the residues 170–256 of p62 (Fig. 7, F and G). The regions of TRIM21 binding p62 excluded its SPRY domain (Fig. 7, D and E). Thus, TRIM21 interacts with multiple regulators and effectors of autophagy.
Figure 7.
TRIM21 interacts with autophagy regulators and effectors. (A and B) Coimmunoprecipitation analyses of GFP-TRIM21 (T21) with Myc-ULK1 (A) and Flag-Beclin 1 (B) in HEK293 cells extracts. (C) GST pull-down analysis of binding between radiolabeled Myc-TRIM21 and GST-mAtg8s. (top) Autoradiogram of pull-down products. (bottom) CBB-stained SDS-polyacrylamide gel with GST-mAtg8s. (D) TRIM21 domains and deletion constructs used. (E) GST pull-down analysis of binding between radiolabeled Myc-TRIM21 deletion mutants and GST-GABARAP and GST-p62. Asterisks and squares denote presence or absence of Myc-TRIM21, respectively. (F) p62 domains and deletion constructs used. (G) GST pull-down analysis of interaction between radiolabeled Myc-TRIM21 and GST-tagged p62. Data representative of three or more experiments. IP, immunoprecipitation; WB, Western blotting.
TRIM21 is a regulator receptor for autophagic degradation of activated IRF3
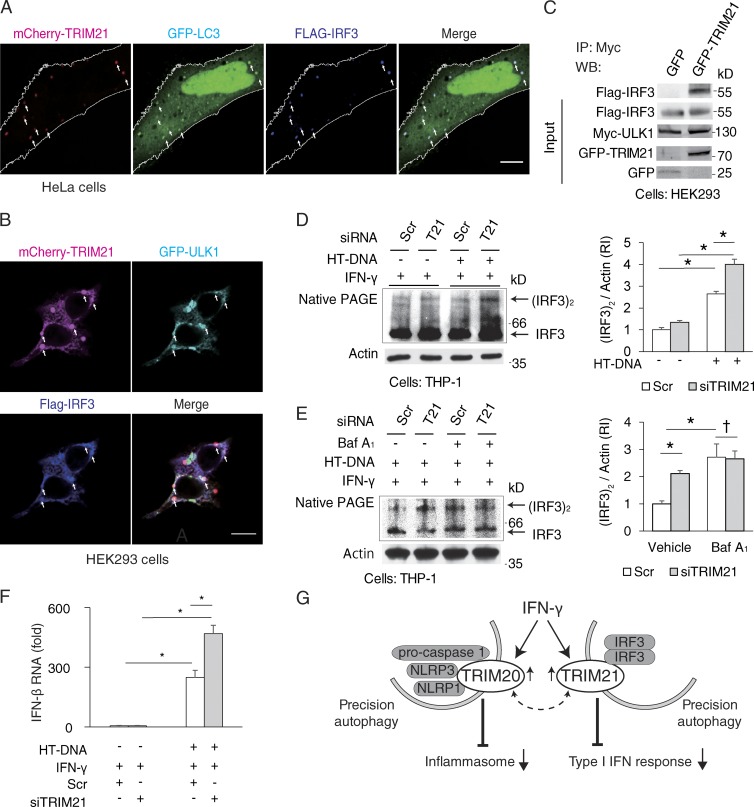
TRIM21 is known to interact with the transcription factor IRF3 (IFN regulatory factor 3) through its SPRY domain (Higgs et al., 2008). It has been proposed that TRIM21 can suppress type I IFN response (Higgs et al., 2008; Espinosa et al., 2009; Yoshimi et al., 2009; Zhang et al., 2013), albeit an activation effect (McEwan et al., 2013), has also been reported. The proposed mechanism for negative regulation of IRF3 is mainly focused on proteasomal degradation of IRF3 (Saitoh et al., 2006; Higgs et al., 2008). However, autophagy is also known to play a suppressive role on type I IFN (Jounai et al., 2007; Saitoh et al., 2009; Mathew et al., 2014; Deretic et al., 2015). We thus wondered whether TRIM21 could cause autophagic degradation of IRF3, analogous to what we observed with TRIM20 and NLRP3. IRF3 colocalized with TRIM21 in LC3-positive dots (Fig. 8 A). Furthermore, IRF3+ TRIM21+ profiles were also ULK1 positive (Fig. 8 B). Moreover, IRF3 was found in protein complexes with ULK1 when TRIM21 was present (Fig. 8 C).
Figure 8.
TRIM21 promotes autophagic degradation of IRF3 dimers and attenuates type I IFN production. (A) Confocal microscopy of HeLa cells coexpressing mCherry-TRIM21, Flag-IRF3, and GFP-LC3B in the presence of bafilomycin A1. White outline, cell boundary. Arrows; colocalization. (B) Confocal microscopy of HEK293 cells coexpressing mCherry-TRIM21, Flag-IRF3, and GFP-ULK1. Arrows, colocalization. (C) Co-immunoprecipitation analysis of IRF3-ULK1 complexes in the presence and absence of TRIM21. Lysates from HEK293 cells transiently expressing Myc-ULK1, Flag-IRF3, and either GFP-TRIM20 or GFP were immunoprecipitated with anti-Myc, and immunoblots were probed as indicated. (D) Levels of dimerized IRF3 were assessed by native PAGE of extracts from THP-1 cells subjected to TRIM21 or control knockdowns and stimulated for 12h by herring testis DNA (HT-DNA) transfected into the cells in the presence of 200 U/ml IFN-γ. (E) Effects of autophagy inhibition (bafilomycin A1) on TRIM21-dependent IRF3 dimer degradation in THP-1 cells. (F) Effects of TRIM21 knockdown on IFN-β mRNA levels after stimulation of THP-1 cells with IFN-γ and HT-DNA. (G) Model of TRIMs’ roles in regulation of inflammation by precision autophagy. TRIM20 targets the inflammasome components for autophagic degradation, whereas TRIM21 targets IRF3 dimer, to suppress inflammasome activity and type I IFN response, respectively. TRIM20 and TRIM21, responding to IFN-γ, directly bind their respective cargo and recruit autophagic machinery to execute degradation. Dashed arrow, cooperation between TRIM20 and TRIM21 may play a role in autophagic responses to IFN-γ. Bars, 10 µm. Data, means ± SE; n ≥ 3; *, P < 0.05; †, P ≥ 0.05 (ANOVA). IP, immunoprecipitation; Scr, scrambled; WB, Western blot.
Cytosolic DNA (during viral infection, e.g., with HIV) induces type I IFN response through endogenous second messenger (cyclic guanosine monophosphate–AMP) by using its adaptor protein STING that results in IRF3 dimerization/activation (Gao et al., 2013). It is the dimerized form of IRF3 that activates type I IFN responses (Takahasi et al., 2003). A knockdown of TRIM21 increased levels of IRF3 dimers in IFN-γ–treated cells stimulated with double-stranded DNA (Herring testis [HT]-DNA) transfected into the cells (Fig. 8 D) but not in cells treated with HT-DNA alone, i.e., in the absence of IFN-γ (Fig. S5 D), in keeping with the role of TRIM21 in acting as an effector of IFN-γ. A knockdown of TRIM21 also increased IRF3 dimers in cells infected with a single-cycle infection HIV-1 virus under conditions when cells were treated with IFN-γ (Fig. S5 E). Bafilomycin A1 protected dimerized IRF3 from degradation; this protection was no longer apparent in cells knocked down for TRIM21 (Fig. 8 E), indicating that dimerized IRF3 was routed for autophagic degradation by TRIM21. As a physiologically relevant consequence, knockdown of TRIM21 resulted in increased levels of IFN-β expression after DNA transfection or infection with HIV-1 (Fig. 8 F and Fig. S5 F). These data show that TRIM21 acts as a platform for IRF3 degradation, connecting it with the autophagic regulators (ULK1) and and effectors such as mAtg8s (Fig. S5 G). A knockdown of TRIM21 resulted in increased levels of IFN-β response to LPS (Fig. S5 H), in keeping with the proposed autophagic targeting of IKKβ (Niida et al., 2010) within a parallel pathway to IRF3-dependent activation of type I IFN responses.
Collectively, our findings show that multiple TRIMs participate in autophagic response to IFN-γ. Specifically, TRIM20 and TRIM21 organize autophagic apparatus to degrade their cognate targets and down-regulate responses via inflammasome/IL-1β and IRF3/type I IFN (Fig. 8 G). Tapering of such responses may be essential to prevent excessive inflammation.
Discussion
Our findings show that a subset of TRIMs act as receptors and regulators for selective autophagy targeting components of the inflammasome and type I IFN response systems. TRIM20 recognizes the inflammasome components, NLRP1, NLRP3, and pro–caspase 1, and leads to their autophagic degradation. A similar principle is at work with TRIM21, which targets activated (dimerized) IRF3 for autophagy. Not only do TRIM20 and TRIM21 directly bind their respective cargo, but they also recruit autophagic machinery thus coordinating target recognition with assembly of the autophagic apparatus and initiation of autophagy. These studies increase the repertoire of currently known autophagic receptors (Johansen and Lamark, 2011; Birgisdottir et al., 2013) and expand the target receptor role of TRIMs in autophagy, previously indicted only for TRIM5α (Mandell et al., 2014). Thus, direct target recognition and assembly of autophagic machinery to conduct a process referred to as precision autophagy (Deretic et al., 2015) is a more general feature of the TRIM family of proteins.
The recognition of cognate targets by TRIM20 and TRIM21 is reminiscent of direct retroviral capsid recognition by TRIM5α (Stremlau et al., 2006), which, as recently shown (Mandell et al., 2014) leads to autophagic degradation of HIV. The principles of precision autophagy (Deretic et al., 2015) may differ fundamentally from targeting of a variety of ubiquitinated cargo earmarked for autophagy by ubiquitin-binding receptors (Stolz et al., 2014). Incidentally, TRIM20 does not possess the RING E3 ubiquitin ligase domain and does not bind p62 (Mandell et al., 2014). The absence of a RING domain and absence of binding to p62 underscores the ubiquitin-independent nature of target recognition by TRIM20. However, engagement of other Sequestosome 1–like receptors, a class (Deretic et al., 2013) of ubiquitin and galectin recognizing receptors (Gomes and Dikic, 2014; Randow and Youle, 2014) may not be ruled out as well as a nontargeting role for ubiquitination in stabilizing autophagy initiation complexes (Shi and Kehrl, 2010; Nazio et al., 2013; Chauhan et al., 2015). Furthermore, inclusion of additional cytoplasmic material along with specific targets during TRIM-directed autophagy may not be ruled out.
Importantly, our findings indicate that substrate recognition by TRIM20 also directs precision autophagy machinery assembled by TRIM20. Thus, in their role in autophagy, TRIM20 and TRIM21 act not only as receptors for autophagy but also as platforms for assembly of regulators (ULK1 and Beclin 1) and effectors (mAtg8s; p62 in the case of TRIM21), into initiation complexes. The presence in TRIM20 complexes of ATG16L1 may reflect direct association or reinforcement of indirect links between ULK1 and ATG16L1 (Gammoh et al., 2013; Nishimura et al., 2013). Other TRIMs may function similarly, as observed with TRIM5α and preliminarily with TRIM6, TRIM17, TRIM22, TRIM49, and TRIM55 (Mandell et al., 2014). The concept of platforms for assembly of autophagic machinery in mammalian cells also extends to generic, starvation-induced autophagy, which utilizes exocyst components specifically endowed with Exo84 (Bodemann et al., 2011). However, TRIM engagement with autophagy may entail other mechanisms, as for example TRIM28 has multiple (both positive and negative) proposed mechanisms of action (Barde et al., 2013; Yang et al., 2013; Pineda et al., 2015), whereas the mechanism of autophagy induction for TRIM13 in response to the ER stress has not been fully delineated (Tomar et al., 2012), although it shows a relationship with p62 and DFCP, an ER-derived autophagy precursor compartment termed omegasome (Axe et al., 2008).
A further major biological finding reported here is that TRIMs are mediators of IFN-γ–induced autophagy. The engagement of multiple TRIMs reveled in our screen should not be surprising, as multiple systems can trigger IFN-γ–induced autophagy, such as the previously described death-associated protein kinase phosphorylation of Beclin 1 (Inbal et al., 2002; Zalckvar et al., 2009) and immunity related GTPase–dependent induction of autophagy (Gutierrez et al., 2004), which has recently been shown to act through a coassembly of ULK1 and Beclin 1 (Chauhan et al., 2015). Additional upstream mechanisms may be controlled by TRIMs detected in our screen, as in the case of TRIM8, which is known to be inducible by IFN-γ (Toniato et al., 2002). TRIM8 activates TAK1 (Li et al., 2011), which is proposed to occur through K63 polyubiquitination. TAK1, in turn, activates AMPK-dependent autophagy (Kanayama et al., 2004; Herrero-Martín et al., 2009; Criollo et al., 2011) by phosphorylating AMPK (Xie et al., 2006). Hence, TRIM8 affects upstream pathways known to activate autophagy. This may explain why TRIM8 was identified as a hit in our IFN-γ–dependent autophagy induction screen. Furthermore, it is likely that TRIMs, known to hetero-oligomerize (Bell et al., 2012) as supported by our observations with TRIM20 and TRIM21, cooperate in IFN-γ induction of autophagy.
The finding that TRIM20 is a mediator of IFN-γ suppression of inflammasome activation provides a mechanism for this important IFN-γ effect in prevention of excessive inflammasome activation and associated pathology in infectious and autoimmune diseases (Minguela et al., 2007; Nandi and Behar, 2011), for which a satisfactory definition has been lacking albeit indirect mechanisms have been proposed (Mishra et al., 2013). The TRIM20-dependent direct recognition and autophagic degradation of the inflammasome components NLRP3, pro–caspase 1, and NLRP1, differs from the previous studies of indirect effect on inflammasome activation via mitophagy (Nakahira et al., 2011; Zhou et al., 2011), and is more akin to the proposed autophagic degradation of AIM2, a sensory component of the DNA-reactive specialized inflammasome, albeit AIM2 has been proposed to be eliminated by a ubiquitin-tag recognizing receptor (Shi et al., 2012). We furthermore demonstrated that FMF disease-associated mutations in the PRY/SPRY domain of TRIM20 (Masters et al., 2009), alter the capacity of TRIM20 to direct autophagic degradation of inflammasome components. These mutations reduced the binding of ULK1, thus explaining in part how the common mutations associated with FMF work. We propose that IFN-γ–TRIM20 autophagy axis normally suppress excessive inflammasome and IL-1β activation and that this ability is blunted by common disease-associated TRIM20 polymorphisms occurring in FMF.
The reported TRIM21-dependent suppression of type I IFN activation by autophagic degradation of IRF3 dimers mirrors the action of TRIM20 in suppressing inflammasome activation. TRIM21, an autoantigen associated with Sjögren syndrome and systemic lupus erythematosus, suppresses type I IFN response (Higgs et al., 2008; Espinosa et al., 2009; Yoshimi et al., 2009; Zhang et al., 2013), albeit this has been ascribed to proteasomal degradation of IRF3 (Higgs et al., 2008) and IRF7 (Higgs et al., 2010). Nevertheless, type I IFN can also be activated by NF-kB, and autophagy has been implicated in degradation of the upstream NF-kB activating kinase, IKKβ (Niida et al., 2010). The TRIM21-directed autophagic degradation of activated IRF3 shown here complements the action of TRIM21 on NF-kB (Niida et al., 2010). Although the mechanism is not fully known, activation of type I IFN system is one major feature of Sjögren syndrome and systemic lupus erythematosus (Banchereau and Pascual, 2006). We thus raise the possibility that perturbations of IFN-γ–TRIM21 autophagy axis may cause activation of type I IFN in autoimmune diseases.
Our findings reported here broaden the concept of TRIMs acting as autophagic receptors and as platforms for assembly of autophagy initiation complexes. Our findings also link cargo recognition by a TRIM, acting as an autophagic receptor, with the function of the same TRIM in the assembly of autophagic machinery triggering the execution of autophagy of a very specific cytoplasmic targets. This brand of autophagy, termed precision autophagy, is guided by TRIMs and has important biological functions. For example, the TRIM20- and TRIM21-precision autophagy uncovered here balances key innate immunity responses, potentially serving as a guardian against excessive inflammation, which in turn may cause pathology during autoimmune processes or in infections causing cytokine storms. We propose that the large family of TRIMs with 70 members in humans endows cells with a precision in deploying autophagy.
Materials and methods
Cells, plasmids, siRNA, and transfection
THP-1, HeLa, and HEK293T cells were obtained from ATCC. Human peripheral blood monocytes were from StemCell Technologies or from healthy individual donors and cultured as described previously (Gutierrez et al., 2004). THP-1 cells were differentiated with 50 nM phorbol 12-myristate 13-acetate (PMA) overnight before use. Full-length human TRIM20 was synthesized, and TRIM21 was purchased from DNASU, and both were cloned by PCR into pDONR221. The TRIMs mutants were generated by site-directed mutagenesis and confirmed by sequencing. pENTR or pDONR221 vectors were generated by BP cloning, and expression vectors were made by the LR reaction (Gateway; Invitrogen). Other plasmids used were Beclin 1 and its deletion mutants (from B. Levine, University of Texas Southwestern Medical Center, Dallas, TX), ULK1 (from S. Tooze, London Research Institute, Cancer Research UK, London, England, UK), ATG16L1 and its deletion mutants (from R. Xavier, Massachusetts General Hospital, Boston, MA), pCI-Caspase 1 (from K. Fitzgerald, University of Massachusetts, Worcester, MA), IRF3 (DNASU), pUNO1-hNLRP3a, and pUNO1-hNLRP1 (InvivoGen). siRNAs were from GE Healthcare and were delivered to cells by either RNAiMAX (Life Technologies) or nucleoporation (Amaxa). Plasmid transfections were performed by either calcium phosphate or nucleoporation (Amaxa). HT-DNA (Sigma-Aldrich) was transfected as described previously (Gao et al., 2013).
Bacterial and viral infection
For infection studies, Escherichia coli strain LF82 (Lapaquette et al., 2010) was infected at MOI of 1:20. Single-cycle infection HIV-1 viruses were generated as previously described (Mandell et al., 2014) and were infected to undifferentiated THP-1 cells (Gao et al., 2013).
Antibodies and reagents
Antibodies used were: Flag (Sigma-Aldrich), HA (Roche), LC3 (Sigma-Aldrich), AMPK, ULK1 p-Ser 317, and p-Ser 555 (Cell Signaling Technology), NLRP1 (Cell Signaling Technology), NLRP3 (Adipogen), Caspase-1, and ULK1 (Santa Cruz Biotechnology, Inc.), and GFP, IRF3, Myc, and Actin (Abcam). To determine autophagic activity by immunoblotting, cells were cultured in the presence of bafilomycin A1, and lysates were subjected to immunoblotting as described previously (Mizushima et al., 2010). The reagents used were Ultrapure LPS (InvivoGen), IFN-γ (PeproTech), Cytotoxic LDH assay (Promega), and TO-PRO-3 Iodide (Life Technologies). Immunoblotting and immunostaining were conducted as previously described (Kyei et al., 2009). FAM-YVAD-FMK stainings (FLICA; ImmunoChemistry Technologies) were performed according to the manufacturer’s instructions.
IL-1β measurement
For IL-1β secretion, THP-1 cells that had been subjected to the differentiation with 50 nM PMA overnight were treated with 2.5 µg/ml LPS for 2 h and then treated with 20 µM nigericin for 30 min. IL-1β measurements were performed using HEK-Blue IL-1β cells (InvivoGen).
TRIM family screen
THP-1 cells were cultured in 96-well plates containing SMARTpool siRNA (GE Healthcare), RNAiMAX (Life Technologies), and PMA. Culture media were changed after overnight incubation, and 48 h after plating, cells were treated with IFN-γ or vehicle for 4 h, and then fixed and stained to detect endogenous LC3 (Alexa Fluor 488 as a fluorochrome) and nuclei (Hoechst 33342). Plates with cells were subjected to HC analysis for image acquisition and data processing. Two separate siRNA screen for induced autophagy were performed with the cutoff (>3 SDs change relative to the mean of stimulated control) for hits.
HC image analysis
HC imaging and analysis was performed using a Cellomics VTI HCS scanner and iDEV software (Thermo Fisher Scientific). Automated epifluorescence image collection was performed until a minimum of 500 cells per well per siRNA knockdown per plate was acquired. Epifluorescence images were machine analyzed using present scanning parameters and object mask definitions. Hoechst 33342 staining were used to automatically detect cellular outlines based on background staining of the cytoplasm, and the mean count of LC3 puncta per cell was determined. Autophagy induction with IFN-γ resulted in a Z′ value of 0.87.
HC analysis of puncta in subpopulations of transfected cells
HeLa and THP-1 cells were transfected with plasmids or siRNA and cultured in full media for overnight (plasmids) or 48 h (siRNA). Cells were then fixed and stained to detect, LC3 (Alexa Fluor 488 or 568 as fluorochromes), GFP, and nuclei. HC imaging and analysis was performed using a Cellomics VTI HCS scanner and iDEV software (Thermo Fisher Scientific). >200 cells were analyzed in more than quadruplicate manner using a 20× objective. Hoechst 33342 staining were used to automatically detect cellular outlines based on background Hoechst staining, and the mean total count or area of punctate of LC3, or TRIM20 per cell was determined. For subpopulation analyses, cells that have above the threshold of the background fluorescence were gated as successfully transfected ones.
Fluorescence confocal microscopy
Fluorescence confocal microscopy was performed as described previously (Kyei et al., 2009). Images were acquired using a microscope (META; Carl Zeiss) equipped with a 63×/1.4 NA oil objective, camera (LSM META; Carl Zeiss), and AIM software (Carl Zeiss). Fluorochromes associated with secondary antibodies were Alexa Fluor 488, 568, or 647.
IRF3 dimerization assay and quantitative RT-PCR
Detection of IRF3 dimerization was performed by native PAGE as previously described (Takahasi et al., 2003). Quantitative RT-PCR was performed as previously described (Kimura et al., 2013) using the following primer sets: ULK1, 5′-AGATGTTCCAGCACCGTGAG and AATGCACAGCTTGCACTTGG-3′; BECN1, 5′-GGAGAACCTCAGCCGAAGAC-3′ and 5′-ACGTTGAGCTGAGTGTCCAG-3′; ACTIN, 5′-GGGCATGGGTCAGAAGGATT-3′ and 5′-TCGATGGGGTACTTCAGGGT-3′; TRIM1, 5′-AAGAATGTGACGAGTTGGTAGAG-3′ and ATGAGGACTGTTGACCGTTC-3′; TRIM5, 5′-CATGCCTCACTGCAAACCAC-3′ and 5′-GGTAACTGATCCGGCACACA-3′; TRIM8, 5′-ATCCTGATGGACAGGACCCA-3′ and 5′-CTCCTTCTTGGCCACTTCGT-3′; TRIM16, 5′-GTAAGCCCACGAACACAAATG-3′ and 5′-TCCAGCCCTGAAACTTCTATTC-3′; TRIM20, 5′-CTGAGTCAGGAGCACCAAGG-3′ and 5′-GCTGCTCCTCCCCTGATTTT-3′; TRIM21, 5′-CAGTCTCGGAAACACCGTGA-3′ and 5′-AATGCCACCTGGAGCTTCTC-3′; TRIM22, 5′-CTCGACCTGCTTATCCGTATTT-3′ and 5′-CTCAGCACAAGGGCTACTATG-3′; TRIM28, 5′-CCATACTGTGCGCTCTACTG-3′ and 5′-GGTTCATGCTTGTGTACGTTG-3′; TRIM56, 5′-TTCTTCGTCAATGGGCTGCT-3′ and 5′-AAGTCATCGGCACAGTCCAG-3′; and TRIM65, 5′-GATCTACCTGAACTTGCCTCTG-3′ and 5′-GAGGAGGGAGGAATCTGTCT-3′. For IFN-β and GAPDH, TaqMan probes and real-time PCR master mixes were used.
Coimmunoprecipitation and GST pull-down
Coimmunoprecipitations were performed as previously described (Kyei et al., 2009) with slight modification. In brief, cells were lysed with NP-40 buffer (Life Technologies) containing 1 mM PMSF and protease inhibitor cocktail (Roche) for 45 min followed by centrifugation. Supernatants were incubated for 2 h with antibodies at 4°C. The immune complexes were captured with Dynabeads (Life Technologies). Immunoprecipitates were washed three times with PBS, eluted with Laemmli SDS-PAGE sample buffer, and subjected to immunoblot analysis.
GST and GST-tagged proteins were expressed in Escherichia coli BL21 (DE3) or SoluBL21 (Amsbio). GST and GST fusion proteins were purified and immobilized on glutathione-coupled Sepharose beads (Glutathione-Sepharose 4 Fast Flow; GE Healthcare), and pull-down assays with in vitro translated [35S]-labeled proteins were performed as described previously (Pankiv et al., 2007). The [35S]-labeled proteins were produced using the TNT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]l-methionine. The proteins were eluted from washed beads by boiling for 5 min in SDS-PAGE gel loading buffer, separated by SDS-PAGE, and radiolabeled proteins were detected in a bioimaging analyzer (BAS-5000; Fujifilm).
Peptide array overlay assay
Peptide arrays were synthesized on cellulose membrane using a MultiPep automated peptide synthesizer (INTAVIS Bioanalytical Instruments AG) as described previously (Kramer et al., 1996). Interaction analyses between peptide and recombinant protein were probed by overlaying the membranes with recombinant protein, and bound proteins were detected with HRP-conjugated anti-GST antibody (clone RPN1236; GE Healthcare).
Statistical analyses
Either a two-tailed Student’s t test or analysis of variance (ANOVA) was used. Statistical significance was defined as P < 0.05.
Online supplemental material
Fig. S1 shows IFN-γ dose response for LC3 puncta formation; induction of autophagy in primary human monocyte–derived macrophages; knockdown efficacy and effects; time course for IFN-γ; averages and range for screen data in Fig. 1 B; induction of TRIM20 expression by IFN-γ; and induction of autophagy by TRIM20 overexpression. Fig. S2 shows TRIM20 interaction with endogenous ULK1 and Beclin 1; mapping of Beclin 1 regions interacting with TRIM20; colocalization of TRIM20 with GABARAP and partial juxtaposition with LC3B; and controls for GST pull-downs with GABARAP. Fig. S3 shows interaction of NLRP3 with TRIM20; LPS and IFN-γ effects on TRIM20-dependent degradation of NLRP3; knockdown efficacy controls; bafilomycin A1 protection of TRIM20 degradation in the presence of NLRP3; endogenous interaction of NLRP3 and ULK1 is mediated by TRIM20; and small amounts of AMPK are in TRIM20 complexes independent of NLRP3. Fig. S4 shows IL1β secretion and controls; images for FLICA; and effects of TRIM20 FMF-associated polymorphisms on number of TRIM20 and LC3 puncta. Fig. S5 shows expression study for TRIM21; presence of TRIM20 and TRIM21 in common protein complexes; knockdown efficacy; absence of effects on IRF3 dimer levels in the absence of IFN-γ; effects of TRIM21 on levels of dimerized IRF3 in HIV1 infection; TRIM21 effects on LPS-induced IFN-β levels; and model of TRIM21 function in precision autophagy of IRF3 dimers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503023/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201503023.dv.
Supplementary Material
Acknowledgments
We thank K. Fitzgerald, B. Levine, R. Xavier, and S. Tooze for constructs.
This work was supported by grants AI042999 and AI111935 from the National Institutes of Health and a grant from the Bill and Melinda Gates Foundation and in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health through grant UL1 TR000041. T. Kimura was supported by Manpei Suzuki Diabetes Foundation and Uehara Memorial Foundation. T. Johansen was supported by grant 196898 from the Norwegian Research Council and grant 71043-PR-2006-0320 from the Norwegian Cancer Society.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMPK
- AMP-activated protein kinase
- ANOVA
- analysis of variance
- CBB
- Coomassie brilliant blue
- CCD
- coiled-coil domain
- FLICA
- fluorochrome-labeled inhibitor of caspases
- FMF
- familial Mediterranean fever
- GABARAP
- γ-aminobutyric acid receptor–associated protein
- HC
- high content
- HT
- Herring testis
- LDH
- lactate dehydrogenase
- LPS
- lipopolysaccharide
- mAtg8
- mammalian Atg8
- MDM
- monocyte-derived macrophage
- PMA
- phorbol 12-myristate 13-acetate
- PYD
- pyrin domain
- TRIM
- tripartite motif
References
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J., and Pascual V.. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 25:383–392. 10.1016/j.immuni.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Barde I., Rauwel B., Marin-Florez R.M., Corsinotti A., Laurenti E., Verp S., Offner S., Marquis J., Kapopoulou A., Vanicek J., and Trono D.. 2013. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 340:350–353. 10.1126/science.1232398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F.G., Horvath G., Stutz A., Alnemri E.S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B.G., Fitzgerald K.A., et al. 2009. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183:787–791. 10.4049/jimmunol.0901363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.L., Malyukova A., Holien J.K., Koach J., Parker M.W., Kavallaris M., Marshall G.M., and Cheung B.B.. 2012. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS ONE. 7:e37470 10.1371/journal.pone.0037470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., and Johansen T.. 2013. The LIR motif - crucial for selective autophagy. J. Cell Sci. 126:3237–3247. [DOI] [PubMed] [Google Scholar]
- Bodemann B.O., Orvedahl A., Cheng T., Ram R.R., Ou Y.H., Formstecher E., Maiti M., Hazelett C.C., Wauson E.M., Balakireva M., et al. 2011. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 144:253–267. 10.1016/j.cell.2010.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthagena L., Bergamaschi A., Luna J.M., David A., Uchil P.D., Margottin-Goguet F., Mothes W., Hazan U., Transy C., Pancino G., and Nisole S.. 2009. Human TRIM gene expression in response to interferons. PLoS ONE. 4:e4894 10.1371/journal.pone.0004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J.J., Wood G., Masters S.L., Richard K., Park G., Smith B.J., and Kastner D.L.. 2006. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1β production. Proc. Natl. Acad. Sci. USA. 103:9982–9987. 10.1073/pnas.0602081103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J.J., Cho Y.H., Lee G.S., Cheng J., Liu P.P., Feigenbaum L., Katz S.I., and Kastner D.L.. 2011. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 34:755–768. 10.1016/j.immuni.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., and Tooze S.A.. 2009. Evolution of Atg1 function and regulation. Autophagy. 5:758–765. 10.4161/auto.8709 [DOI] [PubMed] [Google Scholar]
- Chauhan S., Mandell M.A., and Deretic V.. 2015. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell. 58:507–521. 10.1016/j.molcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A., Niso-Santano M., Malik S.A., Michaud M., Morselli E., Mariño G., Lachkar S., Arkhipenko A.V., Harper F., Pierron G., et al. 2011. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 30:4908–4920. 10.1038/emboj.2011.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., and Akira S.. 2013. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13:722–737. 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Kimura T., Timmins G., Moseley P., Chauhan S., and Mandell M.. 2015. Immunologic manifestations of autophagy. J. Clin. Invest. 125:75–84. 10.1172/JCI73945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan D.F., Shackelford D.B., Mihaylova M.M., Gelino S., Kohnz R.A., Mair W., Vasquez D.S., Joshi A., Gwinn D.M., Taylor R., et al. 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 331:456–461. 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F.J., Sjöstrand M., Eloranta M.L., Ní Gabhann J., Winqvist O., et al. 2009. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23–Th17 pathway. J. Exp. Med. 206:1661–1671. 10.1084/jem.20090585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabri M., Stenger S., Shin D.M., Yuk J.M., Liu P.T., Realegeno S., Lee H.M., Krutzik S.R., Schenk M., Sieling P.A., et al. 2011. Vitamin D is required for IFN-γ-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 3:104ra102 10.1126/scitranslmed.3003045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frake R.A., Ricketts T., Menzies F.M., and Rubinsztein D.C.. 2015. Autophagy and neurodegeneration. J. Clin. Invest. 125:65–74. 10.1172/JCI73944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French FMF Consortium 1997. A candidate gene for familial Mediterranean fever. Nat. Genet. 17:25–31. 10.1038/ng0997-25 [DOI] [PubMed] [Google Scholar]
- Gammoh N., Florey O., Overholtzer M., and Jiang X.. 2013. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat. Struct. Mol. Biol. 20:144–149. 10.1038/nsmb.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.T., Du F., Aroh C., Yan N., Sun L., and Chen Z.J.. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 341:903–906. 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P., and Dinarello C.A.. 1988. IL-1 induces IL-1. III. Specific inhibition of IL-1 production by IFN-γ. J. Immunol. 140:4238–4244. [PubMed] [Google Scholar]
- Gomes L.C., and Dikic I.. 2014. Autophagy in antimicrobial immunity. Mol. Cell. 54:224–233. 10.1016/j.molcel.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., and Deretic V.. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 119:753–766. 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- He C., and Levine B.. 2010. The Beclin 1 interactome. Curr. Opin. Cell Biol. 22:140–149. 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Martín G., Høyer-Hansen M., García-García C., Fumarola C., Farkas T., López-Rivas A., and Jäättelä M.. 2009. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 28:677–685. 10.1038/emboj.2009.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs R., Ní Gabhann J., Ben Larbi N., Breen E.P., Fitzgerald K.A., and Jefferies C.A.. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780–1786. 10.4049/jimmunol.181.3.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs R., Lazzari E., Wynne C., Ní Gabhann J., Espinosa A., Wahren-Herlenius M., and Jefferies C.A.. 2010. Self protection from anti-viral responses—Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-Like receptors. PLoS ONE. 5:e11776 10.1371/journal.pone.0011776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal B., Bialik S., Sabanay I., Shani G., and Kimchi A.. 2002. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell Biol. 157:455–468. 10.1083/jcb.200109094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., and Lamark T.. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 7:279–296. 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., and Okuda K.. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA. 104:14050–14055. 10.1073/pnas.0704014104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A., Seth R.B., Sun L., Ea C.K., Hong M., Shaito A., Chiu Y.H., Deng L., and Chen Z.J.. 2004. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell. 15:535–548. 10.1016/j.molcel.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2011. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol. Med. 3:513–527. 10.1002/emmm.201100160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific C.M., and Debnath J.. 2015. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 25:37–45. 10.1016/j.tcb.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.M., Strack S., Wild F., Hanashima A., Gasch A., Brohm K., Reischl M., Carnio S., Labeit D., Sandri M., et al. 2014. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 10:123–136. 10.4161/auto.26841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim Y.C., Fang C., Russell R.C., Kim J.H., Fan W., Liu R., Zhong Q., and Guan K.L.. 2013. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 152:290–303. 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Takahashi A., Takabatake Y., Namba T., Yamamoto T., Kaimori J.Y., Matsui I., Kitamura H., Niimura F., Matsusaka T., et al. 2013. Autophagy protects kidney proximal tubule epithelial cells from mitochondrial metabolic stress. Autophagy. 9:1876–1886. 10.4161/auto.25418 [DOI] [PubMed] [Google Scholar]
- Konno H., Konno K., and Barber G.N.. 2013. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 155:688–698. 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R.M., Roberts E.F., Um S.L., Börsch-Haubold A.G., Watson S.P., Fisher M.J., and Jakubowski J.A.. 1996. p38 mitogen-activated protein kinase phosphorylates cytosolic phospholipase A2 (cPLA2) in thrombin-stimulated platelets. Evidence that proline-directed phosphorylation is not required for mobilization of arachidonic acid by cPLA2. J. Biol. Chem. 271:27723–27729. 10.1074/jbc.271.44.27723 [DOI] [PubMed] [Google Scholar]
- Kroemer G. 2015. Autophagy: a druggable process that is deregulated in aging and human disease. J. Clin. Invest. 125:1–4. 10.1172/JCI78652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei G.B., Dinkins C., Davis A.S., Roberts E., Singh S.B., Dong C., Wu L., Kominami E., Ueno T., Yamamoto A., et al. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255–268. 10.1083/jcb.200903070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapaquette P., Glasser A.L., Huett A., Xavier R.J., and Darfeuille-Michaud A.. 2010. Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12:99–113. 10.1111/j.1462-5822.2009.01381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yan J., Mao A.P., Li C., Ran Y., Shu H.B., and Wang Y.Y.. 2011. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. USA. 108:19341–19346. 10.1073/pnas.1110946108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., and Levine B.. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Liang Q., Seo G.J., Choi Y.J., Kwak M.J., Ge J., Rodgers M.A., Shi M., Leslie B.J., Hopfner K.P., Ha T., et al. 2014. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 15:228–238. 10.1016/j.chom.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Galluzzi L., Zitvogel L., and Kroemer G.. 2013. Autophagy and cellular immune responses. Immunity. 39:211–227. 10.1016/j.immuni.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., Yamamoto A., Hamasaki M., Noda T., Isaka Y., and Yoshimori T.. 2013. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32:2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C., Silvestri G., Münch J., Kirchhoff F., Simonsen A., et al. 2014. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell. 30:394–409. 10.1016/j.devcel.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S.L., Simon A., Aksentijevich I., and Kastner D.L.. 2009. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu. Rev. Immunol. 27:621–668. 10.1146/annurev.immunol.25.022106.141627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Khor S., Hackett S.R., Rabinowitz J.D., Perlman D.H., and White E.. 2014. Functional role of autophagy-mediated proteome remodeling in cell survival signaling and innate immunity. Mol. Cell. 55:916–930. 10.1016/j.molcel.2014.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan W.A., Tam J.C., Watkinson R.E., Bidgood S.R., Mallery D.L., and James L.C.. 2013. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol. 14:327–336. 10.1038/ni.2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinzer U., Quartier P., Alexandra J.F., Hentgen V., Retornaz F., and Koné-Paut I.. 2011. Interleukin-1 targeting drugs in familial Mediterranean fever: a case series and a review of the literature. Semin. Arthritis Rheum. 41:265–271. 10.1016/j.semarthrit.2010.11.003 [DOI] [PubMed] [Google Scholar]
- Minguela A., Pastor S., Mi W., Richardson J.A., and Ward E.S.. 2007. Feedback regulation of murine autoimmunity via dominant anti-inflammatory effects of interferon γ. J. Immunol. 178:134–144. 10.4049/jimmunol.178.1.134 [DOI] [PubMed] [Google Scholar]
- Mishra B.B., Rathinam V.A., Martens G.W., Martinot A.J., Kornfeld H., Fitzgerald K.A., and Sassetti C.M.. 2013. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat. Immunol. 14:52–60. 10.1038/ni.2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., and Yoshimori T.. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116:1679–1688. 10.1242/jcs.00381 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Levine B.. 2010. Methods in mammalian autophagy research. Cell. 140:313–326. 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., and Ohsumi Y.. 2011. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27:107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12:222–230. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi B., and Behar S.M.. 2011. Regulation of neutrophils by interferon-γ limits lung inflammation during tuberculosis infection. J. Exp. Med. 208:2251–2262. 10.1084/jem.20110919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazio F., Strappazzon F., Antonioli M., Bielli P., Cianfanelli V., Bordi M., Gretzmeier C., Dengjel J., Piacentini M., Fimia G.M., and Cecconi F.. 2013. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 15:406–416. 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- Niida M., Tanaka M., and Kamitani T.. 2010. Downregulation of active IKK β by Ro52-mediated autophagy. Mol. Immunol. 47:2378–2387. 10.1016/j.molimm.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Kaizuka T., Cadwell K., Sahani M.H., Saitoh T., Akira S., Virgin H.W., and Mizushima N.. 2013. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 14:284–291. 10.1038/embor.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omenetti A., Carta S., Delfino L., Martini A., Gattorno M., and Rubartelli A.. 2014. Increased NLRP3-dependent interleukin 1β secretion in patients with familial Mediterranean fever: correlation with MEFV genotype. Ann. Rheum. Dis. 73:462–469. 10.1136/annrheumdis-2012-202774 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Papin S., Cuenin S., Agostini L., Martinon F., Werner S., Beer H.D., Grütter C., Grütter M., and Tschopp J.. 2007. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1β processing. Cell Death Differ. 14:1457–1466. 10.1038/sj.cdd.4402142 [DOI] [PubMed] [Google Scholar]
- Pineda C.T., Ramanathan S., Fon Tacer K., Weon J.L., Potts M.B., Ou Y.H., White M.A., and Potts P.R.. 2015. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 160:715–728. 10.1016/j.cell.2015.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V., Rybina S., Gerbal F., Delort F., Vicart P., Baldacci G., and Karsenti E.. 2013. MURF2B, a novel LC3-binding protein, participates with MURF2A in the switch between autophagy and ubiquitin proteasome system during differentiation of C2C12 muscle cells. PLoS ONE. 8:e76140 10.1371/journal.pone.0076140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J.D., and White E.. 2010. Autophagy and metabolism. Science. 330:1344–1348. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F., and Youle R.J.. 2014. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 15:403–411. 10.1016/j.chom.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151. 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Codogno P., and Levine B.. 2012. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11:709–730. 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R.C., Tian Y., Yuan H., Park H.W., Chang Y.Y., Kim J., Kim H., Neufeld T.P., Dillin A., and Guan K.L.. 2013. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15:741–750. 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K.P., and Yamaoka S.. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598–605. 10.1038/ni1347 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 456:264–268. 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Hayashi T., Takahara K., Satoh T., Lee H., Matsunaga K., Kageyama S., Omori H., Noda T., et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. USA. 106:20842–20846. 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., and Tschopp J.. 2010. The inflammasomes. Cell. 140:821–832. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Shi C.S., and Kehrl J.H.. 2010. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 3:ra42 10.1126/scisignal.2000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.S., Shenderov K., Huang N.N., Kabat J., Abu-Asab M., Fitzgerald K.A., Sher A., and Kehrl J.H.. 2012. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 13:255–263. 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., and Tooze S.A.. 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 186:773–782. 10.1083/jcb.200907014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D.J., Sundquist W.I., and Sodroski J.. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA. 103:5514–5519. 10.1073/pnas.0509996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K., Suzuki N.N., Horiuchi M., Mori M., Suhara W., Okabe Y., Fukuhara Y., Terasawa H., Akira S., Fujita T., and Inagaki F.. 2003. X-ray crystal structure of IRF-3 and its functional implications. Nat. Struct. Biol. 10:922–927. 10.1038/nsb1001 [DOI] [PubMed] [Google Scholar]
- Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., and Iwasaki A.. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 106:2770–2775. 10.1073/pnas.0807694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International FMF Consortium 1997. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell. 90:797–807. 10.1016/S0092-8674(00)80539-5 [DOI] [PubMed] [Google Scholar]
- Tomar D., Singh R., Singh A.K., Pandya C.D., and Singh R.. 2012. TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim. Biophys. Acta. 1823:316–326. 10.1016/j.bbamcr.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Toniato E., Chen X.P., Losman J., Flati V., Donahue L., and Rothman P.. 2002. TRIM8/GERP RING finger protein interacts with SOCS-1. J. Biol. Chem. 277:37315–37322. 10.1074/jbc.M205900200 [DOI] [PubMed] [Google Scholar]
- Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., and Elazar Z.. 2011. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 20:444–454. 10.1016/j.devcel.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Xie M., Zhang D., Dyck J.R., Li Y., Zhang H., Morishima M., Mann D.L., Taffet G.E., Baldini A., Khoury D.S., and Schneider M.D.. 2006. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc. Natl. Acad. Sci. USA. 103:17378–17383. 10.1073/pnas.0604708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fiskus W., Yong B., Atadja P., Takahashi Y., Pandita T.K., Wang H.G., and Bhalla K.N.. 2013. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. USA. 110:6841–6846. 10.1073/pnas.1217692110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi R., Chang T.H., Wang H., Atsumi T., Morse H.C. III, and Ozato K.. 2009. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J. Immunol. 182:7527–7538. 10.4049/jimmunol.0804121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., and Narendra D.P.. 2011. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12:9–14. 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E., Berissi H., Mizrachy L., Idelchuk Y., Koren I., Eisenstein M., Sabanay H., Pinkas-Kramarski R., and Kimchi A.. 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10:285–292. 10.1038/embor.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Bao M., Lu N., Weng L., Yuan B., and Liu Y.J.. 2013. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 14:172–178. 10.1038/ni.2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Yazdi A.S., Menu P., and Tschopp J.. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature. 469:221–225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.