Abstract
Background
Although Helicobacter pylori infection is necessary for development of gastric adenocarcinoma (GAC), the underlying mechanism remains poorly defined. This study aimed to explore how miR-221 and miR-222 are dysregulated after H. pylori infection and how these 2 miRNAs are involved in pathological development of gastric cancer.
Material/Methods
qRT-PCR analysis was performed to quantify miR-221 and miR-222 expression in patients with H. pylori – induced chronic gastritis, H. pylori-negative healthy controls, and in gastric cancer tissues and the corresponding adjacent normal tissues. Cell models were used to verify the expression profile. Dual luciferase assay was performed to verify putative binding between miR-221 or miR-222 and RECK. A loss-and-gain function study was performed to assess the miR-221/miR-222-RECK axis in gastric cancer cells.
Results
H. pylori infection leads to significantly higher miR-221 and miR-222 expression. MiR-221 and miR-222 can bind the same sequence of RECK 3′UTR, thereby modulating its expression. Through simultaneous regulation over RECK, miR-221 and miR-222 can promote gastric cancer cell growth and invasion.
Conclusions
The miR-221/miR-222-RECK axis might be an important path modulating H. pylori infection-related gastric cancer development.
MeSH Keywords: Helicobacter pylori, MicroRNAs, Stomach Neoplasms
Background
Gastric cancer (GC) currently is the fourth most common malignancy and the second leading cause of cancer-related death in the world [1]. Helicobacter pylori is a microaerophilic pathogen which is associated with a series of gastric diseases such as chronic active gastritis, peptic ulceration, and gastric carcinoma. In fact, H. pylori is classified as a group 1 carcinogen for gastric cancer [2]. Although H. pylori infection is necessary for development of gastric adenocarcinoma (GAC), the underlying mechanism remains poorly defined [3].
Previous studies observed that H. pylori infection is associated with altered miRNAs expression, which might be involved in pathological development of gastric cancer [4]. For example, H. pylori infection leads to downregulated miR-203, miR-204, and miR-449, which promote proliferation and invasion of gastric cancer cells [5–7]. H. pylori infection also results in upregulated miR-21, miR-221, miR-222, and miR-223, which are related to proliferation and apoptosis of cancer cells [8–10]. However, due to the wide regulative network of miRNAs, the biological functions of some dysregulated miRNAs are still not fully understood.
RECK (reversion-inducing cysteine-rich protein with Kazal motifs) is a tumor-suppressor gene in gastric cancer [11]. Previous studies reported that several dysregulated miRNAs, including miR-21, miR-25, miR-374, and miR-222, can promote cancer development through inhibiting RECK [9,10,12,13]. Therefore, it is evident that RECK is an important tumor-suppressor gene in gastric cancer. However, whether other miRNAs are involved in regulating RECK in gastric cancer is not clear. Therefore, in this study, we explored the association between H. pylori infection and changes in miR-221 and miR-222 expression. Then we explored the regulation of miR-221 and miR-222 over RECK and whether this axis is involved in cancer cell growth and invasion.
Material and Methods
Human specimens
All of the participants were recruited from the First People’s Hospital of Chengdu, China. This study was approved by the ethics committee of the hospital. The gastric mucosal specimens were obtained from 20 patients who had gastroscope examination, among which 10 cases had H. pylori-induced chronic gastritis (mean age: 42 years; 6 men and 4 women) and 10 H. pylori-negative healthy controls (mean age: 38 years; 5 men and 5 women). Informed consent was obtained all participants before the study. The H. pylori infection was confirmed by positive results of bacterial culture, C13-urea breath test, and histologic testing at the same time.
Gastric cancer tissues and the corresponding adjacent normal tissues were obtained from 8 patients with primary gastric cancer who underwent radical gastrectomy. Informed consent was obtained all participants before resection. The tumor stage was assessed based on the International Union Against Cancer’s (UIAC) tumor-node-metastasis (TNM) system (6 patients were stage Ib and 2 were stage II).
Cells and bacteria culture
HEK293T cells and human GAC cell line AGS was obtained from ATCC. Immortalized normal human gastric epithelial cell line GES-1 and human GAC cell lines BGC-823 and SGC-7901 were obtained from Shanghai Institute of Cell Biology, China Academy of Sciences (Shanghai, China). HEK293T cells were cultured in DMEM medium, and other cells were cultured in RPMI-1640 medium supplemented with 10% FBS at 37°C in 5% CO2 in a 25-cm2 flask.
The wild-type H. pylori strains 26695 were obtained from ATCC. The bacteria were grown on CDC anaerobe blood agar plates (BD) and were incubated at 37°C for 2 days in an anaerobic jar containing a gas mixture of 5% O2, 10% CO2, and 85% N2 (DU Scientific).
When the cells reached 80% confluency, the medium was replaced with antibiotic-free medium and H. pylori was added to the flask at a multiplicity of infection of 100:1. At 18 h after H. pylori induction, the cells were lysed to extract total RNA.
Cell transfection and agents used
AGS and SGC-7901 cells were transfected with 50 nM miR-221 or miR-222 mimics, 100 nM antagomiR-221 or antagomiR-222, co-transfected with 40 nM miR-221 and 40 nM miR-222 mimics, co-transfected with 75 nM antagomiR-221 and 75 nM antagomiR-222, or corresponding negative controls (RiboBio, China) using Lipofectamine RNAiMAX (Invitrogen, USA). AGS and SGC-7901 cells with miR-221 or miR-222 overexpression were further transfected with pcDNA3.1-RECK (without 3′UTR) or pcDNA3.1 empty vector using Lipofectamine 2000 reagent (Invitrogen).
qRT-PCR analysis of miR-221 and miR-222 expression
Total RNAs in tissues and cell samples were extracted using the mirVana PARIS Kit (Ambion, USA). The cDNA was reversely transcribed using TaqMan MicroRNA Reverse Transcription Kit. To determine miR-221 and miR-222 expression levels, qRT-PCR was performed using TaqMan MicroRNA Assays (Applied Biosystems). U6 snRNA was used as an internal control.
CCK-8 assay of cell viability
After transfection, cells were plated in 96-well plates at a density of 5×103 cells/well. Cells were cultured for 48 h and then cell viability was measured by Cell Counting Kit-8 (CCK-8) (Dojindo) according to the manufacturer’s instruction. Each test was performed in triplicate.
Flow cytometry analysis of cell apoptosis
At 48 h after treatments, cells were plated in 6-well plates at a density of 4×105 cells/well. The ratio of apoptotic cells was measured using Fluorescein Active Caspase 3 Staining Kit (ab65613, Abcam) with a flow cytometer (FACSCalibur, BD Biosciences). The results were analyzed using CellQuest (BD Biosciences).
Transwell analysis of cell invasion
We suspended 1×105 cells in 200 μL serum-free RPMI-1640 medium and seeded them into the upper chamber of the transwell insert chamber coated with Matrigel (BD Biosciences). Culture medium in the lower chamber was supplemented with 20% FBS as the chemoattractant. At 24 h after incubation in a cell incubator, cells on the top surface of the insert were removed and the cells on the bottom surface were fixed with 4% polyoxymethylene and stained with 0.1% crystal violet for 20 min. Cell counting was performed at 100× magnification under a microscope. Each experiment was performed in triplicate.
Western blot analysis of RECK expression
Cells were lysed by protein lysate (Pierce) and the protein concentration was measured by BCA protein assay kit (Pierce). The lysates were separated on 10% SDS-PAGE and then transferred onto a PVDF membrane. The membranes were blocked with 5% nonfat dry milk and were incubated with primary antibodies for RECK (1:1000, ab88249, Abcam), GAPDH (1:2000, ab125247, Abcam) at 4°C overnight. Membranes were washed and incubated with corresponding horseradish peroxidase-conjugated secondary antibodies (1:10000, anti-mouse IgG (HRP), ab6728, Abcam; 1:10000, anti-rabbit IgG (HRP) ab191866, Abcam). The protein signals were visualized using ECL Western blotting substrate (Pierce) and the signal intensity was determined by using Image-J software.
Dual luciferase assay
Putative binding between miR-221/miR-222 and human RECK mRNA were predicted in TargetScan 6.2. Since miR-221 and miR-222 have the same binding site with RECK 3′-UTR, only 1 mutant RECK 3′-UTR sequence was designed. The wild-type and mutant RECK 3′UTR sequences were chemically synthesized and inserted into pGL3 promoter vector to construct the recombinant reporters pGL3-RECK-wt and pGL3-RECK-mut, respectively. HEK293T and SGC-7901 cells were co-transfected with the reporters and miR-221 or miR-222 mimics. At 24 h after transfection, luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Promega, USA). Firefly luciferase activity was normalized to that of Renilla luciferase.
Statistical analysis
Data analysis was performed using SPSS 17.0. The results are expressed as mean ±SD. Group difference was assessed using Student’s t-test. P<0.05 was considered as statistically significant. Statistical significances are indicated by asterisks (* P<0.05, ** P<0.01, *** p<0.001).
Results
H. pylori infection leads to increased miR-221 and miR-222 expression
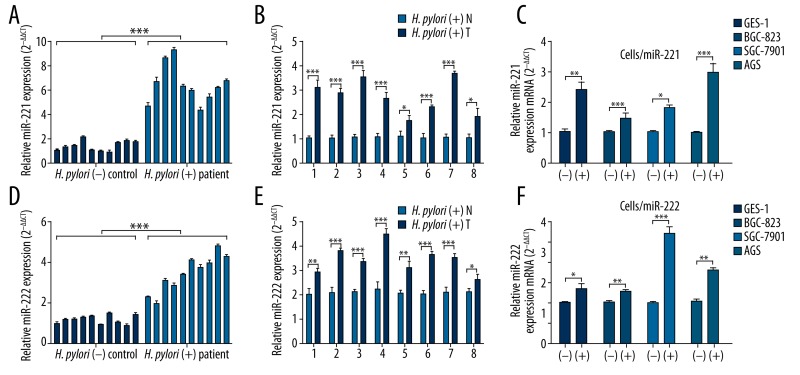
To explore the association between H. pylori infection and miR-221 and miR-222 expression, we firstly assessed miR-221 and miR-222 expression profile in both patients with H. pylori-induced chronic gastritis and gastric cancer. qRT-PCR analysis showed that patients with H. pylori-positive chronic gastritis had significantly higher miR-221 and miR-222 expression compared with H. pylori-negative healthy controls (Figure 1A, 1D). Since H. pylori infection is considered as a cause of gastric cancer, we also compared miR-221 and miR-222 expression between H. pylori-positive gastric cancer tissues and matched adjacent normal gastric mucosa tissues. The results showed that miR-221 and miR-222 expression in the cancer tissues were consistently higher than in adjacent normal tissues in the 8 patients (Figure 1B, 1E). To further confirm that H. pylori infection directly contributes to increased miR-221 and miR-222 expression, we quantified expression of these 2 miRNAs in GES-1, BGC-823, SGC-7901, and AGS cells before and after H. pylori infection. The results showed that H. pylori infection directly led to significantly higher miR-221 and miR-222 expression (Figure 1C, 1F). These results suggest that H. pylori infection can lead to increased miR-221 and miR-222 expression.
Figure 1.
H. pylori infection leads to increased miR-221 and miR-222 expression. (A, D) qRT-PCR analysis of miR-221 (A) and miR-222 (D) expression among 10 patients with H. pylori-positive chronic gastritis and 10 H. pylori-negative healthy controls. (B, E) qRT-PCR analysis of miR-221 (B) and miR-222 (D) expression in H. pylori positive tumor tissues (T) and adjacent normal (N) tissues obtained from 8 gastric cancer patients. (C, F) qRT-PCR analysis of miR-221 (C) and miR-222 (F) expression in GES-1, BGC-823, SGC-7901, and AGS cells before (−) and after H. pylori infection (+). Values are the average of triple determinations with the S.D. indicated by error bars. * P<0.05, ** P<0.01 and *** P<0.001.
Both miR-221 and miR-222 can modulate growth, apoptosis and invasion of gastric cancer cells
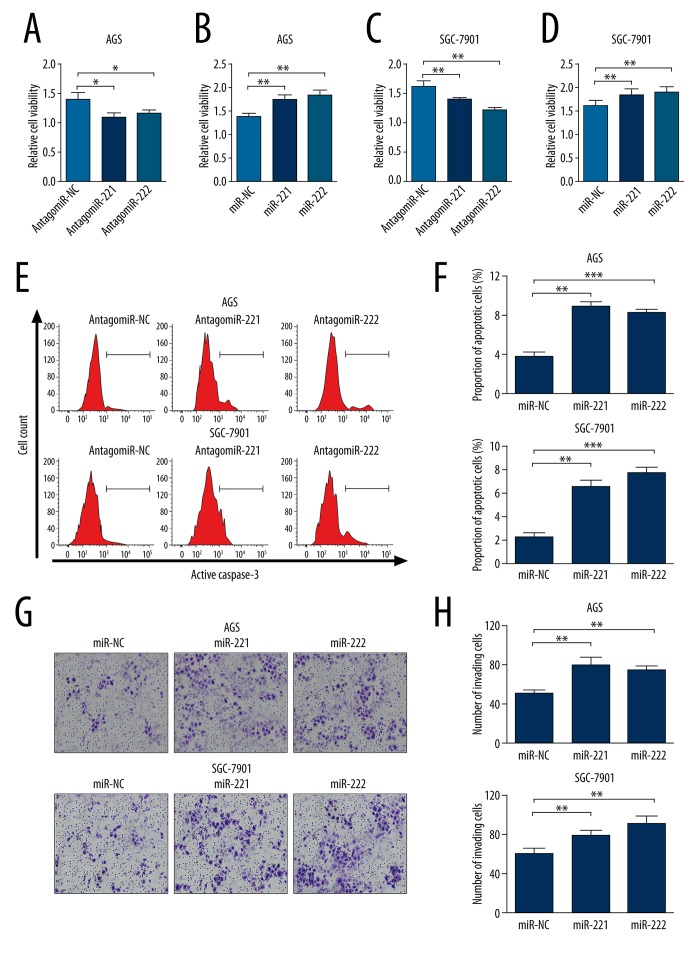
To explore the role of miR-221 and miR-222 in gastric cancer, AGS and SGC-7901 cells were transfected with miR-221 mimics, miR-222 mimics, antagomiR-221 or antagomiR-222 for overexpression and knockdown, respectively. CCK-8 assay showed that knockdown of miR-221 or miR-222 in both AGS and SGC-7901 cells significantly inhibited cell growth (Figure 2A, 2C). In contrast, miR-221 or miR-222 overexpression in both AGS and SGC-7901 cells significantly promoted cell growth (Figure 2B, 2D). In both AGS and SGC-7901 cells, knockdown of endogenous miR-221 or miR-222 resulted in significantly increased proportion of apoptotic cells during cultivation (Figure 2E, 2F). In addition, miR-221 or miR-222 overexpression significantly promoted cell invasion in both AGS and SGC-7901 cells (Figure 2G, 2H). These results suggest that both miR-221 and miR-222 can modulate growth, apoptosis, and invasion of gastric cancer cells.
Figure 2.
Both miR-221 and miR-222 can modulate growth, apoptosis, and invasion of gastric cancer cells. (A–D) CCK-8 assay of cell proliferation of AGS (A, B) and SGC-7901 (C, D) cells with knockdown (A, C) and overexpression (B, D) of miR-221 or miR-222. (E) Representative images of flow cytometry analysis of apoptotic AGS and SGC-7901 cells after knockdown of endogenous miR-221 or miR-222. (F) Quantification of the ratio of apoptotic cells shown in Figure E. (G) Transwell analysis of cell invasion of AGS and SGC-7901 cells with miR-221 or miR-222 overexpression. (H) Quantification of the number of invading cells showed in Figure G. Values are the average of triple determinations with the S.D. indicated by error bars. * P<0.05, ** P<0.01 and *** P<0.001.
Both miR-221 and miR-222 directly target RECK and regulate its expression
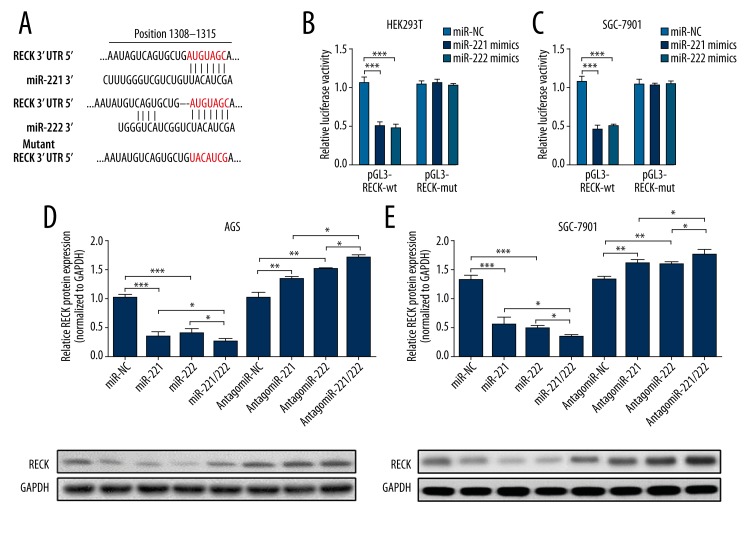
Through searching and comparison in TargetScan 6.2, we identified RECK as a common target of miR-221 and miR-222. Since miR-221 and miR-222 have the same binding sites with RECK 3′UTR, only 1 wild-type reporter (pGL3-RECK-wt) and 1 mutant reporter (pGL3-RECK-mut) carrying wild-type and mutant RECK 3′UTR sequence, respectively, were designed (Figure 3A). Both miR-221 and miR-222 could significantly inhibit luciferase activity of pGL3-RECK-wt in both HEK-293T and SGC-7901 cells, but had no significant inhibiting effect on pGL3-RECK-mut (Figure 3B, 3C). In AGS and SGC-7901 cells, miR-221 and miR-222 mimics could significantly reduce RECK protein expression. Simultaneously, transfection of miR-221 and miR-222 showed additive effect in inhibiting RECK expression (Figure 3D, 3E). On the contrary, inhibiting endogenous miR-221 and miR-222 significantly promoted RECK protein expression. Knockdown of miR-221 and miR-222 simultaneously showed stronger effect in inducing RECK expression than knockdown of either alone (Figure 3D, 3E). These results suggest that both miR-221 and miR-222 directly target RECK and regulate its expression.
Figure 3.
Both miR-221 and miR-222 directly target RECK and regulate its expression. (A) Predicted binding between miR-221 or miR-222 and RECK. A mutant 3′UTR sequence of RECK without specific binding with miR-221 and miR-222 was designed. (B, C) Dual luciferase assay of HEK293T (B) and SGC-7901 (C) cells co-transfected with pGL3-RECK-wt/mut and miR-221 or miR-222 mimics. (D, E) Western blot analysis of RECK expression at protein level in AGS (D) and SGC-7901 (E) cells transfected with miR-221, miR-222, miR-221, miR-222, antagomiR-221, antagomiR-107, or antagomiR-221 and antagomiR-222. Values are the average of triple determinations with the S.D. indicated by error bars. * P<0.05, ** P<0.01 and *** P<0.001.
Both miR-221 and miR-222 can modulate growth, apoptosis, and invasion of gastric cancer cells through RECK
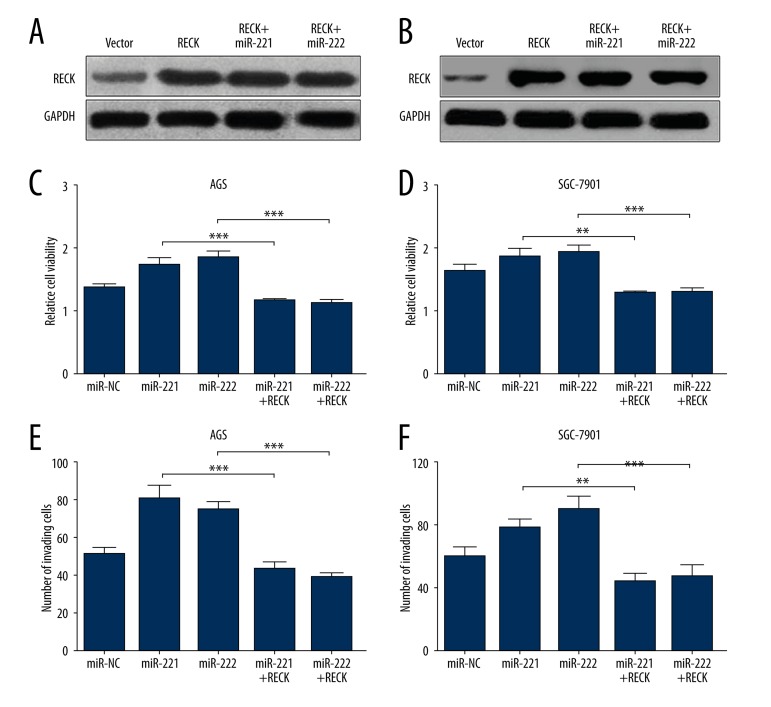
Previous studies reported that RECK is an important protein modulating invasion and metastasis of colorectal cancer cells and also regulating growth of gastric cancer cells [14]. Therefore, we further explored the role of the miR-221/miR-222-RECK axis in gastric cancer cells. AGS and SGC-7901 cells were first transfected with PCDNA3.1-RECK for overexpression. miR-221 or miR-222 had no effect on RECK protein expression of this vector (Figure 4A, 4B). Overexpression of RECK could significantly reverse the miR-221- or miR-222-induced higher cell growth rate (Figure 4C, 4D). In addition, enforced RECK expression also abrogated miR-221- or miR-222-induced stronger cell invasion (Figure 4E, 4F).
Figure 4.
Both miR-221 and miR-222 can modulate growth, apoptosis, and invasion of gastric cancer cells through RECK. (A) Western blot analysis of RECK protein expression in AGS (A) and SGC-7901 (B) cells transfected with PCDNA3.1-RECK with or without miR-221 or miR-222. (C, D) CCK-8 assay of cell proliferation of AGS (C) and SGC-7901 (D) cells transfected with PCDNA3.1-RECK with or without miR-221 or miR-222 overexpression. (E, F) Transwell analysis of cell invasion of AGS (E) and SGC-7901 (F) cells transfected with PCDNA3.1-RECK with or without miR-221 or miR-222 overexpression.
Discussion
Although accumulating evidence shows that H. pylori infection is associated with dysregulated miRNAs expression that contribute to pathological development of gastric cancer [4], the role of the miRNAs are not fully understood. In the current study, we observed that both miR-221 and miR-222 were upregulated after H. pylori infection. This finding is consistent with a previous study which reported that miR-221 and miR-222 are both upregulated in gastric cancer tissue-derived mesenchymal stem cells [15]. However, how they are involved in the pathological development of gastric cancer is not quite clear. A recent study found that high serum miR-222 level is significantly correlated with clinical stages and lymph nodes metastasis in gastric cancer [16]. Elevated miR-222 significantly inhibits RECK expression, thereby promoting gastric cancer cell proliferation [9]. In fact, miR-221 and miR-222 share a part of the same sequence, suggesting they may have some common targets. For example, a recent study found that miR-221 and miR-222 can also simultaneously target PTEN and regulate gastric carcinoma cell proliferation and radioresistance [17]. However, whether these 2 miRNAs have other targets or other common targets in gastric cancer is not clear.
RECK is generally viewed as a tumor suppressor and is downregulated in several types of cancers, including gastric cancer [18–20]. Aberrant methylation of RECK gene is also considered as a marker for early diagnosis and treatment of peritoneal metastasis of gastric cancer [20]. This membrane-anchored glycoprotein acts as a negative regulator of matrix metalloproteinase-9, a key enzyme regulating tumor invasion and metastasis [21]. It can also inhibit tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation [22].
Previous studies reported that several miRNAs can inhibit RECK expression and thus promote gastric cancer development, including miR-21, miR-25, miR-222, and miR-374 [9,10,12,13]. These findings suggest that RECK is an important tumor-suppressing protein in gastric cancer. Therefore, it is necessary to fully determine its upstream regulators. Through prediction in online informatics databases, we observed that both miR-221 and miR-222 have the same putative binding sites with RECK. Because RECK was verified to be a target of miR-222 and due to its well established tumor suppressor role, we decided to further study whether it is a common target of miR-221 and miR-222. By performing dual luciferase assay, we confirmed that both miR-221 and miR-222 can effectively bind to 3′-UTR of RECK. Western blot analysis also confirmed that both miR-221 and miR-222 could effectively inhibit RECK expression at the protein level. Furthermore, simultaneous overexpression or inhibition of miR-221 and miR-222 had stronger effect than either alone, suggesting they may have some synergic effects.
Because the regulative role of miR-221 and miR-222 over RECK expression is confirmed, we further verified the biological function of miR-221/miR-222-RECK axis in gastric cancer cells. By performing loss-and-gain analysis, we confirmed that the miR-221/miR-222-RECK axis can directly regulate proliferation and invasion of gastric cancer cells.
Conclusions
H. pylori infection results in significantly increased miR-221 and miR-222 expression, both of which can effectively bind to 3′-UTR of RECK. The miR-221/miR-222-RECK axis might be an important path modulating H. pylori infection-related gastric cancer development. These findings might provide some useful evidence about the molecular mechanism of gastric cancer development due to H. pylori infection.
Footnotes
Source of support: This study was supported by the Key Clinical Specialty Discipline Construction Program
Reference
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Asaka M, Sepulveda AR, Sugiyama T, Graham DY. Gastric Cancer. In: Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: physiology and genetics. Washington (DC): 2001. [Google Scholar]
- 3.Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148(4):719–31.e3. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Wang Q, Liu H, et al. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett. 2010;297(2):137–43. doi: 10.1016/j.canlet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Xu G, Yin C, et al. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget. 2014;5(22):11631–40. doi: 10.18632/oncotarget.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Li L, Su J, Zhang G. Decreased miR-204 in H. pylori-associated gastric cancer promotes cancer cell proliferation and invasion by targeting SOX4. PloS One. 2014;9(7):e101457. doi: 10.1371/journal.pone.0101457. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, Chen Y, Zhang B, Liu G. Increased microRNA-223 in Helicobacter pylori-associated gastric cancer contributed to cancer cell proliferation and migration. Biosci Biotechnol Biochem. 2014;78(4):602–8. doi: 10.1080/09168451.2014.895661. [DOI] [PubMed] [Google Scholar]
- 9.Li N, Tang B, Zhu ED, et al. Increased miR-222 in H. pylori-associated gastric cancer correlated with tumor progression by promoting cancer cell proliferation and targeting RECK. FEBS Lett. 2012;586(6):722–28. doi: 10.1016/j.febslet.2012.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88(12):1358–66. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 11.Song SY, Son HJ, Nam E, et al. Expression of reversion-inducing-cysteine-rich protein with Kazal motifs (RECK) as a prognostic indicator in gastric cancer. Eur J Cancer. 2006;42(1):101–8. doi: 10.1016/j.ejca.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Wang Y, Yang L, et al. MiR-25 promotes gastric cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2014;385(1–2):207–13. doi: 10.1007/s11010-013-1829-x. [DOI] [PubMed] [Google Scholar]
- 13.Xie J, Tan ZH, Tang X, et al. MiR-374b-5p suppresses RECK expression and promotes gastric cancer cell invasion and metastasis. World J Gastroenterol. 2014;20(46):17439–47. doi: 10.3748/wjg.v20.i46.17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark JC, Thomas DM, Choong PF, Dass CR. RECK – a newly discovered inhibitor of metastasis with prognostic significance in multiple forms of cancer. Cancer Metastasis Rev. 2007;26(3–4):675–83. doi: 10.1007/s10555-007-9093-8. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Zhao C, Shi H, et al. Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: novel biomarkers and a mechanism for gastric cancer. Br J Cancer. 2014;110(5):1199–210. doi: 10.1038/bjc.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Z, Qian F, Yang X, et al. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31(9):164. doi: 10.1007/s12032-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 17.Chun-Zhi Z, Lei H, An-Ling Z, et al. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noda M, Oh J, Takahashi R, et al. RECK: a novel suppressor of malignancy linking oncogenic signaling to extracellular matrix remodeling. Cancer Metastasis Rev. 2003;22(2–3):167–75. doi: 10.1023/a:1023043315031. [DOI] [PubMed] [Google Scholar]
- 19.Murai R, Yoshida Y, Muraguchi T, et al. A novel screen using the Reck tumor suppressor gene promoter detects both conventional and metastasis-suppressing anticancer drugs. Oncotarget. 2010;1(4):252–64. doi: 10.18632/oncotarget.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du YY, Dai DQ, Yang Z. Role of RECK methylation in gastric cancer and its clinical significance. World J Gastroenterol. 2010;16(7):904–8. doi: 10.3748/wjg.v16.i7.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi C, Sheng Z, Horan TP, et al. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad Sci USA. 1998;95(22):13221–26. doi: 10.1073/pnas.95.22.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong KJ, Wu DC, Cheng KH, et al. RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation. J Cell Physiol. 2014;229(2):191–201. doi: 10.1002/jcp.24434. [DOI] [PubMed] [Google Scholar]