Abstract
Purpose
The enteric microbiome is known to play a major role in healthy gut homeostasis and several disease states. It may also contribute to both the intestinal recovery and complications that occur in patients with short bowel syndrome. The extent and nature of alterations to the gut microbiota following intestinal resection, however, are not well studied in a controlled setting. The purpose of this investigation is to characterize the effects of massive small bowel resection on the murine enteric microflora.
Methods
Wild-type C57BL6 mice, following a week of acclamation to a liquid rodent diet, underwent either 50 % proximal small bowel resection (SBR) or a sham operation. Mice were sacrificed, and enteric contents from the small bowel, cecum, and stool were harvested at 7 and 90 days post-operatively. DNA was isolated, and the V3–V5 regions of the 16s rRNA gene amplified and pyrosequenced on a Roche 454 platform. Sequences were clustered into operation taxonomic units and classified. Communities were then analyzed for diversity and phylogenic composition.
Results
In the long-term group, the microbes inhabiting the ileum of mice undergoing SBR and sham operation differed significantly at the genus level (p<0.001). Small bowel contents collected before and after SBR also differed significantly (p= 0.006). This was driven by an increase in Lactobacillus and decrease in Enterobacteriaceae species in mice undergoing SBR. No difference was seen in the long-term stool or in stool, cecal, or ileal contents in the short-term. No difference in microbial community diversity was found in any group.
Conclusion
Bowel resection induces long-term changes in the microbial community of the murine ileum, but not at more distal sites of the gastrointestinal tract. The increase in Lactobacillus encountered small bowel of resected mice correlates with limited previous studies. These changes may reflect an adaptive response of the microbiota to maximize energy extraction, but further studies are needed to establish the role played by this altered community.
Keywords: Short bowel syndrome, Gut microbiome, Intestinal adaptation, Bacterial overgrowth
Introduction
Short bowel syndrome (SBS) is an important morbidity associated with surgical removal of a large portion of the small intestine. In children, most cases occur as a consequence of necrotizing enterocolitis, a devastating necroinflammatory condition of the small and large bowels. Individuals with SBS rely on parenteral nutrition for variable durations, and the extent of reliance is difficult to predict. Specifically, dependence is only moderately (and inversely) correlated with residual bowel length following resection. Indeed, over a 3-year course, only 50 % of SBS patients achieved enteral independence (defined as lack of reliance of parenteral nutrition for sustenance), while 25 % required intestinal transplantation and 25 % died.1 Thus, bowel adaptation is a key factor in the health and survival of patients with SBS.
Components of the gut microbial community and small bowel bacterial overgrowth are thought to contribute to recurrent blood stream infections and sepsis,2 vitamin deficiencies,3 and failure to wean from parenteral nutrition 4 that are often encountered in SBS patients. Emerging data are establishing the role of an altered gut microbial community in diseases such as inflammatory bowel disease,5 obesity,6 malnutrition,7 diabetes, and cancer.8 Additionally, the role that a healthy microbiota plays in normal gut metabolic, trophic, and immune function is increasingly recognized.9
The characteristics and function of the gut microbiome following massive bowel resection are largely unstudied. The purpose of the present study was to characterize the changes that occur in the murine intestinal microbiota following massive small bowel resection (SBR).
Materials and Methods
Animals
C57BL6 mice were obtained from Jackson Laboratories (Bar Harbor, ME) at 5 weeks of age. The mice were individually housed on arrival in an animal facility with a 12-h light/dark cycle. This study was approved by the Washington University Animal Studies Committee (Protocol 20130308) in accordance with the National Institute of Health laboratory animal care and use guidelines.
Diets and Operation
At 7 weeks of age, mice were placed on a standard liquid diet (LD; PMI Micro-Stabilized Rodent Liquid Diet LD 101, TestDiet; 35 % kcal fat). Mice were kept on this diet for 1 week prior to operation, as studies have shown microbial responses to dietary changes to occur over a small number of days.10 At 8 weeks of age, they underwent either a 50 % proximal SBR or a sham operation (transection and anastomosis only).11 Non-operative controls were not used, as we were primarily interested in assessing the role of intestinal resection, and the sham-operated mice are subjected to the same laparotomy and anesthetic conditions. Additionally, a previous study using a porcine model showed only very small differences in the microbiota between sham and non-operated pigs.12
Briefly, animals undergoing SBR underwent transection of the small intestine between 12 cm proximal to the ileocecal junction and at 1–2 cm distal to the ligament of Treitz after ligation of the involved mesentery. The proximal and distal resection margins were anastomosed end-to-end with interrupted 9–0 nylon suture. Mice were injected with 2 ml normal saline intraperitoneally after abdominal closure and kept in an incubator at 37 ° C with free access to drinking water overnight. They were removed from the incubator, individually housed, and returned to the standard LD for the remainder of the experimental period. Mice are typically fed this diet preoperatively and for 1 week following resection, as standard rodent chow results in increased intestinal obstruction and mortality. In this study, we maintained mice on this diet throughout the experimental period to minimize diet-related effects on the microbiota in the middle of the study period. As per our standard protocol, no perioperative antibiotics were administered.
Experimental Design and Sample Collection
Mice were killed at post-operative day (POD) 7 (arm 1) or 90 (arm 2). For all mice, stool samples were collected after conducting an ethanol anal swab at the following times: before placement on LD, day of operation, and day of harvest. Stool was flash-frozen in an ethanol-dry ice slurry upon collection and immediately stored at −80 °C until analyzed. Small bowel luminal contents were also collected from the ileum of all mice at the time of harvest. After removal of the small intestine, the ileum was flushed with 1.5 ml of sterile phosphate-buffered saline (PBS) into a sterile conical tube. This tube was centrifuged at 14,000g for 10 min and the supernatant aspirated. Additionally, on the day of operation, the distal 3 cm of the resected small bowel of mice undergoing SBR was similarly flushed and pelleted to provide samples of small bowel lumen contents on the day of operation. Cecal contents were collected at the time of harvest by sharply opening the cecum and placing it in 1.5 ml sterile BPS, vigorously vortexing, removing the cecal tissue, and pelleting. These pellets were frozen at −80 °C until analyzed. Total DNA was extracted from all enteric content samples using QIAamp DNA Stool Mini Kit (Qiagen, Valencia CA) with additional bead-beating at the time of lysis.
Sample Sequencing and Sequence Data Processing
The V3–V5 region of the 16S rRNA gene was amplified using primers 357F (5′-CCTACGGGAGGCAGCAG -3′) and 926R (5′-CCGTCAATTCMTTTRAGT -3′). Primers also contained an adaptor sequence and one of 96 tags unique to each sample. PCR was performed with the following conditions: 30 cycles of 95 °C at 2 min, 50 °C at 0.5 min, and 72 °C at 5 min. Amplicons were purified, pooled at equimolar concentrations, and pyrosequenced on the Roche 454 Titanium platform using a protocol developed by the Human Microbiome Project.13 The 16s rRNA gene data was submitted to the Sequence Read Archives (SRA) database.
Data processing and quality control (QC) were performed according to standardized protocols developed by the Human Microbiome Project.13 In brief, samples were demultiplexed by sample barcode, allowing one mismatch per barcode. Reads were filtered to remove samples with average quality score <35 and/or read length less <200 nt. Chimeric sequences were removed using Chimera-Slayer. Following initial QC, samples with a read depth <1,000 were resequenced and reprocessed. Samples passing QC were then classified from the phylum to the genus level using the Ribosomal Database Project (RDP) Naive Bayesian Classifier (version 2.2, training set 6).14 Taxa assigned with <0.5 confidence were reassigned to the next higher taxonomic level in which the classification threshold was >0.5.
Data Analysis
After initial sequence data processing, a taxonomical matrix was constructed with row as genera and column as subjects. The taxonomical matrix is rarefied to the minimal number of reads in the matrix using vegan community ecology package15 before any further analysis. We use multi-dimensional scaling (MDS) to explore the microbiome data structure. MDS is an ordination technique, which aims to discover the data pattern in N-dimensional spaces. For microbiome data, it allows the investigator to identify the subject relationships based on the bacterial composition and abundance. Bray-Curtis dissimilarity is used to calculate the pair-wised dissimilarity. Data visualization was performed using MASS package.16
Permutational multivariate analysis of variance (PERMANOVA) is used for formal statistical testing whether the bacterial community structure differs between different variables. PERMANOVA partitions the Bray-Curtis dissimilarity matrix among sources of variation and use permutation test with pseudo-F ratios to obtain the p values. To extract the genera that contribute to the difference between two bacterial communities, we performed Metastats analysis.17 Metastats is a statistical method based on Fisher’s exact test developed for the HMP study. p values from the multiple comparison are adjusted by FDR approach. The genera are considered to be significantly different if (1) p<0.1 and (2) the mean relative abundance for a given genus is at least 1 % in one group. Wilcoxon rank sum test is used to test the differences on Shannon diversity and richness between two groups. These are two different methods of comparing species diversity in a given community. The richness score simply relates the total number of species present. The Shannon index typically ranges from 1.5 to 3.5 and additionally takes into account the relative abundance of each species, reflecting the degree of uncertainty that a member picked at random can be assigned to a specific species.
Quantitative 16s PCR
Total bacterial quantification was done by quantitative real-time PCR of 16s ribosomal small subunit (ssu16s) DNA as target. Custom Taqman assay for ssu16s rDNA target was developed using 5′-AAACTCAAATGAATTGACGGGG-3′ as forward, 5′-TCGTTGCGGGACTTAACCC-3′ as reverse, and 6FAM-ACGCGAAGAACCTTAC as probe. Ten-microliter reactions were prepared using 3 μL of DNA (concentrations of DNA adjusted to ~20 ng/μl), 5 μL TaqMan 2X Environmental Master Mix, 0.5 μL 20X primer-probe mix, and 1.5 μL nuclease-free H2O (all reagents from Applied Biosystems, Inc.). Amplifications were performed for 40 cycles in a 7500 Fast Real-Time PCR System (Applied Biosystems, Inc.). Copy numbers for each transcript in each sample were calculated using 7500 Fast Real-Time PCR System Sequence Detection Software v. 1.3.1 (Applied Biosystems, Inc.) against a known copy number standard curve. A standard curve was generated using serial 10-fold dilutions of known copies of a plasmid DNA as copy number standards.
Results
Survival, Weight Change, and Adaptation
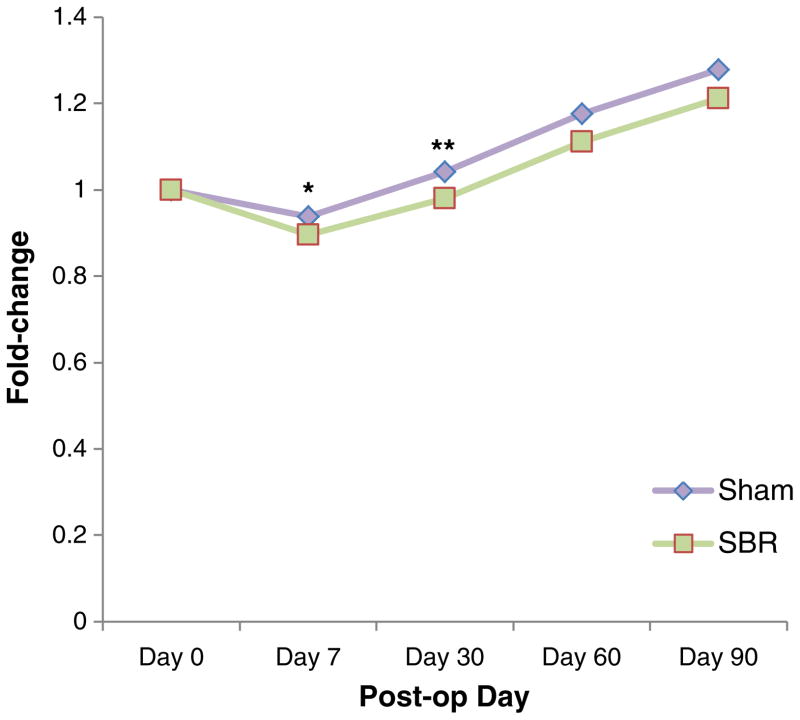
Six (43 %) of the 14 mice undergoing SBR and ten (83 %) of the 12 mice undergoing sham operation survived to POD 90 (arm 2). In the short-term arm (arm 1), ten (77 %) of the 13 from the SBR group and each of the seven sham group survived to POD 7. Both SBR and sham-operated mice lost weight in the first week of the study, but SBR mice experienced greater weight loss (Fig. 1). SBR mice dropped to 89 % of original weight vs 94 % in shams at POD 7 in arm 2 (p=0.005) and 86 vs 93 % in arm 1 (p=0.001) of the study. Both SBR and sham mice eventually regained and surpassed pre-operative body weight, but the sham mice remained statistically heavier than SBR mice until POD 60 when these differences ceased to be significant.
Fig. 1.
Weight change following small bowel resection. Linear curve of changes in weight following small bowel resection and sham operations expressed as fold change from pre-operative weight (*p=0.005; **p= 0.03)
Mice undergoing SBR in both short- and long-term arms of the study displayed expected histological adaptation. In the short-term arm of the study, villus height was 43 % greater in SBR than sham groups (p=0.002), and in the long-term group, villus height was 41 % greater (p=0.01).
Sequencing Depth
In total, 1.3 million high-quality reads targeting V3–V5 regions were produced. The average read depth was 7,510 (1,727) reads/sample. At this depth, we identified 9 phyla and 218 genera. Firmicutes was the most abundant phylum, accounting for 64.0 % of the total bacterial community in the mice intestine. Total reads of 24.2 % were unclassified at a genus level.
SBR vs Sham Comparisons
Diversity
There were no statistically significant differences in the diversity scores of the stool, cecal, or ileal contents when comparing SBR to sham-operated mice in either the short- or long-term arm of the study (Table 1).
Table 1.
Comparisons of richness, diversity, and community make up between SBR and sham-operated mice
| Experimental arm | Sample type | Richness (±standard error)
|
Shannon diversity (±standard error)
|
Community structure | ||||
|---|---|---|---|---|---|---|---|---|
| Sham | SBR | p | Sham | SBR | p | p | ||
| Short-term (arm 1) | POD 0 stool | 49.7±3.3 | 48±1.8 | 0.3 | 2.4±0.1 | 2.4±0.1 | 0.3 | 0.17 |
| POD 7 stool | 54.7±6.4 | 53.2±5.2 | 0.7 | 2.4±0.2 | 2.5±0.2 | 0.5 | 0.78 | |
| Cecum | 56.3±5.8 | 53.1±5.8 | 0.3 | 2.5±0.1 | 2.6±0.2 | 0.7 | 0.83 | |
| POD 7 SB contents | 46.4±15.9 | 42±10.5 | 0.6 | 1.8±0.3 | 1.7±0.6 | 0.5 | 0.63 | |
| Long-term (arm 2) | POD 0 stool | 47.8±3.4 | 47.7±2.4 | 0.9 | 2.2±0.1 | 2.2±0.1 | 0.3 | 0.62 |
| POD 90 stool | 53.1±5.1 | 51.8±3.3 | 0.6 | 2.4±0.2 | 2.4±0.1 | 0.9 | 0.3 | |
| Cecum | 53.8±6.2 | 49.2±4.6 | 0.1 | 2.5±0.3 | 2.5±0.1 | 0.5 | 0.09 | |
| POD 90 SB contents | 44.2±10.2 | 47±8.9 | 0.6 | 1.7±0.3 | 1.9±0.4 | 0.3 | 0.001* | |
p < 0.05
Community Comparisons
Comparisons of microbial communities were carried out at both the phylum and genus levels. As Supplemental Fig. 1a demonstrates, there were no significant community differences between sham and SBR groups at the phylum level at any site of the sampled gastrointestinal tract in arm 1. In arm 2, (Fig. S1b), stool and cecal contents did not significantly differ, but the ileal contents did differ (p=0.03). Figure S1c shows that the phylum breakdown in the sampled small bowel difference was driven by a decrease in Proteobacteria and a decrease in Actinobacteria in the small bowel of SBR mice relative to sham mice at 90 days post-operation.
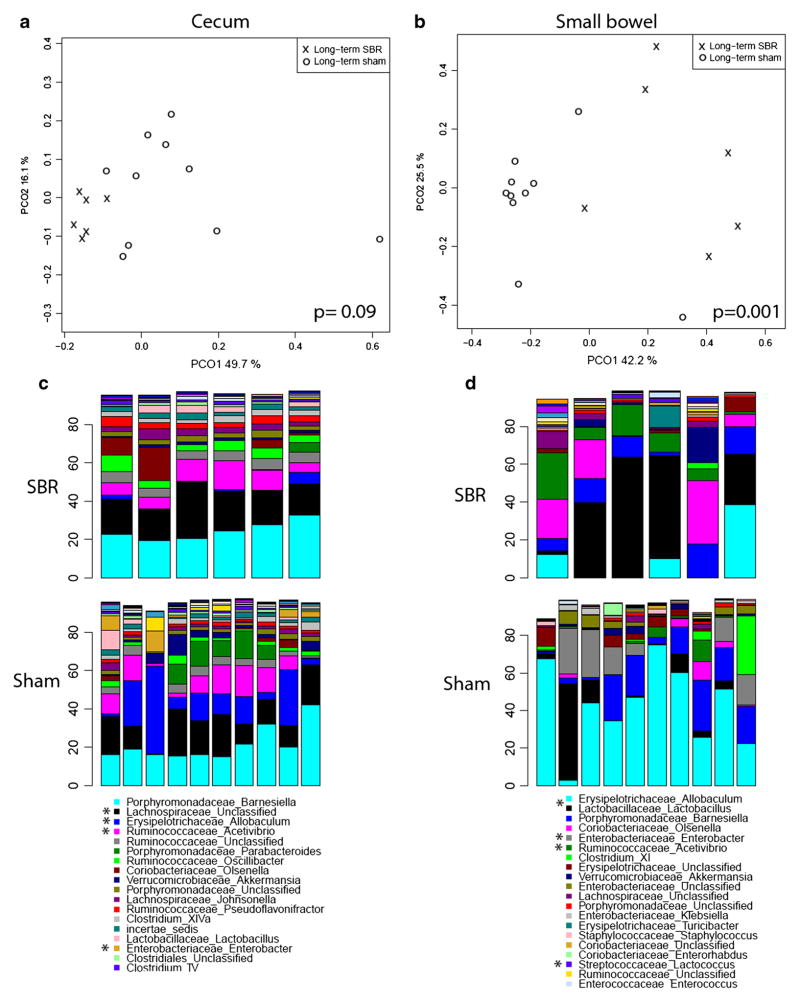
The p values for genus level comparisons are shown in Table 1. The microbial communities inhabiting the ileum, cecum, and stool did not significantly differ between SBR and sham groups at POD 7. Similarly, in the long-term group, the stool communities did not differ between the sham and SBR groups at POD 90. The community of the cecal contents, however, differ significantly (p=0.09) and the ileal contents did differ significantly (p=0.001). The principal coordinate analysis (PCoA) of these communities is shown in Fig. 2a, b. Figure 2c, d shows the predominant genera composing the cecum and ileal contents of these groups. The trend seen in cecal contents was driven primarily by a relative increase in Lachnospiraceae and Ruminococcaceae, as well as a decrease in Allobaculum, Enterobacter, and Parabacteroides spp. relative to the sham-operated mice. There was no significant difference in total bacterial biomass in the cecal contents of sham and SBR mice (4.03×106 vs 4.94×106 copies of 16s DNA per microliter total DNA, p=0.34). The community differences noticed in the ileal contents were largely attributable to a significant increase in Lactococcus and Acetivibrio spp. in mice undergoing SBR, along with relative decreases in Enterobacteriaceae (including Klebsiella and Enterobacter) and Allobaculum.
Fig. 2.
Principal coordinate analysis comparing community structure of cecal contents (a) and small bowel contents (b) of sham vs SBR mice at post-operative day 90. Genus-level comparisons of bacterial communities at same time points in cecal contents (c) and small bowel contents (d). Asterisk denotes genus with significantly different representation in sham vs SBR
Pre- vs Post-operative Comparisons
Diversity
Table 2 shows the diversity differences between pre- and postoperative stool of mice that underwent small bowel resection. As opposed to the sham vs SBR comparisons, significant changes in diversity exist when comparing pre-operative to post-operative stool samples. In arm 1, there was a significant increase in the diversity of the stool microbial community from POD 0 to POD 7 in the SBR, but not in the sham group. In arm 2, there was a significant increase in the diversity of the stool microbial community in both SBR and sham-operated mice from POD 0 to POD 90. When performing these temporal comparisons with a more conservative Bonferroni approach, however, only the stool from sham-operated mice in the long-term arm of the study showed an increase in diversity over time.
Table 2.
Comparison of diversity and community structure of pre- vs post-operative stool samples in Sham and SBR groups for long (arm 1) and short-term (arm 2) arms of the study. Adjusted p values for diversity comparisons represent conservative calculations using Bonferroni correction
| Group | Richness (±standard error)
|
Shannon diversity (±standard error)
|
Community structure | ||||||
|---|---|---|---|---|---|---|---|---|---|
| POD 0 stool | Harvest stool | p | Adjusted p | value POD 0 stool | Harvest stool | p | Adjusted p value | p value | |
| Arm 1 Sham | 49.7±3.3 | 54.7±6.4 | 0.12 | 0.48 | 2.4±0.1 | 2.4±0.2 | 0.7 | 0.28 | 0.05 |
| Arm 1 SBR | 48±1.8 | 53.2±5.2 | 0.02 | 0.08 | 2.4±0.1 | 2.5±0.2 | 0.03 | 0.12 | 0.04 |
| Arm 2 Sham | 47.8±3.4 | 53.1±5.1 | 0.02 | 0.08 | 2.2±0.1 | 2.4±0.2 | 0.01 | 0.04 | 0.001 |
| Arm 2 SBR | 47.7±2.4 | 51.8±3.3 | 0.05 | 0.20 | 2.2±0.1 | 2.4±0.1 | 0.06 | 0.24 | 0.04 |
Community Comparisons
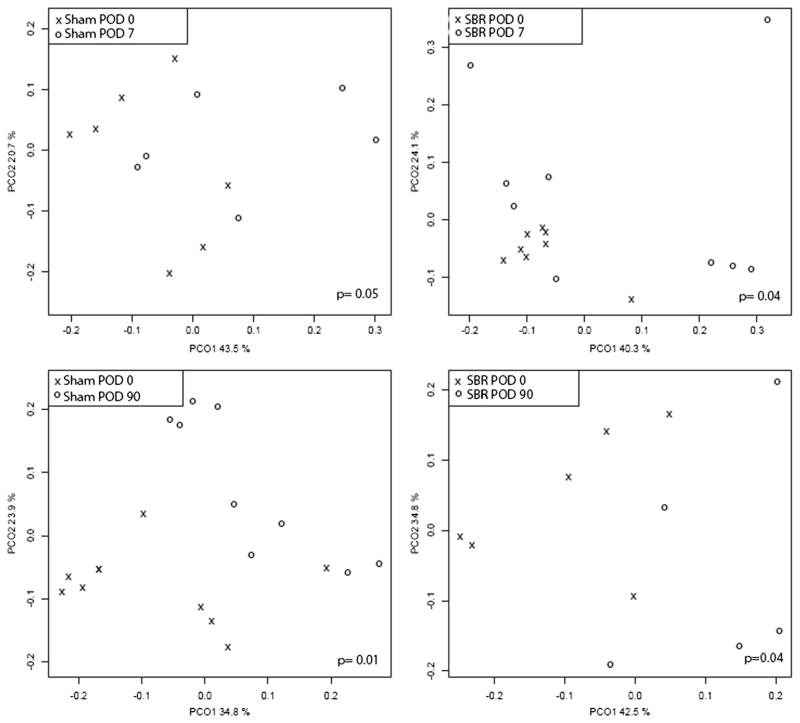
Because of the nature of sample collection, only stool samples could be compared pre- and post-operatively in both sham and SBR mice. In mice undergoing SBR, however, enteric contents from the distal most portion of the resection specimen (distal jejunum/proximal ileum) could be compared to contents just distal to the anastomosis (proximal ileum) at time of harvest. The community composition stool samples differed significantly between POD 0 and the day of harvest in both study arms and in both sham and SBR groups (Table 2). Principal coordinate plots for these comparisons are shown in Fig. 3.
Fig. 3.
Principal coordinate analysis plots representing community comparison of pre- vs post-operative stool samples in sham and SBR groups
Long-term community changes in stool of mice undergoing SBR were driven by an increase in Lactobacillus and Lachnospiraceae. In the short-term SBR group, Lachnospiraceae also increased from POD 0 to POD 7, as did Enterococcus spp. A decrease in several bacteria in the Ruminococcaceae family was also seen.
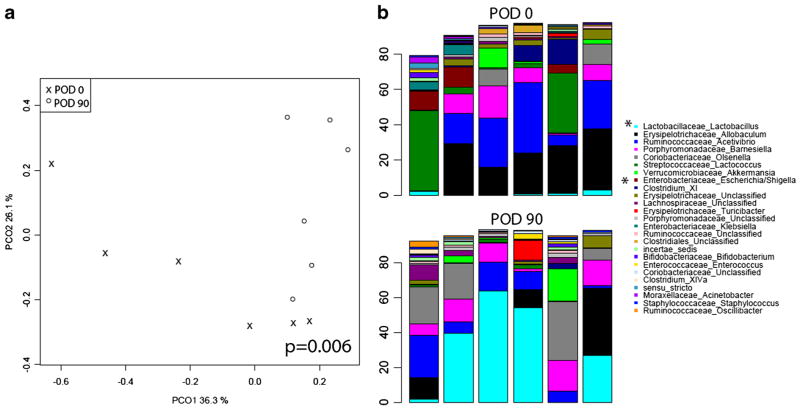
The small bowel contents collected pre- and post-operatively in mice undergoing SBR did display significant long-term community changes (p=<0.05). Figure 4 shows the PCoA analysis plot of this group, as well as the significant genera driving the changes. This was driven mainly by an increase in the Lactobacillus genera and decrease in certain Enterobacteriaceae (Escherichia) following small bowel resection.
Fig. 4.
Principal coordinate analysis (a) comparison at genus level of bacterial community in small bowel contents on POD 0 vs POD 90. Top 25 genera represented in small bowel contents at POD 90 (top) vs POD 0 (bottom) (b). Asterisk denotes genera with significant differences between groups
Discussion
The effects of massive intestinal resection on the gut microbiome have not been well studied in animal models. In this comprehensive description of such changes in a murine proximal small bowel resection model, we determined that the small bowel contents of mice undergoing SBR and sham operation differ significantly by POD 90. Surprisingly, we did not find differences in the overall bacterial diversity throughout the GI tract as a result of bowel resection. Recent studies of both porcine model small bowel resection12 and murine ileocecal resection18 displayed decreased phylogenetic diversity in the resected area compared to sham-operated animals. However, unlike these studies, we did not use pre-operative antibiotics. Antibiotics have been shown to exert profound effects on the gut microbiome19 and predominantly diminish diversity.20 Furthermore, in the porcine study,12 75 % of the small bowel was resected, whereas we removed only 50 %. Because the mice in our study gained weight after a period of intestinal adaptation, it is likely that our model retains sufficient intestinal length to avoid dysbiosis.
In our study, we did not find significant differences in the microbial community’s diversity or overall structure in the first week following small bowel resection, the time period that coincides with structural adaptation of the remnant bowel. This suggests that structural adaptation does not require a shift in prominence of a particular family of microbes. It is certainly possible that microbe-expressed genes and microbe-host interactions may contribute to structural adaptation, but these questions are beyond the scope of this study and a topic for future investigation.
The long-term differences in the community structure of cecal and small bowel contents of SBR mice were driven primarily by increases in Ruminococcus and Lachnospiraceae families and Lactococcus and Acetivibrio genera, as well as decreases in the genera Enterobacter, Klebsiella, Parabacteroides, and Allobaculum. Additionally, mice undergoing SBR experienced an increase in the proportion of Lactobacillus spp. and decrease in Escherichia spp. over time. These are not bacteria that typically overgrow in SBS,21 although increased growth of Lactobacillus has been reported.22 The overgrowth of potential pathogens such as Enterobacter, Escherichia, and Shigella has been reported as possible contributing factor to the increased incidence of bloodstream infection in SBS patients.2 Additionally, overgrowth of such organisms is associated negatively with bowel adaptation.23 The fact that these organisms became less prominent in our study is somewhat unexpected, but the possibility exists that a 50 % SBR is insufficient to induce such changes, as these mice neither develop dilated loops of bowel nor require parenteral nutrition for survival.
In fact, the long-term changes to the small bowel and cecal microflora found in this study more likely reflect an appropriately adapted community of organisms in response to bowel resection. Lactobacillus spp. promote innate immunity in the murine gastrointestinal tract24 and in rat models of SBS; Lactobacillus administration decreases bacterial translocation25 and promotes intestinal adaptation.26 There are scant data, however, in support of Lactobacilli in the management of human SBS.
It is difficult to interpret the significance of the increase in members of the Lachnospiraceae and Ruminococcus families following SBR. Several Lachnospiraceae produce butyrate, and the increase in this family could be related to colonic environmental factors.
Our data demonstrate long-term changes in the murine small bowel and proximal colonic microbiota in response to massive small bowel resection, but the significance of these changes remains unknown. First, a number of mice in long-term SBR arm of this study died and were excluded from analysis, and this could have biased the results. Obstruction, anastomotic leak, or failure to adapt or another reason contributed to these deaths is not known. In future studies, more frequent sampling and analysis of stool contents prior to death could help elucidate whether such events contain a common microbial community pattern. Additionally, commercially available mice likely harbor a unique flora that is not ideal for drawing conclusions about human microbiota. Further characterization in a “humanized” murine model would likely be both feasible27 and more relevant.
Additionally, in addition to characterizing the microbial environment in SBR, it is also necessary to determine the impact of altering this environment. Altered microbiota in obese individuals can “transmit” obesity to lean animals.6 Similarly, in a murine model of Roux-en-Y gastric bypass, metabolic changes are transmissible to non-operated germ-free animals by stool.28 SBR reduces energy expenditure and delays recovery of lean body mass compared to body fat stores.29 Studies utilizing germ-free and selectively colonized animals will help determine what role changes to the micro-flora play in these alterations and might outline the possible role of altering bacterial communities to optimize management of patients following massive intestinal resection.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11605-014-2631-0) contains supplementary material, which is available to authorized users.
Contributor Information
J. Sommovilla, Email: sommovillaj@wudosis.wustl.edu, Department of Surgery, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
Y. Zhou, Department of Pediatrics, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA. Department of Genetics, The Genome Institute, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
R. C. Sun, Department of Surgery, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
P. M. Choi, Department of Surgery, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
J. Diaz-Miron, Department of Surgery, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
N. Shaikh, Department of Pediatrics, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
E. Sodergren, Department of Genetics, The Genome Institute, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
B. B. Warner, Department of Pediatrics, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
G. M. Weinstock, Department of Genetics, The Genome Institute, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
P. I. Tarr, Department of Pediatrics, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
B. W. Warner, Department of Surgery, Washington University, St. Louis School of Medicine; St. Louis Children’s Hospital, 660 S Euclid Ave., Campus Box 8109, St. Louis, MO 63110, USA
References
- 1.Squires RH, et al. Natural history of pediatric intestinal failure: initial report from the Pediatric Intestinal Failure Consortium. J Pediatr. 2012;161(4):723–8. e2. doi: 10.1016/j.jpeds.2012.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole CR, et al. The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial overgrowth, enteral feeding, and inflammatory and immune responses. J Pediatr. 2010;156(6):941–7. 947e1. doi: 10.1016/j.jpeds.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sentongo TA, Azzam R, Charrow J. Vitamin B12 status, methylmalonic acidemia, and bacterial overgrowth in short bowel syndrome. J Pediatr Gastroenterol Nutr. 2009;48(4):495–7. doi: 10.1097/MPG.0b013e31817f9e5b. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman SS, et al. Influence of bacterial overgrowth and intestinal inflammation on duration of parenteral nutrition in children with short bowel syndrome. J Pediatr. 1997;131(3):356–61. doi: 10.1016/s0022-3476(97)80058-3. [DOI] [PubMed] [Google Scholar]
- 5.Dicksved J, et al. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2(7):716–27. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 6.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kau AL, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134(2):479–82. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 9.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361(9356):512–9. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmrath MA, et al. Intestinal adaptation following massive small bowel resection in the mouse. J Am Coll Surg. 1996;183(5):441–9. [PubMed] [Google Scholar]
- 12.Lapthorne S, et al. Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes. 2013;4(3):212–21. doi: 10.4161/gmic.24372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Human Microbiome Project, C. A framework for human microbiome research. Nature. 2012;486(7402):215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oksanen JBG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H. Vegan: community ecology package. 2013 [Google Scholar]
- 16.Venables WN, Ripley BD, Venables WN. Statistics and computing. 4. New York: Springer; 2002. Modern applied statistics with S; p. xi.p. 495. [Google Scholar]
- 17.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devine AA, et al. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. PLoS One. 2013;8(8):e73140. doi: 10.1371/journal.pone.0073140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartosch S, et al. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70(6):3575–81. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dibaise JK, Young RJ, Vanderhoof JA. Enteric microbial flora, bacterial overgrowth, and short-bowel syndrome. Clin Gastroenterol Hepatol. 2006;4(1):11–20. doi: 10.1016/j.cgh.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Bouhnik Y, et al. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol. 1999;94(5):1327–31. doi: 10.1111/j.1572-0241.1999.01016.x. [DOI] [PubMed] [Google Scholar]
- 23.Cole CR, Ziegler TR. Small bowel bacterial overgrowth: a negative factor in gut adaptation in pediatric SBS. Curr Gastroenterol Rep. 2007;9(6):456–62. doi: 10.1007/s11894-007-0059-3. [DOI] [PubMed] [Google Scholar]
- 24.Galdeano CM, Perdigon G. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol. 2006;13(2):219–26. doi: 10.1128/CVI.13.2.219-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eizaguirre I, et al. Probiotic supplementation reduces the risk of bacterial translocation in experimental short bowel syndrome. J Pediatr Surg. 2002;37(5):699–702. doi: 10.1053/jpsu.2002.32256. [DOI] [PubMed] [Google Scholar]
- 26.Tolga Muftuoglu MA, et al. Effects of probiotics on experimental short-bowel syndrome. Am J Surg. 2011;202(4):461–8. doi: 10.1016/j.amjsurg.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Hirayama K. Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp Anim. 1999;48(4):219–27. doi: 10.1538/expanim.48.219. [DOI] [PubMed] [Google Scholar]
- 28.Liou AP, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5(178):178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tantemsapya N, et al. Body composition and metabolic changes associated with massive intestinal resection in mice. J Pediatr Surg. 2008;43(1):14–9. doi: 10.1016/j.jpedsurg.2007.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.