Abstract
Latinos represent a growing proportion of HIV cases in North Carolina (NC). Understanding how immigrants are involved in local HIV transmission is important to guide interventions. We used phylogenetics to characterize Latino involvement in local HIV transmission chains. Transmission clusters were identified from maximum-likelihood phylogenies constructed with HIV pol sequences from 177 Latinos and 1,496 non-Latinos receiving care in NC. Highly supported clusters involving one or more Latinos were characterized. Migration data were obtained from interviews and chart review. Factors associated with cluster membership were identified using log-binomial regression. Most Latinos were male (76%), immigrants (83%), and had HIV-1B (99%). Immigrants were more likely to report heterosexual risk (67% vs. 23%) than U.S.-born Latinos (p < 0.01). We identified 32 clusters that included one or more Latinos; these involved 53 Latinos (30%) and 41 non-Latinos. Immigrant and U.S.-born Latinos were equally likely to be in clusters, but immigrants were more likely to be in clusters with another Latino (78% vs. 29%; p = 0.02). Cluster composition by ethnicity and risk behavior varied by cluster size; larger clusters contained fewer immigrants and more men who have sex with men (MSM). Factors associated with immigrant membership in local transmission clusters included age <30 years [RR 2.34 (95% CI 1.47–3.73)], Mexican origin [RR 2.55 (95% CI 1.29–6.88)], and residing in the United States longer before diagnosis [RR 1.53 (95% CI 1.09–2.15), per 10 years]. While some Latinos immigrate with HIV infection, many immigrants are involved in transmission networks after arrival, particularly MSM. HIV testing and prevention interventions must consider this heterogeneity and may be better targeted by integrating phylogenetic analyses.
Introduction
Latinos are the largest and one of the fastest growing ethnic minority groups in the United States1 and are disproportionately affected by HIV.2 In North Carolina (NC), as in other areas of the southeastern United States, the Latino population has increased dramatically. The state experienced a nearly 400% increase in the Latino population from 1990 to 2000, one of highest increases among all U.S. states.3 The group more than doubled in size between 2000 and 2010; an estimated 800,120 persons identifying as Latino resided in NC in 2010 (8.4% of the total NC population, which increased from 4.7% in 2000).4 In tandem with increased Latino population growth in NC, Latinos represent an increasing proportion of the local HIV epidemic. From 1995 to 2009, new HIV cases among Latinos rose from 1% to 8% of cases reported to the state.5,6
The extent that Latinos are involved in local HIV transmission networks, through local HIV acquisition, or immigration with infection acquired abroad followed by onward transmission in the region remains unclear. In our previous analysis of HIV-infected Latinos receiving care in NC, most Latino immigrants were diagnosed with HIV within 10 years of U.S. arrival.7 Latinos were also significantly more likely to present to care with advanced HIV disease compared to non-Latinos,8 implying a long period between HIV acquisition and clinical presentation. While these findings suggest that many infections may be acquired abroad, only phylogenetic analyses can provide a better estimation of the degree immigrants may be involved in local transmission. The process of migration has been hypothesized to increase the risk of HIV acquisition through the loss of previously stable social and sexual networks and engagement in riskier behaviors in the United States.9,10 In our prior HIV phylogenetic studies in NC, Latinos were less likely to be identified in larger local transmission networks (n ≥ 3 individuals) compared to non-Latinos,11 suggesting either importation of infections, incomplete sampling among this group, or differing network characteristics (i.e., fewer concurrent partnerships).
In this study, we integrated behavioral, migration, and HIV sequence data to elucidate HIV transmission patterns among HIV-infected Latinos in NC. Our overall objective was to characterize HIV transmission patterns to assess Latino involvement in local HIV transmission chains and determine if transmission occurs predominately within the ethnic group or is integrated with non-Latinos. Elucidating HIV transmission networks involving Latinos will inform targeted HIV testing and prevention efforts among this vulnerable group.
Materials and Methods
Study design and population
We conducted a cross-sectional analysis of 177 Latinos and 1,496 non-Latinos who received care at University of North Carolina (UNC) clinics between 1999 and 2011 and who had an HIV pol sequence available for analysis. Latino/Hispanic ethnicity was self-identified (defined as a person of Mexican, Puerto Rican, Cuban, South or Central American, or other Spanish culture or origin, regardless of race). Of the Latino patients, 102 participated in face-to-face interviews conducted from 2010 to 2011 to collect behavioral and migration history. Most of these Latino patients (70%) were also enrolled in the UNC Center for AIDS Research HIV Clinical Cohort (UCHCC). The remaining 75 Latinos were included from the UCHCC database but not interviewed. The interview data collection has been described previously.7 In brief, interviews were conducted in Spanish or English by a native culturally competent interviewer with extensive experience working with the local Latino population. Questions spanned HIV testing, partnerships, risk behaviors, and migration history, including birth location, immigration year, return visits, and location of HIV testing. Latinos who were not interviewed were similar to the interviewees by immigrant status, sex, and initial CD4 counts but were more likely to have an earlier year of diagnosis [median 2001 (IQR 1998–2004) vs. 2007 (IQR 2002–2008)].
Clinical and demographic data were abstracted from institutional databases and medical charts using standardized methods. Patients with reported transmission risk behaviors were categorized as heterosexual, men who have sex with men (MSM) [without regard to number of experiences disclosed or sexual orientation], injection drug use (IDU), or other/unknown. Participants reporting IDU in addition to heterosexual or MSM risk were categorized as IDU. Non-Latinos were selected as references from the UCHCC; all had received care at UNC from 1999 to 2011 and had an HIV pol sequence available for analysis.
HIV-1 sequences and phylogenetic inference
Full length protease (PR) and partial reverse transcriptase (RT) sequences were extracted from genotypes obtained during clinical care. Most assays were HIV GenoSure or GenoSure Plus (Laboratory Corporation of America, Research Triangle Park, NC) or the TRUGENE HIV-1 assay (Siemens Healthcare Diagnostics, Tarrytown, NY). Sequences were sampled from 1997 to 2011. In the event that a patient had multiple genotypes, only the oldest sequence was included in the analysis. Subtypes were identified with the Subtype Classification and Evolutionary Algorithms.12 Mutations associated with transmitted drug resistance (TDR) were identified using the 2009 standardized surveillance list from the World Health Organization.13 Sequences were aligned with the MUSCLE algorithm,14 and edited manually15 with stripping of gapped positions. Maximum-likelihood (ML) phylogenies were initially reconstructed using FastTree16 with the general time reversible model of nucleotide substitution. Statistical support of clades was initially assessed with local support values [Shimodaira–Hasegawa (SH) test] in FastTree.
Transmission clusters were defined as clades that included at least one sequence from a Latino patient, with short genetic distances (≤1.5% mean intracluster genetic distance difference), and supported by SH test ≥90% and by confirmatory analyses. We reconstructed the initial ML tree using references available in the Los Alamos National Laboratory (LANL) HIV database (www.hiv.lanl.gov) using Viroblast17 to identify the top 10 related sequences for each study sequence (n = 244 unique LANL sequences; sequences believed to be from the same study population were excluded). Any cluster formed by study sequences that was then disrupted by inclusion of the references was not considered robust. The clusters were further confirmed using the more computationally intensive RAxML v.7.0.418 to construct an ML tree under the same model conditions. Trees were reconstructed first using the complete sequences and then evaluating the third-base codon position only (to assess false clustering due to convergent evolution if sequences share similar drug resistance mutations). Bootstrapping of 1,000 replicates was also performed on the complete sequence tree. Clusters with topology similar to the initially reconstructed clusters and involved in at least one sequences from a Latino patient were analyzed.
Statistical analyses
Our primary outcome was whether a Latino patient was a member of any transmission cluster. We assessed cluster composition by race/ethnicity, immigrant status, transmission risk group, and sex for both small clusters “pairs” (n = 2 members) and larger clusters (n ≥ 3 members). For immigrant Latinos, factors associated with membership in “local” clusters were evaluated. Local clusters were considered larger clusters (≥3 members) or pairs that involved U.S.-born Latinos, non-Latinos, or immigrants whose partnerships were reported to have formed after immigration (therefore probable transmission in NC) and remained with high branch support after inclusion of the LANL references. Differences in categorical variables were tested with the Pearson's χ2 test and continuous variables with the Kruskal–Wallis test. An exploratory predictive log binomial model was fit to the data, using variables found to be associated (p < 0.20) with the outcome in bivariable analyses. Data were analyzed using Stata v.11.0 (StataCorp Corp., College Station, TX).
Results
Study population characteristics
Latinos were predominantly immigrant men with reported heterosexual risk. Overall, 147 (83%) Latinos were immigrants and 30 (17%) were U.S. born; 76% were men (Table 1). Most Latinos (73%) were antiretroviral (ART) naive at the time of sequence sampling [median sample year 2006 (IQR 2002–2009)] and 62% were sampled within a year following diagnosis [median year of diagnosis 2004 (IQR 2000–2007)]. The median age at diagnosis was 30 years (IQR 25–39). Immigrants were more likely to have lower CD4 counts at entry (median 190 vs. 301 cells/mm3; p = 0.01) and report heterosexual risk (67% vs. 23%; p < 0.001) than U.S.-born Latinos.
Table 1.
Characteristics of Study Population of 177 Latinos and the Reference Group of 1,496 Non-Latinos Enrolled in the UNC Center for AIDS Research HIV Clinical Cohort
| Latino group | Non-Latino reference group | ||
|---|---|---|---|
| Variable | Immigrant n (%) | U.S. born n (%) | n (%) |
| Total | 147 | 30 | 1496 |
| Age <30 years at diagnosis | 69 (47) | 14 (47) | 555 (37) |
| Male sex | 109 (74) | 26 (87) | 1,056 (71) |
| Risk group | |||
| MSM | 39 (27) | 17 (57) | 604 (40) |
| Heterosexual | 98 (67) | 7 (23) | 627 (42) |
| IDU | 2 (1) | 6 (20) | 178 (12) |
| Other/unknown | 8 (5) | 0 (0) | 87 (6) |
| Race/ethnicity | |||
| Latino | 147 | 29 | — |
| White | — | — | 501 (33) |
| Black | — | — | 956 (64) |
| Other | — | — | 39 (3) |
| HIV diagnosis year ≥2000 | 116 (79) | 19 (63) | 714 (48) |
| HIV sequence sampling year ≥2005 | 88 (60) | 21 (70) | 700 (47) |
| ART-naive sequencea | 104 (73) | 17 (59) | 691 (46) |
| Transmitted drug resistanceb | 9 (9) | 1 (6) | 61 (9) |
| CD4 count <200 cells/mm3 at clinic entry | 77 (53) | 8 (27) | 582 (39) |
| Region of originc | |||
| Mexico | 93 (65) | — | — |
| Central Americad | 45 (32) | — | — |
| South America/Caribbeane | 6 (4) | — | — |
At time of sequence sampling.
Among ART-naive sequences.
Region of origin unavailable for three patients.
Includes Costa Rica (2), El Salvador (13), Guatemala (9), Honduras (18), and Panama (3).
Includes Columbia (2), Peru (2), and Dominican Republic (2).
MSM, men who have sex with men; IDU, injection drug use; ART, antiretroviral therapy.
The reference group of non-Latinos (n = 1496) was predominantly male (71%); 64% were black and 33% white. Less than half (46%) were ART naive at sampling and these patients included a larger proportion of patients diagnosed before 2000 [median year 1999 (IQR 1994–2004)]. TDR prevalence was similar across groups (9%).
Among the 147 Latino immigrants, 65% originated in Mexico and 32% in Central America (Table 1). The country of origin was unavailable in the medical records for three immigrants who were not interviewed. The median age at immigration was 25 years (IQR 19–31) and the median year of first immigration was 1998 (IQR 1991–2002). Most (87%) were diagnosed in NC and a median of 6 years (IQR 2–10) elapsed from immigration to diagnosis. Only five patients were diagnosed outside the United States. Among the subset of the immigrants interviewed (n = 87), 74% had not returned to their country of origin since first immigration and most (69%) perceived becoming HIV infected in the United States.
Composition of transmission clusters involving Latinos
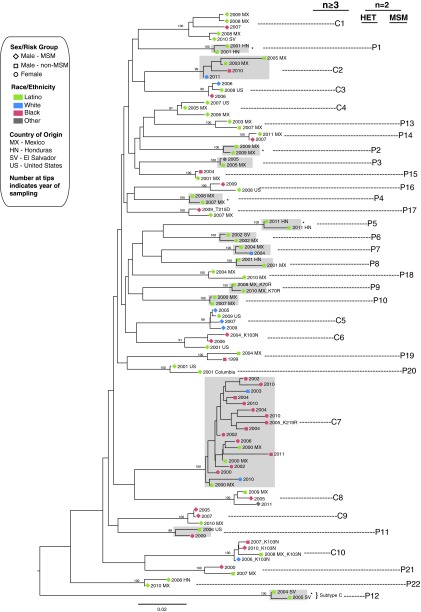
Sequences from 53 (30%; 46 immigrant and 7 U.S. born) Latinos and 41 non-Latinos were identified in 32 discrete clusters containing at least one Latino sequence on the phylogenetic trees identified by FastTree and confirmed with RAxML (Fig. 1). A similar topology was noted when analyzing the third codon position (data not shown) indicating no false clustering due to similar drug resistance mutations. One additional cluster (n = 2 sequences; bootstrap 92% RAxML) was noted in initial analyses but excluded because of disruption by the addition of LANL references. While most clusters were small (22 were pairs; labeled P1–P22 in Fig. 1), there were 10 larger clusters (C1–C10 in Fig. 1; five were triplets, three had n = 4, one had n = 5, and one had n = 18 members). Nearly all sequences from Latinos and those involved in clusters (99%) were subtype B; two were subtype C (P12). There was no significant difference in cluster membership by immigrant status (31% immigrants vs. 23% U.S. born were in clusters; p = 0.67). However, immigrants in clusters were more likely to group with another Latino (78% vs. 29%; p = 0.02) including 13 of 22 sequences in small clusters (pairs), 69% (9/13) of which were heterosexual pairing.
FIG. 1.
Maximum-likelihood phylogenetic tree showing the HIV transmission clusters (n = 32) involving ≥1 Latino sequences from initial tree built with sequences from 177 Latinos and 1496 non-Latinos (only clusters involving Latino sequences with bootstrap support >90% and supported by confirmatory analyses are shown here). Nodes defining clusters are labeled with bootstrap support generated by RAxML. Larger clusters (n ≥ 3 members) are labeled C1–C10. Small cluster “pairs” (n = 2 members) that are heterosexual (HET) are labeled P1–P12 and men who have sex with men (MSM) are labeled P13–P22). Sequences with transmitted drug resistance (TDR) are labeled with identified mutations. *Cluster pairs (n = 5) involving partners immigrating from the same country and where partnerships began prior to immigration. Clusters involving women (n = 14) are highlighted in gray.
Among immigrants, all Latinas were in a cluster (n = 11) and a similar proportion of MSM and heterosexual men (75% and 68%, respectively) were in clusters with other Latinos; overall, 76% (35/46) were in a cluster with another immigrant. Cluster composition by race/ethnicity and risk group varied by cluster size (Table 2). Small clusters (“pairs”; n = 2 members) had a lower proportion of MSM and a higher proportion of immigrants compared to large clusters (n ≥ 3). Of the 44 patients involved in the 22 pairs, 35 (80%) were Latinos, 91% of these were immigrants, and 73% were men. The nine non-Latinos members in these pairs included six men (all black) and three women (one black, one white, and one Native American).
Table 2.
Composition of Small (n = 2 Members) and Large Clusters (n ≥ 3 Members Clusters) Involving Latinos Compared to Latinos Not Found in Clusters
| Clusters with ≥1 Latino | ||||
|---|---|---|---|---|
| Variable | No cluster | Small (pairs; n = 2) | Large (n ≥ 3) | Pa |
| Total clusters, n | — | 22 | 10 | |
| Age ≤30 years at diagnosis, mean % | 43% | 59% | 60% | 0.81 |
| Female, mean % | 25% | 27% | 12% | 0.19 |
| MSM, mean % | 27% | 34% | 72% | 0.05 |
| Race/ethnicity, mean % | ||||
| Latino | 100% | 80% | 43% | 0.001 |
| Black | — | 16% | 37% | 0.04 |
| White | — | 2% | 17% | 0.05 |
| Latino Immigrant, mean % | 81% | 73% | 31% | 0.003 |
p-value represents comparisons between small and large clusters calculated with Kruskal–Wallis rank test.
MSM, men who have sex with men.
Most pairs (12/22; 54%) were male–female pairs reporting heterosexual risk (P1–P12 in Fig. 1). These heterosexual pairs were predominantly composed of immigrants (9/12; 75%) and most were known partners; in seven of these the partners shared the same country of origin and in five the partnerships began prior to immigration and HIV diagnosis. These five pairs (P1, P2, P4, P5, and P12) were considered potential “nonlocal” clusters because HIV transmission may have occurred outside the United States prior to immigration. In the remaining 10 pairs (P13–P22) both individuals were men (including 14 Latinos and six black men), in six pairs both members endorsed MSM risk, in three clusters one man reported MSM risk and the other heterosexual risk (two were Latino), and in one both men reported heterosexual risk (one was Latino). In total, 71% (10/14) of Latino men in these male-only clusters reported MSM risk.
Ten larger clusters (≥3 members; C1–C10) were identified, which included sequences from 18 Latinos and 32 non-Latinos (23 black, 8 white, and 1 other). Immigrants who were in large clusters were more likely to have lived in the United States longer prior to diagnosis compared to immigrants who were not members of clusters or pairs [median 10 years (IQR 5–13) vs. 6 years (IQR 2–10); p = 0.03]. These larger clusters also included a higher proportion of Latino MSM compared to Latinos who were not found in any cluster (mean % MSM 72% in large clusters vs. 27% not in any cluster; p = 0.02). Only two larger clusters (C2 and C7) were composed exclusively of heterosexuals. One of these clusters had 18 members (C7), 13 of whom were women. Two clusters composed only of men included a Latino man reporting heterosexual risk (C8 and C10). In total, 12% (5/42) of Latino men in any pair or cluster reported heterosexual risk but clustered with MSM or other men (behavioral data from three of these Latino men were obtained from an interview). One larger cluster of ART-naive MSM involved TDR (K103N mutation) among all four members (C10).
Latino immigrants in local HIV transmission clusters
Most of the Latino transmission clusters (85%) were considered local clusters and were further evaluated to explore factors associated with immigrant membership in local transmission chains (either through onward HIV transmission or acquisition). In the bivariable analysis, immigrants in local clusters were more likely to be diagnosed in NC (97% vs. 83%; p = 0.04), report MSM risk (44% vs. 21%; p = 0.005), reside in the United States longer before diagnosis (median 9 vs. 6 years; p = 0.01), and be of Mexican origin (86% vs. 57%; p = 0.002) (Table 3). CD4 count, RNA viral load, diagnosis year, or TDR was not associated with local cluster inclusion. In the multivariable model, younger age [risk ratio (RR) 2.34 (95% CI 1.47–3.73)], longer U.S. residence before diagnosis [RR 1.53 (95% CI 1.09–2.15) per 10 years], and originating in Mexico [RR 2.55 (95% CI 1.29–6.88)] remained independently associated with cluster membership among immigrants (Table 3).
Table 3.
Factors Associated with Membership in Local Transmission Clusters Involving Latinos Among 147 Latino Immigrants
| Adjustedb | ||||||
|---|---|---|---|---|---|---|
| Variable | In cluster | Not in cluster | Pa | RR | 95% CI | p |
| Number of patients | 36 | 111 | ||||
| Age <30 years at diagnosis | 21 (58%) | 48 (48%) | 0.12 | 2.34 | 1.47, 3.73 | <0.001 |
| Sex and transmission risk | ||||||
| Female | 6 (17%) | 32 (29%) | 0.02 | Ref | Ref | Ref |
| MSM | 16 (44%) | 23 (21%) | 1.58 | 0.73, 3.40 | 0.24 | |
| Male, non-MSM | 14 (39%) | 56 (50%) | 1.01 | 0.45, 2.22 | 0.99 | |
| Mexican originc | 31 (86%) | 62 (57%) | 0.002 | 2.55 | 1.29, 6.88 | 0.01 |
| Year of HIV diagnosis, median (IQR) | 2005 (2001–2008) | 2004 (1999–2007) | 0.21 | — | — | — |
| Years from migration to HIV diagnosis,d median (IQR) | 9 (4–12) | 6 (2–9) | 0.01 | 1.53e | 1.09, 2.15 | 0.01 |
| Diagnosed in North Carolinaf | 35 (97) | 89 (83) | 0.04 | 3.76 | 0.52, 27.27 | 0.52 |
| CD4 count (cells/mm3),g median (IQR) | 219 (76–384) | 182 (46–356) | 0.23 | — | — | — |
| Viral load (log10 coples/ml),g median (IQR) | 4.9 (4.3–5.2) | 4.7 (4.1–5.3) | 0.31 | — | — | — |
Unadjusted p-value based on Pearson's χ2 test for categorical variables and the Kruskal–Wallis test for continuous variables.
Adjusted log-binomial model includes age at diagnosis, sex and transmission risk, Mexican origin, years from migration to HIV diagnosis, and diagnosed in NC.
Country of origin unavailable for three patients.
Year of immigration missing for 12 patients.
Per 10 year increments elapsed from first migration to diagnosis.
Location of diagnosis missing for four patients.
First available at clinic entry.
RR, risk ratio; MSM, men who have sex with men; ART, antiretroviral therapy.
Discussion
Our primary objective was to better understand HIV transmission patterns among Latinos residing in NC by combining HIV sequence and migration data. We found evidence for ongoing local HIV transmission after arrival for many Latino immigrants. Among the 147 immigrants in our study, 24% were identified in local transmission clusters after excluding those immigrants whose partnerships were reported prior to U.S. arrival. Immigrants in clusters were significantly more likely to be in clusters with other Latinos (76% with another immigrant Latino), indicating within-ethnicity transmission; however, sequences from immigrant and U.S.-born Latinos were equally likely to be in transmission clusters. Based on our multivariable analysis, young Latino immigrants, those of Mexican origin, and those with longer periods of U.S. residence prior to diagnosis were more likely to be involved ongoing HIV transmission locally and thus may benefit from intensified prevention measures. Endorsing sex with men was not a significant predictor in the multivariable analysis.
Although most immigrants reported heterosexual risk, a higher proportion of MSM was identified in transmission clusters. Larger clusters (≥3 members) were also almost entirely composed of MSM; only two Latinas were identified among all the larger clusters. Higher HIV prevalence among established immigrant Latino MSM has been reported compared to U.S.-born and recently arrived immigrant Latino MSM, suggesting HIV acquisition after arrival.19 The phylogenetic analysis in our study not only shows such local transmission, but reveals characteristics of these cluster members. Although transmission directionality cannot be discerned, we identified within ethnicity transmission and clustering with non-Latinos. Larger clusters frequently included black MSM, a group of particular concern in the region as surveillance data reveal increasing HIV incidence rates in this group.2
Phylogenetic cluster analysis can also reveal probable HIV transmission routes that may be unidentified or misclassified by patient history. Therefore, these methods can help assess the degree of nondisclosure of MSM activity, a particular concern among immigrants as homosexuality and HIV remain stigmatized within Latino communities.20,21 In our study, a higher proportion of Latino men, most of whom were Mexican immigrants, reported heterosexual risk than expected compared to U.S. surveillance data in which MSM risk predominates among Mexican-born men.22 We found that 12% of Latino men who were cluster members reported heterosexual risk but were identified in clusters with only men or MSM suggesting such evidence of nondisclosure, although this appears to be minimal. Thus, culturally appropriate prevention programs for immigrants who endorse sex with men and those who endorse sex with women are needed.
Most of the Latino clusters identified in this study likely represent an HIV transmission event that occurred locally and signify groups of ongoing HIV transmission. Five of the 32 clusters (16%) were pairs consisting of known heterosexual partners from the same originating country, and most of these persons reported the partnership predated immigration. All the Latinas in clusters, except for two, were identified in heterosexual pairs. The degree of local HIV transmission could be underestimated by excluding these pairs as “local clusters,” as it is possible that transmission could have occurred prior to, during, or following immigration. The study is also limited by recall or reporting bias, as not all Latinos were interviewed for behavioral data. Our findings are also limited to NC where immigrants largely originate in Mexico.
Transmission patterns may vary by geographic location within the United States and by country/region of origin. Additionally, the limitations inherent to phylogenetic analyses can lead to a limited view of the actual/complete transmission network. Incomplete sampling of persons involved in transmission can result if these people are undiagnosed, receive care at another clinic, or never had a genotype. Unsampled third parties may also be involved in transmission, thereby preventing any inferences that can be made on a direct linkage between two individuals or the directionality of transmission. Our study is strengthened by the stringent criteria used to define clusters, including short branch lengths and high bootstrap values, and by the incorporation of a large number of sequences from non-Latinos from the region.
To date, few epidemiological studies have incorporated phylogenetic analyses to understand HIV transmission patterns among immigrants or migrants23–25 and none has been specifically conducted among immigrant Latinos in the United States. Most prior studies have focused on regions with high HIV diversity, assessing clusters of HIV subtypes to evaluate linkages across borders due to migration25 and importation of non-B subtypes through immigration in Europe.26–28 As subtype B predominates in the Americas, not surprisingly, only two sequences from Latinos in our study were non-B subtypes. Nevertheless, by incorporating migration data, such as originating country and migration dates, inferences can be made as to the degree that HIV transmission may have occurred locally or within ethnic groups in regions dominated by one subtype. Among heterosexual migrants in the Netherlands and former Dutch colonies, the majority with subtype B, phylogenetic analysis revealed that HIV transmission was found to occur primarily within ethnic groups.23
In conclusion, our study reveals a previously undescribed view of HIV transmission among immigrant Latinos in a region characteristic of the southeastern United States. As in many areas of the southeast, new HIV diagnoses continue to be reported among Latinos and other racial/ethnic groups. Ongoing monitoring of phylogenetic and migration data may help better target prevention interventions. Interventions with a continued focus on early testing and linkage to care, particularly among young, Mexican immigrants, that consider the heterogeneity (i.e., diversity in originating countries and risk behaviors) among Latinos are needed. Molecular epidemiology studies conducted in the U.S. southeast and migrant sending communities in Mexico and Central America will also further inform the degree of HIV connection or relatedness between the two regions. Studies with broader geographic sampling in the United States could also be used to help better define population-level HIV transmission patterns among established immigrant and U.S.-born Latinos.
Sequence Data
A subset of sequences analyzed in this study is available in GenBank under accession numbers JX160108–JX161480.
Acknowledgments
We thank patients and staff of the UNC Center for AIDS Research Clinical Cohort, Wake County Human Services and Durham Early Intervention HIV Clinics. This work was supported by the UNC at Chapel Hill Center for AIDS Research (P30AI50410), the National Center for Advancing Translational Sciences, National Institutes of Health (5KL2RR025746-04), and the National Institute of Allergy and Infectious Diseases, National Institutes of Health (K08AI112432-02). The data in this study have been presented previously at the Conference on Retroviruses and Opportunistic Infections, Atlanta, GA, March 3–6, 2013, abstract number I-140.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Pew Hispanic Center: Statistical Profiles of the Hispanics in the United States, 2011. Available from www.pewhispanic.org/2013/02/15/statistical-portrait-of-hispanics-in-the-united-states-2011/ Accessed September10, 2013
- 2.Prejean J, Song R, Hernandez A, et al. : Estimated HIV incidence in the United States, 2006–2009. PLoS One 2011;6(8):e17502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochhar R, Suro R, and Tafoya S: The New Latino South: The Context and Consequences of Rapid Population Growth. Pew Hispanic Center, Washington, DC, 2005 [Google Scholar]
- 4.United States Census Bureau: 2010 Census Briefs: The Hispanic Population. Issued May 2011. Available from www.census.gov/prod/cen2010/briefs/c2010br-04.pdf Accessed September10, 2013
- 5.North Carolina Department of Health and Human Services: 1999 STD Surveillance Report: Available from www.epi.state.nc.us/epi/hiv Accessed March27, 2011
- 6.North Carolina Department of Health and Human Services: Epidemiologic Profile for HIV/STD Prevention & Care Planning: Available from www.epi.state.nc.us/epi/hiv/ Published December 2010. Accessed March27, 2011
- 7.Dennis AM, Wheeler JB, Valera E, et al. : HIV risk behaviors and sociodemographic features of HIV-infected Latinos residing in a new Latino settlement area in the Southeastern United States. AIDS Care 2013;25:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis AM, Napravnik S, Seña AC, and Eron JJ: Late entry to HIV care among Latinos compared with non-Latinos in a Southeastern US cohort. Clin Infect Dis 2011;53(5):480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magis-Rodriguez C, Lemp G, Hernandez MT, et al. : Mexican migrants and their vulnerability to HIV. J Acquir Immune Defic Syndr 2009;51(Suppl 1):S21–25 [DOI] [PubMed] [Google Scholar]
- 10.Prosser AT, Tang T, and Hall HI: HIV in persons born outside the United States, 2007–2010. JAMA 2012;308(6):601–607 [DOI] [PubMed] [Google Scholar]
- 11.Dennis AM, Hue S, Hurt CB, et al. : Phylogenetic insights into regional HIV transmission. AIDS 2012;26(14):1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pond SLK, Posada D, Stawiski E, et al. : An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comp Biol 2009;5(11):e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett DE, Camacho RJ, Otelea D, et al. : drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009;4(3):e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC: MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 2004;32(5):1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall T: BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 98/98/NT. Nucl Acids Symp Ser 1999;41:95–98 [Google Scholar]
- 16.Price MN, Dehal PS, and Arkin AP: FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26(7):1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng W, Nickle DC, Learn GH, et al. : ViroBLAST: A stand-alone BLAST web server for flexible queries of multiple databases and user's datasets. Bioinformatics 2007;23(17):2334–2336 [DOI] [PubMed] [Google Scholar]
- 18.Stamatakis A: RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22(21):2688–2690 [DOI] [PubMed] [Google Scholar]
- 19.Oster AM, Russell K, Wiegand RE, et al. : HIV infection and testing among Latino men who have sex with men in the United States: The role of location of birth and other social determinants. PLoS One 2013;8(9):e73779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz R, Ayala G, Bein E, et al. : The impact of homophobia, poverty, and racism on the mental health of gay and bisexual Latino men: Findings from 3 US cities. Am J Public Health 2001;91(6):927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garland JM, Andrade AS, and Page KR: Unique aspects of the care of HIV-positive Latino patients living in the United States. Curr HIV/AIDS Rep 2010;7(3):107–116 [DOI] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention: HIV Surveillance Report, 2011; Vol. 23 Available from www.cdc.gov/hiv/topics/surveillance/resources/reports/ Published February 2013. Accessed January14, 2014 [Google Scholar]
- 23.Kramer MA, Cornelissen M, Paraskevis D, et al. : HIV transmission patterns among The Netherlands, Suriname, and The Netherlands Antilles: A molecular epidemiological study. AIDS Res Hum Retroviruses 2010;27(2):123–130 [DOI] [PubMed] [Google Scholar]
- 24.Schulter E, Oette M, Balduin M, et al. : HIV prevalence and route of transmission in Turkish immigrants living in North-Rhine Westphalia, Germany. Med Microbiol Immunol 2011;200:219–223 [DOI] [PubMed] [Google Scholar]
- 25.Ansari AA, Mayne AE, Takahashi Y, and Pattanapanyasat K: Incorporation of innate immune effector mechanisms in the formulation of a vaccine against HIV-1. Adv Exp Med Biol 2011;780:143–159 [DOI] [PubMed] [Google Scholar]
- 26.González-Alba JM, Holguín Á, Garcia R, et al. : Molecular surveillance of HIV-1 in Madrid, Spain: A phylogeographic analysis. J Virol 2011;85(20):10755–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Gascun CF, Waters A, Regan CM, et al. : Human immunodeficiency virus type 1 in Ireland: Phylogenetic evidence for risk group-specific subepidemics. AIDS Res Hum Retroviruses 2012;28(9):1073–1081 [DOI] [PubMed] [Google Scholar]
- 28.Parczewski M, Leszczyszyn-Pynka M, Bander D, et al. : HIV-1 subtype D infections among caucasians from Northwestern Poland—phylogenetic and clinical analysis. PLoS One 2012;7(2):e31674. [DOI] [PMC free article] [PubMed] [Google Scholar]