Abstract
In the absence of pathogen attack, organisms usually suppress immune responses to reduce the negative effects of disease resistance. Monoubiquitination of histone variants at specific gene loci is crucial for gene expression, but its involvement in the regulation of plant immunity remains unclear. Here, we show that a rice SWI/SNF2 ATPase gene BRHIS1 is downregulated in response to the rice blast fungal pathogen or to the defense-priming-inducing compound BIT (1,2-benzisothiazol-3(2h)-one,1, 1-dioxide). The BRHIS1-containing complex represses the expression of some disease defense-related genes, including the pathogenesis-related gene OsPBZc and the leucine-rich-repeat (LRR) receptor-like protein kinase gene OsSIRK1. This is achieved through BRHIS1 recruitment to the promoter regions of target genes through specific interaction with monoubiquitinated histone variants H2B.7 and H2A.Xa/H2A.Xb/H2A.3, in the absence of pathogen attack or BIT treatment. Our results show that rice disease defense genes are initially organized in an expression-ready state by specific monoubiquitination of H2A and H2B variants deposited on their promoter regions, but are kept suppressed by the BRHIS1 complex, facilitating the prompt initiation of innate immune responses in response to infection through the stringent regulation of BRHIS1.
Keywords: chromatin remodeling, disease defense, histone H2A and H2B monoubiquitination, priming, SNF2
Introduction
Due to sessility, plants rely on a complex, sophisticated, innate immune system to fight pathogen assaults. The activation of inducible defenses may bring about costs that can negatively affect fitness; therefore, plant immune system is usually suppressed or minimally expressed until induced in response to pathogen attack 1, 2. Like immunity in invertebrate animals, the plant immune system enables the primary pathogen infection to induce lifelong enhanced resistance to the secondary infection. This common immune memory correlates with the so-called cellular priming that renders more rapid and robust responses to secondary attacks to primed cells than to non-primed cells 3, 4. Defense priming can be induced by pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs, respectively), damage-associated molecular patterns (DAMPs), pathogen effectors, wound stimuli, or treatments with some natural or synthetic compounds. This process has been recently proven pivotal to diverse types of systemic plant immunity 3, 5, 6, 7, 8, including systemic acquired resistance (SAR) 8, 9, 10, induced systemic resistance (ISR) 5, 6, 7, 11, the resistance provided by symbiotic fungi 12, β-aminobutyric acid-induced resistance (BABA-IR) 13, and wound-induced resistance 5, 6, 7, 14, and thus enables its promising application in sustainable modern pest management in the field since some priming-inducing compounds have been used as pesticides on the basis of their known plant health- and yield-increasing effects 3, 15.
However, until recently the underlying molecular mechanism of cellular defense priming remains largely unclear. A widely acceptable hypothesis proposes that some dormant cellular signaling components, such as mitogen-activated protein kinases, are activated during defense priming by exposure to secondary biotic or abiotic stresses 3, 4. Additionally, the emerging data have also linked this induced plant immunity to epigenetic modifications, such as histone H3 and H4 acetylation, H3K4 methylation, and H2A.Z—an H2A variant—replacement 16, 17, 18. Monoubiquitination of histone variants, however, has not yet been correlated with plant disease resistance despite its critical role in gene activation 19, 20.
Belonging to the DNA-dependent ATPase family, SWI/SNF2 proteins are responsible for chromatin modification and gene activation 21, 22, suggesting that the epigenetic regulation by SNF2 proteins may play important roles in defense priming. So far, four members of this SNF2 family in Arabidopsis, Photoperiod-Independent Early flowering 1 (PIE1), Splayed (SYD), Brahma (BRM), and Decrease in DNA Methylation 1 (DDM1), have been found to function as chromatin remodellers in the epigenetic control of disease resistance 23. PIE1 17, 18 and BRM 24 are involved in the constitutive repression of SAR, while SYD is associated with the activation of some genes in the JA (jasmonate)/ET (ethylene) signaling pathway and with the resistance against Botrytis cinerea 25. DDM1 seems to play a role in maintenance of the stability of nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins 26, 27, 28. Except for the confirmed role of PIE1 in H2A.Z deposition on the promoters of SA (salicylic acid)-responsive genes 17, the epigenetic molecular mechanisms of the other three SNF2 proteins (SYD, BRM, DDM1) in plant defense are not well understood.
Rice (Oryza sativa L.) is the most important staple food for more than half of the world’s population. Rice blast disease, caused by the fungus Magnaporthe oryzae, annually decreases rice yields by ∼13–30% 29, 30. Probenazole (3-allyloxy-1,2-benzisothiazole-1,1-dioxide, PBZ) is an effective agrochemical widely used to control rice blast disease 31, 32. Previous studies have reported that salicylic acid (SA) acts as a defense signal in the PBZ-induced resistance in adult rice at the 8-leaf stage, but not in young plants at the 4-leaf stage 32, which implies that the PBZ-induced resistance in young plants is SA-independent.
Here, we show that an SNF2 ATPase gene, named BRHIS1, is downregulated by 1,2-benzisothiazol-3(2h)-one,1,1-dioxide (BIT), an active metabolite of PBZ, or by the infection of M. oryzae, in rice seedlings. BRHIS1, interacting with OsTINP1 (a rice homolog of human TGF-β-inducible nuclear protein 1 and yeast NSA2 involved in ribosome biogenesis and cell cycle regulation and proliferation 33), is recruited to the promoter regions of certain antioxidative stress-related and disease defense-related genes, but not to those of SA marker genes, via specific interaction with some monoubiquitinized histone variants deposited on the BRHIS1-targeted promoter regions. This interaction constantly restricts the defense gene expression to a basal level. Upon BIT treatment or pathogen attack, the BRHIS1 suppression is relieved to induce defense priming. Our data suggest the critical role of BRHIS1 in the SA-independent disease resistance of rice.
Results
BRHIS1 encodes a putative RING finger SNF2 ATPase
To elucidate the mechanism of SA-independent defense priming in rice, we performed full-length cDNA suppression subtractive hybridization to identify genes responsive to the defense-priming-inducing compound, BIT 34, using rice seedlings (21-day-old, 4-leaf stage) treated with 0.2 mM BIT by spraying. This approach identified a BIT-downregulated gene Os08g0180300, encoding a putative SWI/SNF2 class ATPase of superfamily 2 helicase 35 (Fig1A and B). Besides the SWI/SNF2 domain, this protein also contains a RING finger domain known to mediate protein–protein interactions 22. There are 39 putative SNF2 family genes in the rice genome 36, but their functions are unclear. As we demonstrate below, Os08g0180300 functions in priming some antioxidative stress-related and disease defense-related genes via direct interaction with several H2A and H2B variants. Therefore, we refer to it as BIT-responsive Histone-interacting SNF2 ATPase 1 (BRHIS1) herein. BRHIS1 is constitutively expressed and localized in the nucleus (Fig EV1A and B), consistent with its presumed functions in chromatin remodeling (see below).
Figure 1.
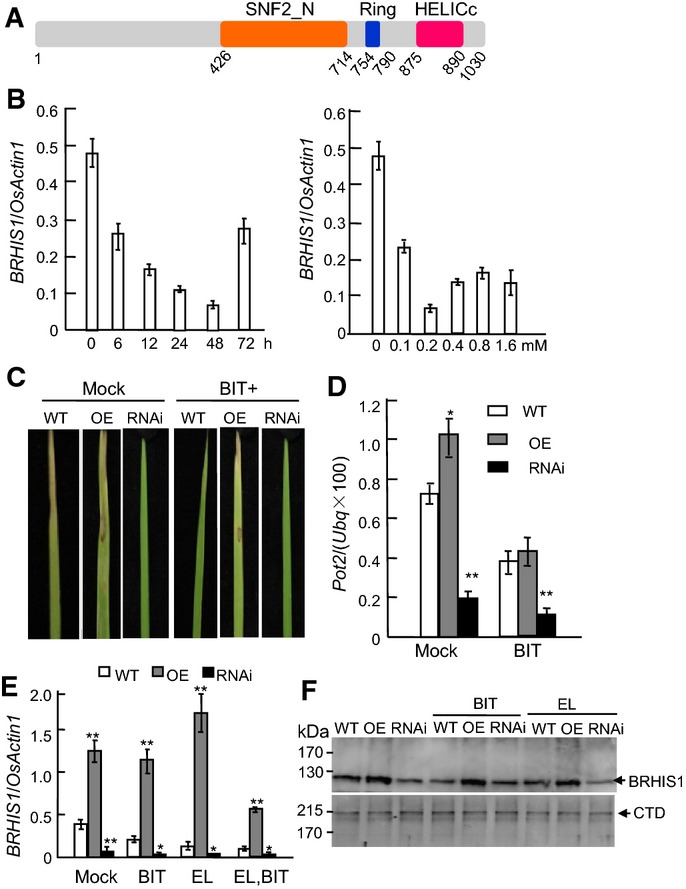
BIT-downregulated RING finger SWI/SNF2 ATPase BRHIS1 is a negative regulator in rice blast resistance
- A Functional domains of BRHIS1. HELICc, helicase superfamily conserved C-terminal domain. Numbers indicate amino acid positions.
- B Time course of BRHIS1 expression in 21-day-old seedlings of wild-type (japonica rice cultivar ZH11) treated with 0.2 mM BIT (left panel), and dose-dependent (right panel) expression of BRHIS1 after BIT treatment for 48 h.
- C Blast resistance phenotypes of the leaves of ZH11 (WT), BRHIS1-OE (OE), and BRHIS1-RNAi (RNAi) 21-day-old seedlings, 6 days after inoculation with incompatible M. oryzae EL0917 (EL). BIT (0.2 mM) and mock (water) were applied to the plants 24 h before fungus inoculation.
- D Growth rates of EL0917 in plants at 6 day post-inoculation were represented as the infection ratios of fungal DNA (Pot2) to rice host Ubiquitin DNA (Ubq) and were quantified by qPCR.
- E, F BRHIS1 mRNA (E) and protein (F) levels in ZH11, the BRHIS1-OE, and the BRHIS1-RNAi plants with or without (mock) the 24-h BIT treatment and/or pathogen inoculation (EL) were determined by qRT–PCR and immunoblotting, respectively. Relative expression values represent the mRNA level of BRHIS1 normalized to that of OsActin1. RNA polymerase II carboxy-terminal domain (CTD) repeat protein was used as a loading control.
Data information: 300 seedlings were pooled for each group. Error bars indicate SD (n = 3), and the significant difference from WT was determined by paired two-tailed t-tests (*P < 0.05; **P < 0.01).

BRHIS1 is constitutively expressed and localized in the nucleus
- The expression pattern of BRHIS1. The expression patterns were based on the Rice Microarray Database RiceXPro (http://ricexpro.dna.affrc.go.jp/).
- A P35S::BRHIS1-CFP construct was introduced into rice mesophyll protoplasts to confirm the nuclear localization of BRHIS1 marked by the nucleus-targeted protein TDR 53 fused with RFP. Scale bars, 10 μm. Representative images from three independent experiments are shown.
BRHIS1 functions as a suppressor of rice blast resistance
To characterize the function of BRHIS1, knockdown and overexpression rice lines were obtained by RNA interference (RNAi) and the ubiquitin promoter (Pubi) driven BRHIS1 cDNA. Since BRHIS1 is BIT-responsive, we explored its potential roles in disease defense priming by assessing the responses of the BRHIS1-RNAi, the BRHIS1-OE, and the wild-type (japonica cultivar Zhonghua11, ZH11) plants to blast fungus. When these 21-day-old seedlings were challenged with the rice blast fungus Magnaporthe oryzae EL0917, the BRHIS1-RNAi plants exhibited the strongest resistance to the pathogen, whereas the BRHIS1-OE plants were mostly susceptible, with or without the BIT (0.2 mM, 24 h) pretreatment (Figs1C and EV2A). To measure fungal growth in the inoculated plants, we used a DNA-based real-time PCR to quantify M. oryzae EL0917 with two primer sets specific to M. oryzae Pot2 37 and rice ubiquitin, respectively. For DNA samples from the infected leaves, the real-time PCR analysis of the infection ratio [MgPot2/(Osubiquitin × 100)] was coincident with their phenotype analysis (Fig1D). In addition, increased resistance to EL0917 correlated with suppressed expression of BRHIS1 mRNA and the protein (Fig1E and F). Taken together, these results suggest that BRHIS1 is a negative regulator of resistance to rice blast pathogen.

BRHIS1 suppresses the resistance to M. oryzae infection by specifically regulating the expression of certain disease defense-related genes in rice seedlings
- Seedlings (21-day-old) of ZH11 (WT), BRHIS1-OE, and BRHIS1-RNAi were treated with 0.2 mM BIT or water (mock) 1 day before incompatible M. oryzae race EL0917 inoculation. Six days after the inoculation, the plants were photographed.
- Expression profiles of three SA marker genes, OsNPR1, OsWRKY03, and OsWRKY71.
- Expression profiles of four OsPBZc homologs in the PR-gene cluster.
- Expression profiles of certain differentially expressed defense-related genes identified by RNA-seq.
Data information: Relative expression values are the ratios of mRNA levels of the genes to that of OsActin1, determined by qRT–PCR. WT, OE, and RNAi indicate ZH11, the transgenic BRHIS1-OE and BRHIS1-RNAi lines, respectively. BIT indicates the 24-h 0.2 mM BIT treatment. EL indicates the M. oryzae EL0917 inoculation. The gene annotation numbers (by http://rapdb.dna.affrc.go.jp) are Os12g0555500 for OsPBZ1, Os12g0555300 for OsPBZ19, Os12g0555100 for OsPBZ15, and Os12g0555000 for OsPBZ14. 300 seedlings were pooled for each group. Error bars indicate SD (n = 3), and the significant difference from the mock or WT was analyzed by paired two-tailed t-tests (*P < 0.05; **P < 0.01).
BRHIS1 regulates the expression of certain defense-related genes
Induction of peroxidase (POD) correlates with disease resistance 38, 39. To determine whether the enhanced resistance of the BRHIS1-RNAi plants is associated with the constitutive activation of peroxidase, the expression of two peroxidase genes, OsPrx41 (NM_001056594.1) and OsPOXgX9 (D16442.1), and POD activities, were investigated. The results showed that both the POD gene expression and POD activities were upregulated in the BRHIS1-RNAi seedlings, indicating that the downregulation of BRHIS1 constitutively activates defense responses (Fig2A). However, neither BRHIS1-RNAi nor BRHIS-OE affected the expression of three SA marker genes, OsNPR1 40, OsWRKY03 41, and OsWRKY71 42 (Fig EV2B), which suggests that the BRHIS1-mediated disease defense priming is SA-independent.
Figure 2.
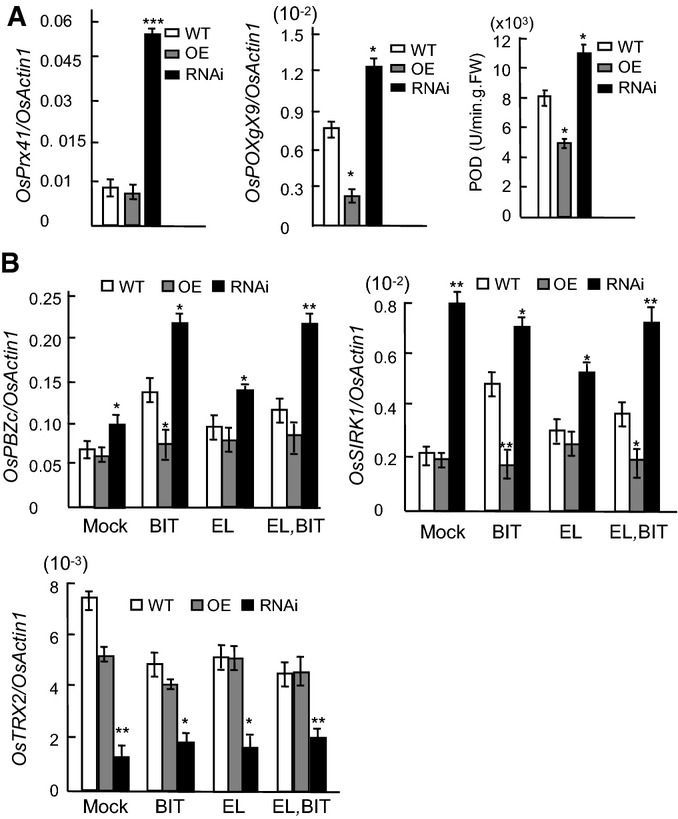
BRHIS1 suppresses defense responses marked by pathogen-attack-induced oxidative burst
- POD activities and POD mRNA levels in the wild-type (ZH11), BRHIS1-OE, and BRHIS1-RNAi plants.
- Expression profiles of two differentially expressed disease defense-related genes, OsPBZc (Os12g0555200), and OsSIRK1 (Os09g0356000), and a differentially expressed antioxidant gene OsTRX2 (Os01g0913000) identified by RNA-seq, in ZH11, BRHIS1-OE, and BRHIS1-RNAi plants with or without (mock) the 24-h BIT treatment and/or M. oryzae EL0917 (EL) inoculation.
Data information: Relative expression values represent mRNA levels of the genes normalized to that of OsActin1. 300 seedlings were pooled for each group. Error bars indicate SD (n = 3), and the significant difference from WT was determined by paired two-tailed t-tests (*P < 0.05; **P < 0.01; ***P < 0.001).
Next, we performed whole transcriptome sequencing (RNA-seq) to screen BRHIS1 target genes using 21-day-old seedlings of the BRHIS1-RNAi and the WT, ZH11. Thirty upregulated (over twofold change) and 33 downregulated (over twofold change) disease defense-related genes were identified in the BRHIS1-RNAi compared with ZH11 (Table EV1, GenBank accession number: SAMN03771629). Two upregulated plant–pathogen interaction-related genes, OsPBZc (Os12g0555200) and OsSIRK1 (Os09g0356000), were selected for further analysis. Our qRT–PCR indicated that both OsPBZc and OsSIRK1 were upregulated in the BIT-treated or fungus-infected ZH11, as well as in the BRHIS1-RNAi (Fig2B). Further qRT–PCR analysis also revealed that none of the other four OsPBZc homologs within the OsPBZc cluster, SA-responsive PBZ1 (Os12g0555500), OsPBZ14 (Os12g0555000), OsPBZ15 (Os12g0555100), and OsPBZ19 (Os12g0555300), were BRHIS1-targeted and responsive to the BIT treatment and the pathogen infection (Fig EV2C). In addition, seven other differentially expressed defense-related genes were also validated by qRT–PCR (FigEV2D).
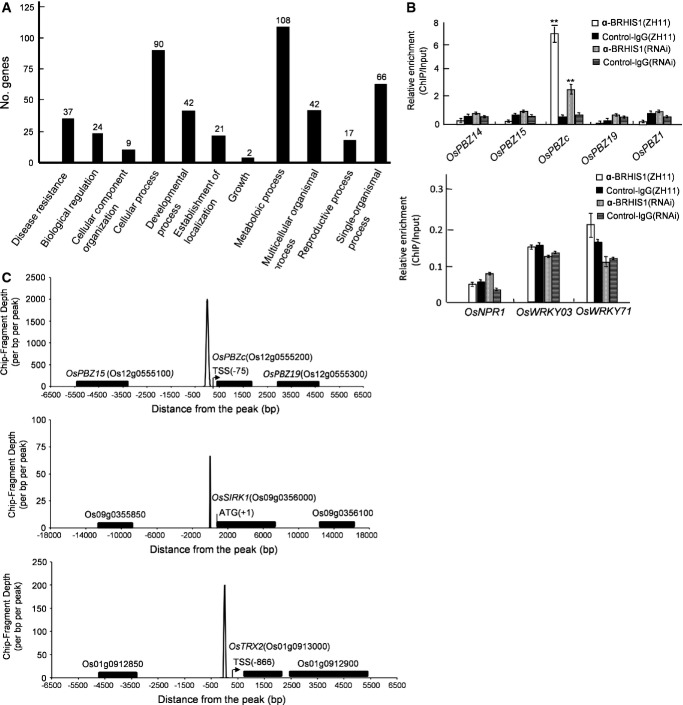
Dependence of the molecular identity of BRHIS1 as a putative SNF2 ATPase suggests that BRHIS1 may act directly on transcription of its target genes as a chromatin-remodeling factor. We further used deep-sequencing immunoprecipitation (ChIP-seq) to identify global BRHIS1 locations in the rice genome (ZH11). This ChIP-seq detected about 400 gene loci, including 37 disease resistance-related genes (FigEV3A, Table EV2, NCBI accession number: SAMN03771629) that contain OsPBZc and OsSIRK1 but no SA marker genes. This observation was confirmed by our chromatin immunoprecipitation (ChIP) assays (Figs3A and EV3B). Our ChIP assays also indicated no BRHIS1 occupancy at the promoter regions of PBZ1, OsPBZ14, OsPBZ15, and OsPBZ19 (FigEV3B). These findings conform to our RNA-seq data and further support our concept that the BRHIS1-involved blast resistance of young rice plants is SA-independent. Distribution of BRHIS1 locations along the OsPBZc and OsSIRK1 promoters was also determined (FigEV3C), consistent with the results of our ChIP-qPCR (Fig3A). To further confirm the reliability of the ChIP-seq data, BRHIS1 locations in the promoter regions of two other disease resistance-related gene analogs, RGA2 (Os12g0489800) and RGA3 (Os04g0111900), were validated by ChIP-qPCR (Fig3B).

BRHIS1 does not bind to the PBZc promoter by itself
Yeast one-hybrid (Y1H) assay showed no direct BRHIS1 binding to the DNA sequence of the OsPBZc promoter region. The bait sequences, PBZc-P1 (−107 to −386 bp), PBZc-P2 (−185 to −436 bp), PBZc-P3 (−287 to −538 bp), PBZc-P4 (−339 bp to −592 bp), PBZc-P5 (−371 bp to −634 bp), and PBZc-P6 (−470 to −660 bp), were cloned into the pAbAi vector. The BRHIS1 cDNA was cloned into the pGADT7 vector and introduced into the bait transformants bearing the constructs containing PBZc-P1 to PBZc-P6. The pGADT7 vector was used as a negative control. 100 and 10−1 indicate the relative dilutions of the applied yeast cells. The PBZc-P1 sequence was self-activated in the Y1H assay. Representative images from three independent experiments are shown.
Figure 3.
BRHIS1 is recruited to the promoter regions of certain disease defense-related and antioxidative stress-related genes
- BRHIS1 ChIP analysis of the OsPBZc and OsSIRK1 promoters in ZH11.
- Two BRHIS1 ChIP-seq positive disease resistance-related gene analogs, RGA2 and RGA3, were also validated by ChIP-qPCR.
- BRHIS1 ChIP analysis of the OsTRX2 promoter in ZH11.
Data information: Normal rabbit control IgG was used as a negative control. BRHIS1-RNAi plants were used as a specificity control for the ChIP performed. Relative enrichment was represented as the normalized ratio of the ChIP DNA to the input genomic DNA at the site. 300 seedlings were pooled for each group. Data that came from three independent experiments were averaged. Error bars indicate SD, and the significant difference determined by paired two-tailed t-tests is shown (*P < 0.05; **P < 0.01; ***P < 0.001).
BRHIS1 is specifically recruited to the OsPBZc promoter, but not to the promoters of four OsPBZc homologs and SA marker genes
- BRHIS1 ChIP-seq positive genes were grouped into 11 major GO (Gene Ontology) pathways. The figures above each column indicate the numbers of genes involved in each GO pathway.
- BRHIS1 ChIP analysis of the promoter regions of OsPBZ14 (−421 to −620 bp), OsPBZ15 (−472 to −723 bp), OsPBZc (−310 to −570 bp), OsPBZ19 (−151 to −386 bp), and OsPBZ1 (−8 to −413 bp) in ZH11 (upper panel). BRHIS1 ChIP analysis of the promoter regions of OsNPR1, OsWRKY03, and OsWRKY71 in ZH11 (bottom panel). 300 seedlings were pooled for each group. Error bars indicate SD (n = 3), and ** indicates the significant difference from the control-IgG-ChIP analyzed by paired two-tailed t-tests (**P < 0.01).
- Distribution of the BRHIS1 ChIP-seq reads along the OsPBZc, OsSIRK1, and OsTRX2 promoters in ZH11 plants. Negative controls in adjacent loci are also shown. 300 seedlings were pooled for each group. Data that came from three independent experiments were averaged.
The combination of ChIP-seq and RNA-seq analyses helped identify 13 BRHIS1-complex-bound target genes (Table EV3). Of these, besides OsPBZc and OsSIRK1, a thioredoxin-encoding gene OsTRX2 (Os01g0913000) was also involved. Given that pathogen infection-triggered oxidative stress may suppress thioredoxin, a key antioxidant, we examined its expression profile by qRT–PCR and validated BRHIS1 recruitment to its promoter by ChIP-qPCR. As we expected, the qRT–PCR data indicated downregulation of OsTRX2 by M. oryzae infection and BRHIS1 silence (Fig2C), and the ChIP assays validated BRHIS1 occupancy at the OsTRX2 promoter (Fig3C), strongly supporting our notion that suppressing BRHIS1 primes defense responses marked by oxidative burst.
BRHIS1 specifically interacts with OsTINP1 and certain histone H2B and H2A variants
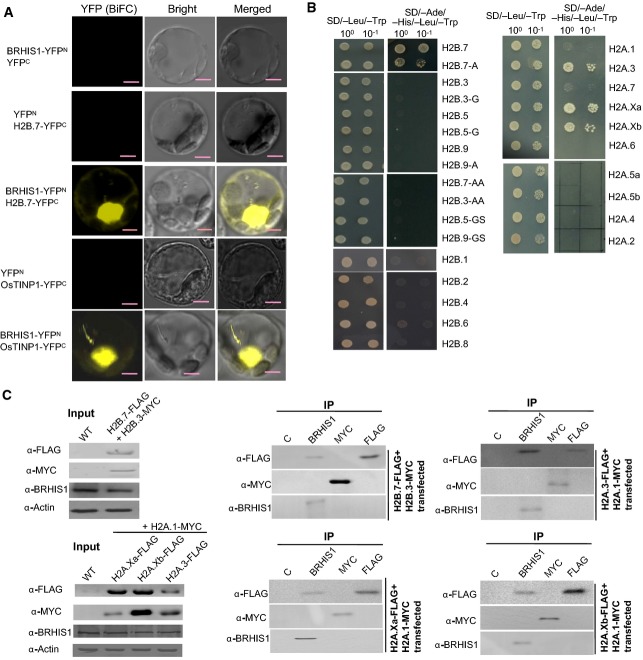
We observed that BRHIS1 did not directly bind to the DNA sequences of the OsPBZc promoter per se, in our yeast one-hybrid assays (FigEV4). This observation suggests that BRHIS1 might interact with some sequence-specific transcriptional activator(s)/repressor(s), or some factor(s) that are specifically located on the promoter regions. To further understand the molecular mechanism of BRHIS1 regulation, we conducted a yeast two-hybrid (Y2H) screening using BRHIS1 as bait and thus identified two BRHIS1-interacting proteins, the histone H2B variant H2B.7 (encoded by Os01g0152900) and the putative transforming growth factor beta inducible nuclear protein OsTINP1 (encoded by Os07g0673100). The in vivo interactions of BRHIS1 with H2B.7 and OsTINP1 in the nucleus were confirmed by bimolecular fluorescence complementation (BiFC) in rice protoplasts (Fig4A). OsTINP1 is a homolog of human TINP1 and yeast NSA2. TINP1 and NSA2 belong to the ribosome S8e superfamily and are involved in ribosome biogenesis and cell cycle regulation and proliferation 33, but the function of OsTINP1 in rice is unknown. OsTINP1 is constitutively expressed, as does BRHIS1 (Fig EV5A), implying the involvement of OsTINP1 in the BRHIS1-containing complex.
Figure 4.
BRHIS1 specifically interacts with OsTINP1 and certain histone H2A and H2B variants
- BiFC assays in rice mesophyll protoplasts confirmed the in vivo BRHIS1-H2B.7 and BRHIS1-OsTINP1 interactions. Scale bars, 5 μm. Representative images from three independent experiments are shown.
- Yeast two-hybrid (Y2H) assays that tested the possible interactions between BRHIS1 and H2B (left panel) or H2A variants (right panel). 100 and 10−1 indicate the relative dilutions of yeast cells for Y2H. H2B.7-A, H2B.3-G, H2B.5-G, H2B.9-A, H2B.7-AA, H2B.3-AA, H2B.5-GS, and H2B.9-GS are mutants with mutations within the conserved H2B domain. Representative images from three independent experiments are shown.
- Co-IP analysis in rice mesophyll protoplasts transiently co-transfected with various constructs expressing FLAG- or MYC-tagged H2A and H2B variants. Immunoprecipitates of BRHIS1, FLAG, MYC, or normal control Rabbit IgG (C) were immunoblotted with anti-FLAG, anti-MYC, or anti-BRHIS1. The protein levels of BRHIS1 and FLAG-tagged- or MYC-tagged H2A or H2B variants in the transfected cells are also shown by Western blot analysis. 100 seedlings were pooled for each group. Representative images from two independent experiments are shown.

Analysis of the BRHIS1-interacting proteins, OsTINP1 and certain histone H2A and H2B variants
- The expression pattern of OsTINP1. The expression patterns were based on the Rice Microarray Database RiceXPro (http://ricexpro.dna.affrc.go.jp/).
- Phylogenetic analysis of rice H2B and H2A variants on the basis of amino acid sequences, using MEGA 4.0 and the neighbor-joining method. The bootstrap test of the inferred phylogeny was performed with 1,000 replications, and the bar indicates 0.1 substitutions per site. The accession numbers for the proteins are as follows: OsH2B.1 (A2XF66); OsH2B.2 (NP_001062114); OsH2B.3 (NP_001042044); OsH2B.4 (NP_001042026); OsH2B.5 (NP_001042053); OsH2B.6 (A2WKT1); OsH2B.7 (NP_001042049); OsH2B.8 (Q9LGH8); OsH2B.9 (NP_001056388); OsH2B.10 (A2WKS3.1); OsH2B.11 (NP_001044754); OsH2A.1 (NP_001059914.1); OsH2A.2 (NP_001059915.1); OsH2A.3 (NP_001175591.1); OsH2A.4 (NP_001055762.1); OsH2A.5a (NP_001043136.1); OsH2A.5b (NP_001051106.1); OsH2A.6 (NP_001054458.1); OsH2A.7 (NP_001066688.1); OsH2A.Xa (NP_001051106.1); OsH2A.Xb (NP_001066920.1).
- Multi-alignment of the amino acid sequences of H2B.7 and three other H2B variants which show the closest phylogenetic relationship to H2B.7, using the DNASTAR software. The conserved H2B domain is indicated. Various mutations created in the H2B variants for interaction tests are also shown.
- Specificity test of the commercially available anti-H2A.X by immunoblotting, using the yeast cells expressing various rice H2A variants.
Given the critical role of histone modification in transcriptional regulation, we drew attention to H2B.7 to unveil the correlation between BRHIS1-mediated chromatin-remodeling events and blast resistance. Database searches identified 11 H2B variants in rice (FigEV5B). H2B.7 and three other H2B variants, H2B.3 (encoded by Os01g0152300), H2B.5 (encoded by Os01g0153300), and H2B.9 (encoded by Os05g0574300), share a highly conserved H2B domain wherein there are only three sequence variations (FigEV5C). To determine whether the BRHIS1-involved interaction is specific to H2B.7 or is a general property of H2B, we created a variety of mutants that have mutations on these three variations (FigEV5C), and tested the interactions between BRHIS1 and all these H2B variants and their mutants. Y2H assay indicated that the mutations in H2B.7 (H2B.7-A, H2B.7-AA) impaired or prevented its interaction with BRHIS1 and that other H2B variants, as well as their mutants, also failed to interact with BRHIS1 (Fig4B). We further demonstrated this specific BRHIS1-H2B.7 interaction in vivo by co-immunoprecipitation (Co-IP) analysis. As shown in Fig4C, immunoprecipitated by anti-BRHIS1 was the FLAG-tagged H2B.7, but not the Myc-tagged H2B.3, in rice protoplasts co-transfected with an H2B.7-FLAG and an H2B.3-Myc fusion constructs.
To further explore the possibility that another histone protein H2A may also interact with BRHIS1, we tested the possible interaction between BRHIS1 and all the ten H2A variants identified by database searches (FigEV5B). Y2H assay revealed that three H2A variants, H2A.3, H2A.Xa, and H2A.Xb, interacted with BRHIS1 (Fig4B). Indeed, in the subsequent Co-IP analysis where rice protoplasts were co-transfected with an H2A.1-Myc and an H2A.3-FLAG, an H2A.Xa-FLAG, or an H2A.Xb-FLAG fusion constructs, the specific in vivo interactions between BRHIS1 and H2A.3, H2A.Xa, or H2A.Xb were confirmed (Fig4C).
The BRHIS1-interacting histone variants are in monoubiquitinated form
Given the critical roles of histone monoubiquitination in gene activation, we performed Co-IP assays to investigate the ubiquitination state of the BRHIS1-interacting histones. In all the tested materials (ZH11, BIT-treated ZH11, and BRHIS1-RNAi), the immunoprecipitated H2B (∼29 kDa) and H2A (∼23 kDa) by anti-BRHIS1 can be detected by both anti-H2B and anti-monoubiquitinated-H2B (uH2B), and both anti-H2A and anti-monoubiquitinated-H2A (uH2A), respectively, indicating that the BRHIS1-interacting histones are in a monoubiquitinated state (Fig5). Since the specific interactions of BRHIS1-H2B.7, BRHIS1-H2A.3, BRHIS1-H2A.Xa, and BRHIS1-H2A.Xb were shown by Y2H and Co-IP analysis done with rice protoplasts, it can be inferred that the coimmunoprecipitated H2B is no other than the H2B.7 and that the coimmunoprecipitated H2A is no other than the H2A.3/H2A.Xa/H2A.Xb. Furthermore, a ChIP assay using a commercially available anti-H2A.X that specifically recognizes rice H2A.3 and H2A.Xb (Fig EV5D) revealed the specific occupancy of H2A.3/H2A.Xb at the PBZc promoter within the OsPBZc cluster (Fig6A), consistent with the BRHIS1 ChIP result (Fig EV3A). These data suggest the histone-mediated selective promoter targeting of the BRHIS1 complex in transcriptional regulation.
Figure 5.

The BRHIS1-interacting histone variants are in a monoubiquitinated state
Co-IP analysis in BRHIS1-RNAi plants (mock-treated) and ZH11 plants with or without (mock) the 24-h BIT treatment. BRHIS1 immunoprecipitates were immunoblotted with antibodies against H2B, monoubiquitinated H2B (uH2B), H2A, or monoubiquitinated H2A (uH2A). Anti-BRHIS1 was used to control for levels. Representative images from three independent experiments are shown.
Figure 6.
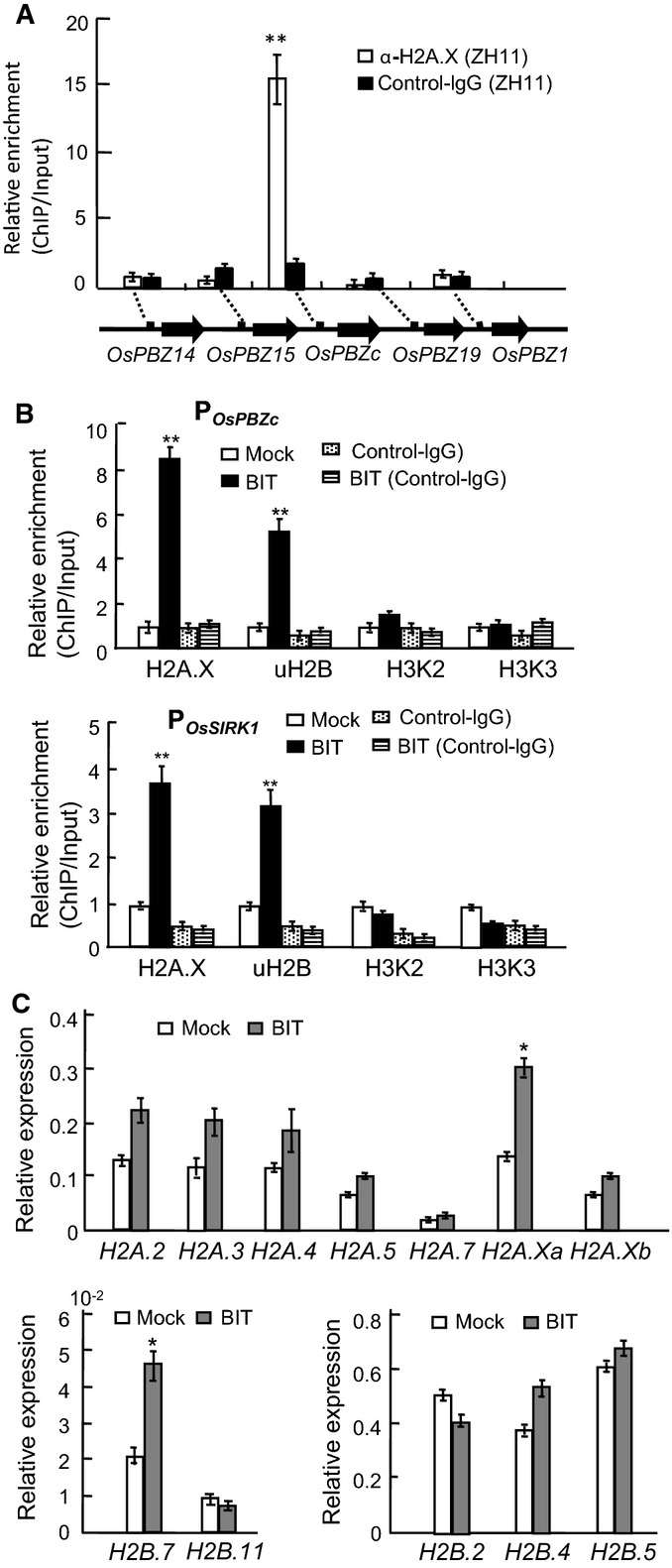
Monoubiquitinated BRHIS1-interacting histone variants mediate the selective regulation of gene expression by the BRHIS1 complex in response to BIT
- ChIP analysis of the promoters of OsPBZc and the other four members of the PR-gene cluster in ZH11, using an anti-H2A.X antibody.
- ChIP analysis of the OsPBZc and OsSIRK1 promoters in ZH11 plants with and without (mock) the 24-h BIT treatment, using anti-H2A.X, anti-uH2B, anti-H3K4/K9me2 (H3K2), or anti-H3K4/K9me3 (H3K3) antibodies.
- Expression analysis of various H2A and H2B variants by qRT–PCR in ZH11 with or without (mock) the 24-h BIT treatment. Relative expression values represent mRNA levels of the genes normalized to that of OsActin1.
Data information: 300 seedlings were pooled for each group. Error bars indicate SD (n = 3), and the significant difference analyzed by paired two-tailed t-tests (*P < 0.05; **P < 0.01) is indicated.
Further ChIP analysis using anti-H2A.X and anti-uH2B showed that the deposition of monoubiquitinated H2A.3/H2A.Xb and H2B.7 on both the OsPBZc and OsSIRK1 promoter regions were significantly increased by BIT treatment (Fig6B), suggesting the monoubiquitinated-histone-variants-modulated gene expression in BIT-induced resistance. However, based on our ChIP analysis using antibodies against H3 lysine-4/lysine-9 dimethylation (H3K4/K9me2) and H3 lysine-4/lysine-9 trimethylation (H3K4/K9me3), we found that H2B monoubiquitination-triggered H3 methylation associated with transcriptionally active chromatin 43 was unaffected in both the OsPBZc and OsSIRK1 promoter regions in BIT-treated ZH11 plants (Fig6B). This finding implicates the independence of on-site H3 methylation in monoubiquitinated-H2B.7-activated gene expression.
The increased H2A.X- and uH2B-based ChIP enrichment in the OsPBZc and OsSIRK1 promoter regions in response to BIT raised a possibility that expression of these histone genes might be affected by BIT signaling. Indeed, qRT–PCR analysis of certain histone variant genes showed that the transcript levels of H2A.Xa and H2B.7, but not those of the other histone variant genes, were upregulated in BIT-treated plants (Fig6C), indicating the BIT-responsive character of H2A.Xa and H2B.7.
Discussion
Although known as “sensitization” as early as 1933 3, defense priming is still an enigma to date. Here, we report that rice employs an SA-independent, specific H2A and H2B monoubiquitination-regulated, disease defense priming negatively operated by BRHIS1, a previously unrecognized SNF2 DNA-dependent ATPase, strongly supporting the hypothesis that epigenetic change sustains long-lasting immune memory in plants.
BRHIS1 and the BRHIS1-interacting monoubiquitinated H2A and H2B variants are transcriptional regulators in age-related innate immunity
SA signaling pathway has long been considered to play a central role in plant defense against pathogens. However, out results show that SA-responsive genes, such as PBZ1, OsNPR1, OsWRKY03, and OsWRKY71, are unresponsive to BRHIS1-mediated blast resistance of rice seedlings, consistent with the published data that SA acts as a defense signal only in adult rice plants 32. These data taken together indicate that the regulation of induced immune responses in rice is age-dependent. We have found that with the help of monoubiquitinated H2B.7 and H2A.3/H2A.Xa/H2A.Xb, BRHIS1 is specifically targeted to given disease defense-related gene loci implicated in blast resistance of rice seedlings. Moreover, BRHIS1-influenced monoubiquitination of H2B.7 and H2A.3/H2A.Xa/H2A.Xb promotes exclusive disease defense-related gene expression in response to pathogen infection in rice seedlings. Our mechanism for monoubiquitinated-histone-involved transcriptional regulation by BRHIS1 provides a model for how innate immunity is age dependently regulated in plants.
Specific interaction of BRHIS1 with certain H2B and H2A variants underlies the selective promoter targeting of BRHIS1
H2A and H2B consist of various variants that may have distinct biological functions 44. Database searches reveal that rice has 10 H2A and 11 H2B variants, but their unique functions are totally unknown, except for the conserved role of H2A.X phosphorylation in meiotic double-strand break formation 45. Here, we show that the transcriptional activation of certain defense-related genes by BRHIS1 is mediated by four rice histone variants, H2B.7, H2A.Xa, H2A.Xb, and H2A.3, thus unveiling a previously unappreciated link between monoubiquitination of histone variants and plant innate immunity. Distinct from the H2A variant H2A.Z reported to play a key role in SAR 17, 18, the BRHIS1-interacting histone variants function in monoubiquitinated form. Of note, H2A.Z deposition suppresses 17, 18, while H2B.7 and H2A.3/H2A.Xa/H2A.Xb monoubiquitinations activate, expression of defense-related genes. In addition, these monoubiquitinated histone variants provide the selective promoter targeting for the BRHIS1 complex, based on our ChIP analysis, which suggests that specific modified histone variants confer the loci selectivity on SWI/SNF complexes along with specific transcriptional activators/repressors 46, 47. Undoubtedly, monoubiquitinated H2B.7 and H2A.3/H2A.Xa/H2A.Xb may have different roles than BRHIS1 recruiting. Given reduced BRHIS1 binding with increasing monoubiquitination of H2B.7 and H2A.3/H2A.Xa/H2A.Xb, we deduce that these histone monoubiquitinations could also get involved in the control of BRHIS1 activities.
A potential cross talk between monoubiquitinated H2A and H2B variants at defense gene loci in BRHIS1-mediated transcriptional regulation
In a broad sense, the roles of H2A and H2B ubiquitinations are deemed distinct 19, and the relationship between H2A and H2B during transcription initiation at the same locus still remains unknown 19. The co-regulation of OsPBZc and OsSIRK1 expression by monoubiquitinated H2A and H2B variants provides a potential cross talk between monoubiquitinated H2A and H2B at these sites, as well as their parallel positive effects on transcriptional activation of given defense-related genes.
A model for BRHIS1-mediated disease defense responses
Here, we propose a BRHIS1-mediated disease defense-response model in young rice plants (Fig7). Under normal growth conditions, monoubiquitinated histone variants, H2A.3/H2A.Xa/H2A.Xb and H2B.7, are deposited on the promoter regions of certain disease defense genes, such as OsPBZc and OsSIRK1. However, for the purpose of minimizing the negative effects of disease resistance, BRHIS1 binds, as a complex, to monoubiquitinated H2A.3/H2A.Xa/H2A.Xb and H2B.7 to blunt their further monoubiquitination and thus constantly suppresses the expression of its target genes (Fig7A). Upon the perception of signals from BIT or pathogen assaults, H2A.Xa and H2B.7 expression is upregulated, whereas BRHIS1 expression is reduced, thereby leading to more deposition of monoubiquitinated H2A.3/H2A.Xa/H2A.Xb and H2B.7, and to the concurrent relief of BRHIS1-complex binding, on the local chromatin. Meanwhile, BRHIS1 activities could be further inhibited with increasing histone monoubiquitination within a possible positive feedback loop. All these steps taken together eventually facilitate and ensure the prompt activation of defense gene expression in disease defense priming (Fig7B). Our working model suggests that plants may establish an expression-ready state at some poised promoters of defense genes, thus facilitating rapid modulation of defense gene expression for induced immune responses.
Figure 7.
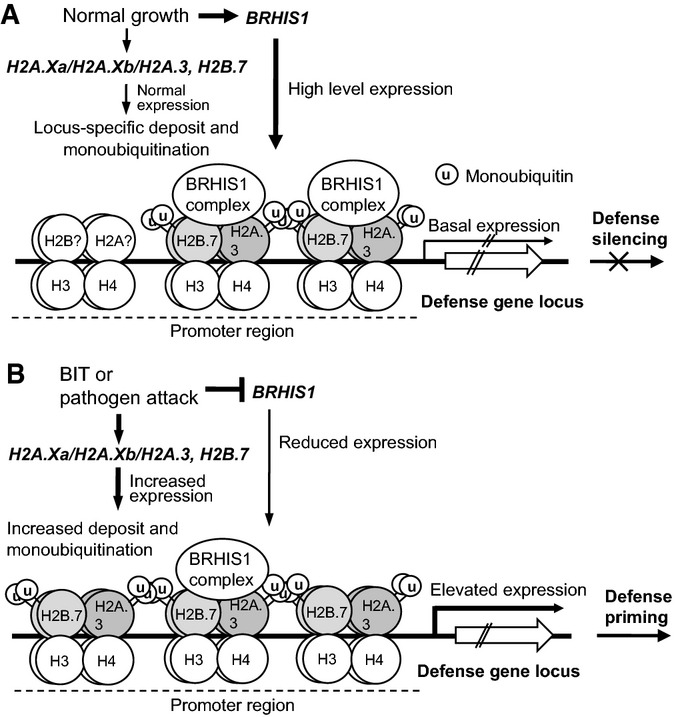
Working model of rice disease defense priming associated with BRHIS1-mediated chromatin remodeling
Simplified chromatin structure of a disease defense-related gene locus, where H2A.3 is used as an example, is shown.
- Under normal growth conditions, BRHIS1 is expressed at a relatively high level, and the BRHIS1 complex, containing OsTINP1 and other possible interactors as well, is recruited to the promoter regions where it targets through binding to most of, if not all, the monoubiquitinated H2B.7 and H2A.3/H2A.Xa/H2A.Xb. This BRHIS1-complex binding obstructs further monoubiquitinations and thus constantly restricts promoter activities to a basal level.
- Upon pathogen attack or BIT treatment, H2A.Xa and H2B.7 expression is upregulated, while BRHIS1 expression is decreased, thus leading to more deposition of H2A.3/H2A.Xa/H2A.Xb and H2B.7, and to the relief of the BRHIS1-complex binding, on the chromatin. The increasing monoubiquitination of these histone variants could also further inhibit BRHIS1 activities. This process significantly improves promoter activities and consequently induces strong expression of certain disease defense genes.
Overall, our data support a novel epigenetic control model for SA-independent disease defense responses wherein BRHIS1 negatively regulates disease defense-relevant chromatin-remodeling events for defense priming.
Materials and Methods
Reagents and antibodies
BIT powder was purchased from Sigma-Aldrich. Anti-BRHIS1 rabbit IgG was raised and purified by Invitrogen with a synthetic peptide corresponding to the N-terminal sequence of BRHIS1, RFPSRSSFGTDNKR. Anti-monoubiquitinated H2B (clone 56), anti-H3K4/K9me2 (07-1843), anti-H3K4/K9me3 (07-992), anti-H2A (ABE327), and anti-monoubiquitinated H2A (clone E6C5) were from Millipore. Anti-RNA polymerase II CTD repeat YSPTSPS (ab5408), anti-H2B (ab1790), anti-H2A.X (ab11175), and anti-MYC (ab9106) were from Abcam. Anti-FLAG (F3165 Mz) was from Sigma-Aldrich.
Full-length cDNA suppression subtractive hybridization (FL-SSH)
The construction and differential screening of the subtracted cDNA library by FL-SSH were described previously 34.
Plant growth and conidial inoculation
Seedlings of japonica rice ZH11 and the transgenic plants were grown in a growth chamber under 14-h-light long-day conditions at 30/25°C day/night cycles. For BIT treatment, BIT solution was sprayed over the 3-week-old rice seedlings grown in growth chambers. Pathogen inoculation with rice blast fungus was performed as described before 48. Magnaporthe oryzae race EL0917, which is virulent for ZH11, was grown on rice polish agar medium for about 10 days at 25°C in the dark. Magnaporthe oryzae spore formation was then induced under blue light for 2–3 days. Spore suspension (1 × 105–5 × 105 conidia per ml) was sprayed onto the rice plants. After inoculation, the plants were incubated at 25°C with saturated humidity for 20 h, and then transferred to a moist vinyl tunnel at 25–30°C. Infection rate of M. oryzae was determined by qPCR as described elsewhere 37, 49. The sequence used for quantifying rice DNA is a single-copy sequence from the rice OsUbiquitin gene. The MoPot2 transposon sequence used for quantifying M. oryzae DNA has about 100 copies in the fungus genome. The infection ratio (Pot2/OsUbi × 100) was calculated from the determined numbers of the target sequences of MoPot2 and OsUbiquitin in each sample. The data for each line were collected from total 144 DNA samples taken from three independent inoculation tests.
Transcriptome sequencing
Total RNAs of ZH11 and BRHIS1-RNAi line were isolated with TRIzol (Invitrogen) according to the manufacturer’s instructions. Then the cDNA libraries for digital gene expression were prepared according to the Illumina sequencing protocol. A virtual library was prepared containing all the possible CATG+17 base-length sequences of the reference gene sequences (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/). A total of 4,888,951 clean tags from ZH11 and 5,611,798 clean tags from BRHIS1-RNAi line were obtained, and mapped to the rice genomic reference sequence (from the japonica cultivar Nipponbare). The number of unambiguous clean tags for each gene was calculated and then normalized to TPM (number of transcripts per million clean tags) 50 (NCBI accession number: SAMN03771629).
ChIP and ChIP-sequencing
ChIP was performed according to Fiil et al 51. Lysed nuclei were sonicated to be in the size range of 0.3–1.5 kb. Supernatant was incubated with anti-BRHIS1, anti-H2A.X, anti-uH2B, anti-H3K4/K9me2, or anti-H3K4/K9me3 antibodies coupled to protein-A agarose beads overnight at 4°C. Part of the eluted DNA was tagged and sequenced (ChIP-seq) by the Illumina sequencing system. Relative enrichment of ChIP DNA to input genomic DNA on the selected sites was estimated by qPCR as described 52. For the ChIP-seq, we merged the reads with overlap before mapping to the reference genome (MSU 7.0 http://rice.plantbiology.msu.edu/), and 6,156,890 (99.59%) reads were merged to a total of 6,182,222 reads pair. Based on the map result of BWA-MEM align method (version: 0.7.4. http://bio-bwa.sourceforge.net/), we filtered the false-positive peak region of mean depth < 10×. After that, we identified disease resistance-related genes with gene annotation, gene ontology (GO), and KEGG pathway analyses. We ran the ChIP-seq data analysis using the online software HOMER (http://homer.salk.edu/homer/ngs/index.html). To obtain the graphical distribution of the BRHIS1 ChIP-seq reads for each sample over the TSS, we calculated the average number of reads on each position from 3,000 bp upstream to 1,000 bp downstream of the TSS of all genes (according to GenBank), normalizing by the total number of reads for each ChIP-Seq experiment. All the ChIP-seq data were collected from three independent experiments (NCBI accession number: SAMN03771629).
Co-IP analysis
The procedure for Co-IP was essentially based on a published protocol 51. Nuclear fractions isolated from rice seedling leaves were incubated with anti-BRHIS1, anti-FLAG, or anti-MYC antibody coupled to protein-A agarose beads (GE Healthcare) overnight at 4°C. The beads were washed three times with IP-B buffer, and bound proteins were eluted with buffer containing 2% SDS and 0.1% β-mercaptoethanol. The samples were analyzed by SDS–PAGE and immunoblotting.
Confocal microscopy
For subcellular localization analysis, the CFP fusion of the full-length BRHIS1, P35S::BRHIS1-CFP, was made in a pUC18-based vector driven by the CaMV 35S promoter. TDR (Tapetum Degeneration Retardation 53) cDNA coding sequences were amplified using the primer pairs TDRRFP-T5F (5′-ATGGGAAGAGGAGACCACCTGCT-3′)/TDRRFP-T5R (5′-ATCAAACGCGAGGTAATGCAGGT-3′) and cloned into a pUC18-based vector containing an RFP gene driven by the CaMV 35S promoter. CFP fluorescence was excited by 458 nm and visualized with a confocal scanning microscope fitted with a 40× water immersion objective (7 DUO; Zeiss).
Y2H screen
The full-length coding sequence of BRHIS1 was cloned into the pGBKT7 vector (Clontech) and transformed into the yeast strain Y187 as bait. The rice cDNA library was constructed from 3-week-old rice seedlings of ZH11. The Y2H screen was performed according to the user’s manual of a Matchmaker Library Construction & Screening Kit (Clontech). The site-directed mutagenesis for H2B.7 and the other H2B variants was carried out by using a QuikChange Site-Directed Mutagenesis kit (Stratagene) with the pGADT7 constructs containing H2B.7 or the other H2B variants as templates.
BiFC assays
For BIFC assay, the constructs, P35S::BRHIS1-YFPN, P35S::H2B.7-YFPC, and P35S::OsTINP1-YFPC, were created and used to transiently transfect rice mesophyll protoplasts from ZH11 with the polyethyleneglycol (PEG)-calcium method. The yellow fluorescent protein (YFP) reconstructed was excited by 514 nm and visualized at 527 nm with a confocal scanning microscope (7 DUO; Zeiss).
Protein analysis
Total protein was extracted from rice seedling leaves, rice protoplasts, or yeast cells in 2× SDS–PAGE buffer for Western blots. Subsequently, separated proteins were transferred onto PVDF membranes (Millipore). After overnight incubation with the primary antibodies in TBS containing 1% BSA and 0.1% Tween-20 at 4°C, membranes were then rinsed with TBS containing 0.1% Tween-20 (TBST), followed by incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature. After TBST washes, Amersham ECL Prime (GE Healthcare) was used to visualize signals on the membranes by ChemiDoc XRS+ (Bio-Rad).
Data availability
The RNA-seq and ChIP-seq data from this publication have been submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) and assigned the identifier SAMN03771629.
Statistics
About 300 seedlings were pooled for each sample. All the data came from three independent experiments. Value represented the average ± the standard deviation of the average (SD). Significant difference was determined by paired two-tailed Student’s t-tests. P < 0.05 was considered significant.
Acknowledgments
We thank Dr. Jun-Xian He and Dr. Letian Chen for comments on the manuscript, Dr. Qing-Hua Pan for kindly providing M. oryzae race EL0917 and technical assistance with conidial inoculation, and Dr. Li-Zhen Tao for kindly providing the vectors for BiFC and subcellular localization analysis. This work was supported by grants from the Ministry of Science and Technology of China (2012AA10A303, 2013CBA01401) and the National Natural Science Foundation of China (30871331 and 30740072).
Author contributions
QZ and YL conceived and designed the experiments. XL, QZ, YJ, and ZJ performed the experiments. XL and QZ analyzed the data. XL, QZ, and YL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
References
- Brown GD, Gordon S. Fungal beta-glucans and mammalian immunity. Immunity. 2003;19:311–315. doi: 10.1016/s1074-7613(03)00233-4. [DOI] [PubMed] [Google Scholar]
- van Hulten M, Pelser M, van Loon LC, Pieterse CM, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. Molecular aspects of defence priming. Trends Plant Sci. 2011;16:524–531. doi: 10.1016/j.tplants.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CM, Mauch-Mani B. Priming in plant-pathogen interactions. Trends Plant Sci. 2002;7:210–216. doi: 10.1016/s1360-1385(02)02244-6. [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJ, Flors V, Garcia-Agustin P, Jakab G, Mauch F, Newman MA, Pieterse CMJ, Poinssot B, Pozo MJ, et al. Priming: getting ready for battle. Mol Plant Microbe Interact. 2006;19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- Conrath U. Priming of induced plant defence responses. Adv Bot Res. 2009;51:361–395. [Google Scholar]
- Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- Kohler A, Schwindling S, Conrath U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 2002;128:1046–1056. doi: 10.1104/pp.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell. 2009;21:944–953. doi: 10.1105/tpc.108.062158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, van Wees SC, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo MJ, Verhage A, García-Andrade J, García JM, Azcón-Aguilar C. Mycorrhizas – Functional Processes and Ecological Impact. Berlin, Heidelberg and New York, NY: Springer-Verlag; 2009. Priming plant defences against pathogens by arbuscular mycorrhizal fungi; pp. 137–149. [Google Scholar]
- Zimmerli L, Jakab G, Metraux JP, Mauch-Mani B. Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta -aminobutyric acid. Proc Natl Acad Sci USA. 2000;97:12920–12925. doi: 10.1073/pnas.230416897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C, Buchala A, Schoonbeek HJ, Metraux JP, Lamotte O. Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 2008;55:555–567. doi: 10.1111/j.1365-313X.2008.03540.x. [DOI] [PubMed] [Google Scholar]
- Beckers GJ, Conrath U. Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol. 2007;10:425–431. doi: 10.1016/j.pbi.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz M, Conrath U, Peterhansel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011;12:50–55. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, Reyes JC. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Takken FL. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14:286–294. doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Heyse KS, Weber SE, Lipps HJ. Histone modifications are specifically relocated during gene activation and nuclear differentiation. BMC Genom. 2009;10:554. doi: 10.1186/1471-2164-10-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Knizewski L, Ginalski K, Jerzmanowski A. Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 2008;13:557–565. doi: 10.1016/j.tplants.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA. Epigenetic control of plant immunity. Mol Plant Pathol. 2010;11:563–576. doi: 10.1111/j.1364-3703.2010.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su Y, Wagner D. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19:403–416. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TL, Kunkel BN, Richards EJ. Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 2002;16:171–182. doi: 10.1101/gad.952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Richards EJ. Gene duplication and hypermutation of the pathogen resistance gene SNC1 in the Arabidopsis bal variant. Genetics. 2009;183:1227–1234. doi: 10.1534/genetics.109.105569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler RS, Leong SA, Teng PS. Rice Blast Disease. Wallingford, UK: CAB International; 1994. [Google Scholar]
- Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol. 2009;27:141–150. doi: 10.1016/j.tibtech.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Tada Y, Yokozeki Y, Akagi H, Hayashi N, Fujimura T, Ichikawa N. Chemical induction of disease resistance in rice is correlated with the expression of a gene encoding a nucleotide binding site and leucine-rich repeats. Plant Mol Biol. 1999;40:847–855. doi: 10.1023/a:1006244323934. [DOI] [PubMed] [Google Scholar]
- Iwai T, Seo S, Mitsuhara I, Ohashi Y. Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol. 2007;48:915–924. doi: 10.1093/pcp/pcm062. [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Jacquier A, Fromont-Racine M. Nsa2 is an unstable, conserved factor required for the maturation of 27 SB pre-rRNAs. J Biol Chem. 2006;281:27099–27108. doi: 10.1074/jbc.M602199200. [DOI] [PubMed] [Google Scholar]
- Zhang QY, Liu YG. A full-length cDNA subtractive hybridization (FLSSH) method and its application in the isolation of inducible expression genes in rice. J S China Agric Univ. 2010;31:34–37. [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang C, Nie P, Lu X, Wang M, Liu W, Yao J, Liu Y, Zhang Q. Characterization and expression analysis of the SNF2 family genes in response to phytohormones and abiotic stresses in rice. Biol Plant. 2011;55:625–633. [Google Scholar]
- Berruyer R, Poussier S, Kankanala P, Mosquera G, Valent B. Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology. 2006;96:346–355. doi: 10.1094/PHYTO-96-0346. [DOI] [PubMed] [Google Scholar]
- Chittoor JM, Leach JE, White FF. Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 1997;10:861–871. doi: 10.1094/MPMI.1997.10.7.861. [DOI] [PubMed] [Google Scholar]
- Liu X, Williams CE, Nemacheck JA, Wang H, Subramanyam S, Zheng C, Chen MS. Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol. 2010;152:985–999. doi: 10.1104/pp.109.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D, et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J. 2007;5:313–324. doi: 10.1111/j.1467-7652.2007.00243.x. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Bai XQ, Qian Q, Wang XJ, Chen MS, Chu CC. OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 2005;15:593–603. doi: 10.1038/sj.cr.7290329. [DOI] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol. 2007;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell. 2013;25:2998–3009. doi: 10.1105/tpc.113.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- Pan QH, Hu ZD, Tanisaka T, Wang L. Fine mapping of the blast resistance gene Pi15, linked to Pii, on rice chromosome 9. Acta Bot Sin. 2003;45:871–877. [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, et al. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc. 2008;2008:t5049. doi: 10.1101/pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Data Availability Statement
The RNA-seq and ChIP-seq data from this publication have been submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) and assigned the identifier SAMN03771629.