Abstract
Peptides featuring the LR(S/T) motif were identified that could specifically target CLL1. Nanomicelles conjugated to CCL1-targeting peptides were loaded with DiI or daunorubicin during synthesis. The resulting nanomicelles were approximately 15 nm in diameter, had high affinity for cells expressing CLL1 and could transport the DiI load to the interior of the cells, including the nucleus. The targeting nanomicelles delivered DiI to LSC isolated from 2 out of 3 clinical specimens, but did not bind to peripheral blood mononuclear cells and normal hematopoietic stem cells. The nanomicelles could be loaded with daunorubicin with up to 5 mg of drug per 20 mg of telodendrimer, a clinically useful degree of drug loading. In conclusion, CLL1-targeting nanomicelles have the potential to be used for targeted drug delivery to leukemia stem cells.
Keywords: leukemia stem cells, nanoparticles, targeted therapy
INTRODUCTION
The concept of cancer stem cells has tremendous implications for the management of cancer [1,2]. Cancer stem cells have been identified in both hematological and solid malignancies, suggesting that existence of cancer stem cells may be a common feature of most malignancies. Cancer stem cells can self-renew and regenerate more cancer cells. Therefore, in order to cure cancer, cancer stem cells must be eradicated. However, cancer stem cells are chemoresistant compared to their progeny cancer cells [3,4]. The goal of this study is to demonstrate the feasibility of using drug-loaded nanoparticles that are engineered to bind with high affinity and specificity to acute myeloid leukemia (AML) stem cells (LSC). Chemoresistance of LSC can be overcome with high-dose chemotherapy followed by bone marrow transplantation. However, high-dose chemotherapy is associated with severe toxicity and therapy-related mortality. Many patients are not eligible for this treatment because of co-morbidities. This is especially true for AML patients, who have a median age at diagnosis of 60 to 65 years. For bone marrow transplantation, autologous bone marrow or stem cells are usually not used because of contamination by LSC. Allogeneic hematopoietic stem cell transplantation is often associated with severe graft-versus-host disease, and is commonly not offered to elderly patients. Therefore, the development of chemotherapy that is specific for LSC is a critical unmet medical need.
Among cancer stem cells, AML LSC have been best characterized, and many of their cell surface molecules are known. For example, the C-type lectin-like molecule-1 (CLL1) is known to be preferentially expressed on most AML LSC. Even though it is normally expressed on CD38+ myeloid progenitors, itis not on CD34+ CD38- hematopoietic stem cells [5,6].
Recently, two of our co-authors (Drs. Luo and Lam) developed a novel biocompatible nanomicelle drug delivery system comprised of a unique amphiphilic polymers called telodendrimers [7]. Telodendrimers consist of cholic acid, lysine and polyethylene glycol (PEG) covalently conjugated together, which imparts the ability to self-assemble into a water-soluble spheroid with a hydrophobic core capable of sequestering many types of drugs. Cholic acid, a primary component of bile acid, possesses a facial amphiphilic structure: a rigid steroid scaffold with four hydrophilic groups on one surface, and hydrophobic methyl groups on the other surface of the scaffold. Lysine is a natural amino acid. PEG is inert and has been used to improve the pharmacokinetics of therapeutic drugs. This nanocarrier system has many attractive characteristics for drug delivery such as high drug loading capacity, narrow polydispersity, well-defined structure, easy chemical modification, physical and chemical stability, biocompatibility and biodegradability.
In this project, we used the phage-display library method and discovered a series of peptides that bind specifically to CLL1. One of these ligands, CLL1-L1, was used to decorate the surface of our nanoplatform to form a novel targeting nanomicelle that we named LSC-targeting nanomicelle. Unlike solid tumors that primarily reside at the extravascular space and are accessible by nanotherapeutics mainly through enhanced permeability and retention effect, LSC and leukemic cells reside primarily inside blood vessels and bone marrow that are directly accessible by nanotherapeutics through intravenous administration. We report herein that targeting nanomicelles displaying CLL1-L1 not only can attach the nanomicelles to the surface of cells expressing CLL1, but, more importantly, can deliver the nanomicelles and their content into the target cells.
MATERIAL AND METHODS
Identification of ligands targeting CLL1
The CLL1 cDNA expression vector was purchased from the American Type Culture Collection (Manassas, VA, USA) and was subcloned to a pcDNA3.1 expression vector. The C7C phage display peptide library was purchased from the New England Biolab (Ipswich, MA, USA). CLL1 was expressed in 5637 (bladder transitional cell carcinoma), A549 (non-small cell lung cancer) and HTB38 (colon cancer) cells for sequential panning to identify peptides that bind to CLL1. At the time when this project was performed, there was no commercially available anti-CLL1 antibody to detect the expression of CLL1. Therefore, we constructed CLL1-RFP (red fluorescence protein) chimeric gene, and cloned into the pcDNA3.1 expression vector in which RFP replaced part of the intracellular domain of CLL1 to allow the monitoring of CLL1 expression. To eliminate any phage that may bind to the confounding cells in vivo, the C7C library was subtracted with whole blood, peripheral blood mononuclear cells (PBMC), normal healthy hematopoietic stem cells left over from allogeneic stem cell transplantation, human umbilical vascular endothelial cells (HUVEC), fibroblasts, 5637, A549 and HTB38 cells, for 4–6 hours with each cell type, before each round of panning (SI, S1). The subtracted phage library was then sequentially panned against 5637, A549 and HTB38 cells expressing CLL1-RFP per manufacturer’s protocol. The library was panned against three different cancer cell types in order to minimize the possibility the isolating artifactual peptides that bind to intrinsic surface molecules on these cells other than CLL1. Each round of panning was performed for only one hour to select those peptides with high affinity to CLL1. After three rounds of panning, 36 clones were selected, amplified and submitted for sequencing. The amino acid sequences were aligned for comparison (Table 1).
Table 1.
Sequences and alignment of peptides from panning against cells expressing CLL1 (a motif of LR(S/T) was observed)
| Sequences of Amino acids |
Number of clones (total:36 clones) |
|---|---|
| 11 | |
| 7 | |
| 3 | |
| 1 | |
| 1 | |
| 1 | |
| 5 | |
| 3 | |
| 2 | |
| 1 | |
| 1 |
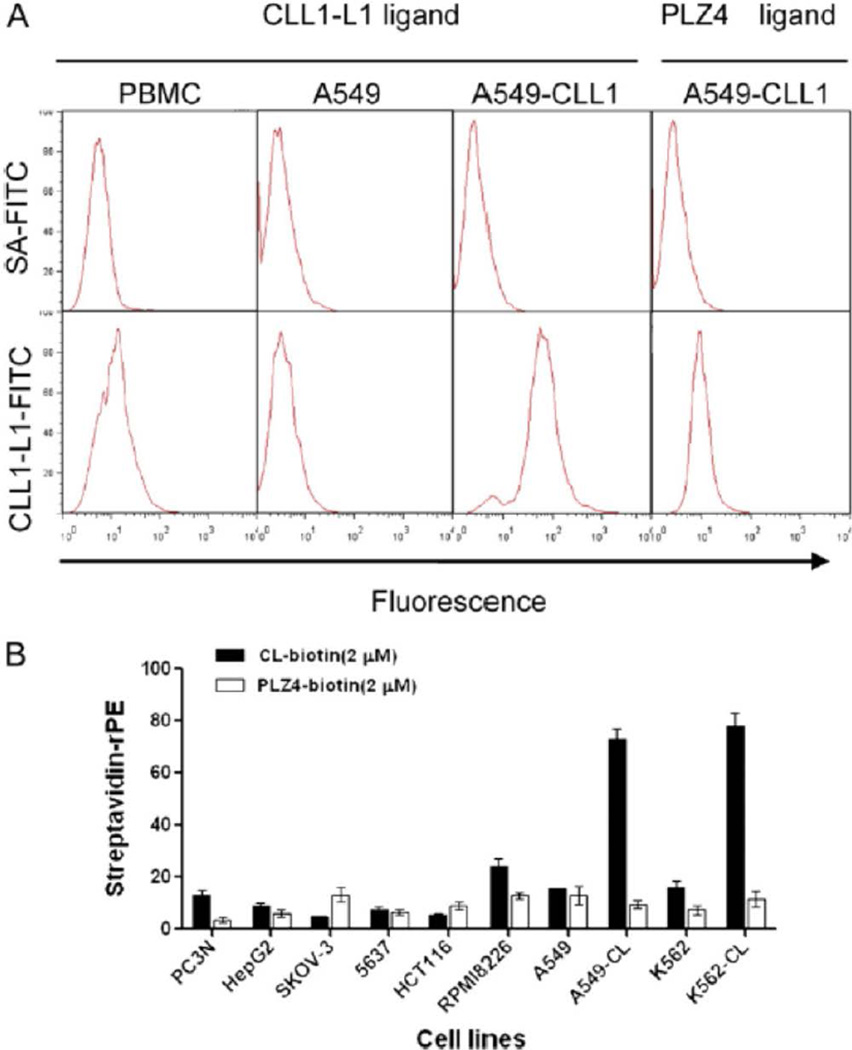
One peptide, CLL1-L1, with the sequence of CDLRSAAVC was synthesized for further analysis. To determine the binding specificity, cell lines were incubated with CLL1-L1 conjugated to biotin and probed with streptavidin (SA)-phycoerythrin (PE) (excitation 480 nm, emission 578 nm) (Figure 1, B). The following cell lines were purchased from the ATCC and used for this study: PC3N (prostate), HepG2 (liver), Skov-3 (ovarian), 5637 (bladder), HCT116 (colon), RPMI8226 (myeloma), A549 (lung), A549 expressing CLL1, 562 (leukemia), and K562 cells expressing CLL1. A bladder cancer–specific ligand, PLZ4, conjugated to biotin was used as the negative control.
Figure 1.
Binding of the CDLRSAAVC peptide to CLL1. (A) Biotinylated CDLRSAAVC peptide was synthesized and used to probe PBMCs, A549 cells transfected with vector alone, and A549 cells expressing CLL1. Specific binding was observed to A549 cells expressing CLL1, but minimal binding was observed with PBMCs or A549 cells transfected with vector alone, or when A549 cells expressing CLL1 were probed with a bladder cancer–specific ligand PLZ4 (far right panel). Cells were probed with SA-FITC (upper panels) or with CLL1-targeting peptide and SA-FITC (lower panels). (B) Binding specificity of the CDLRSAAVC peptide. Cells of various origins were incubated with the biotinylated CLL1-targeting peptide (CL-biotin) or a bladder cancer–specific peptide, PLZ4, as a negative control, and probed with SA-PE.
Synthesis of targeting and non-targeting nanomicelles
The methodology that we used to synthesize CLL1-targeting peptides has been described before, and is detailed in the supplemental information (SI, S2) [8–10]. In brief, a (Alloc)lysine(Fmoc) was coupled onto Rink amide resin in order to introduce anchor groups for the post-cleavage coupling of CLL1 targeting peptides to the telodendrimer molecule. The peptides were synthesized on the lysine(Alloc)-Rink resin sequentially via Fmoc peptide chemistry [11]. After synthesis and cleavage of the lysine(Alloc)-peptide from the solid support, the crude peptide was treated with a buffer of air saturated 50 mM ammonium bicarbonate in the presence of active charcoal, which resulted in formation cyclized peptides via the oxidative coupling of the two cysteines located at the carboxy and amide termini of the peptide. The charcoal was removed by filtration and the solution was lyophilized. Crude peptide was purified by reverse-phase HPLC to at least 95% purity. The molecular weight of separated fractions was characterized by MALDI-TOF MS to confirm the peptide sequence. The purity of the purified peptide was determined by analytical HPLC.
The details of synthesis of nanomicelles have been described before (SI, S3) [7]. In brief, a dendritic core of lysines was synthesized by Fmoc solid-phase chemistry and attached to eight cholic acid (CA) groups via NHS ester chemistry. The octamer, called CA8, was attached to azide-terminated PEG (~5 kD) using solution-phase condensation chemistry to generate a PEG5k-CA8 telodendrimar. To determine the completeness of the coupling, the Kaiser test was performed [12]. The final product was collected by ether precipitation and purified using the repeated dissolve-precipitate procedure in dichloromethane and cold ether to remove the coupling agents and impurities. The final telodendrimer, a white powder, was dissolved in pure water and dialyzed against a large volume of water (MWCO of 3500 Dalton). The telodendrimer solution was then lyophilized to yield white powder. The polydispersity and molecular weight of the telodendrimer was characterized by Gel Permeation Chromatography (GPC) and MALDI-TOF Mass Spectrometry, and the chemical structure was characterized by nuclear magnetic resonance (NMR). The purity of the telodendrimer was measured by analytical HPLC.
Aqueous-phase Click chemistry between azido and alkyne groups, catalyzed by cuprous ion, was used for coupling the alkyne group of CLL1-targeting peptides onto the azide groups at the end of PEG on telodendrimer. The purity of the CLL1-targeting telodendrimer was analyzed using HPLC, and the molecular weight was measured by MALDI-TOF MS.
To load daunorubicin (DNR) or fluorescent dye DiI into nanomicelle, DNR/DiI and telodendrimer (20 mg), at different ratios, was dissolved in chloroform (5 mL) in a 10 mL-flask. The chloroform was removed on a rotaevaporator under vacuum, and further dried under high vacuum for 30 min. One mL of USP saline was then added into the flask. The mixture was vortexed and sonicated for 30 min at room temperature. The final product was analyzed for drug loading capacity with HPLC, nanomicelle size and dispersity with a dynamic light scattering (DLS, Microtrac) particle sizer and transmission electron microscopy (TEM, Philips CM-120). It was filtered through a filter (0.22 µm) before injection into sterilized vials for further studies.
Uptake of drug-loaded nanomicelles by cells expressing CLL1
Since the cytoplasm of adherent cells spreads wider and has a narrower focal plane than that of suspension AML cells, we chose to use adherent A549 cells transfected with a CLL1 expression vector as a model system for easy visualization of nanomicelle-mediated drug delivery. The expression vector was constructed so that the cytoplasmic domain of CLL1 was replaced with green or red fluorescence protein (GFP or RFP) to monitor the expression of CLL1 by green or red fluorescence microscopy. To track the cellular uptake, nanomicelles were loaded with DiI, a red fluorescent dye (excitation: 549 nm, emission: 565 nm). A549 cells transfected with GFP vector alone (control cells), or CLL1-GFP (experimental cells) were cultured in chamber slides overnight. Two types of DiI-loaded nanomicelles were used for this experiment: (1) non-targeting nanomicelles that were not decorated with high-affinity peptides; and (2) targeting nanomicelles that were decorated with CLL1-targeting peptides. A549 expressing GFP and A549 expressing CLL1-GFP were incubated with these two types of nanomicelles for 10, 20, 30 and 60 minutes, and washed with complete culture medium three times to remove unbound nanomicelles. The cells were then examined by fluorescence microscopy.
To further demonstrate that the targeting nanomicelles penetrated into the CLL1-expressing cells, we performed high-resolution live cell imaging with a DeltaVision deconvolution imaging system (AppliedPrecision, WA). A549 expressing GFP and A549 expressing CLL1-GFP were cultured over night on glass bottom culture dishes (MatTek, MA), washed once with complete culture medium, and added with nontargeting or targeting nanomicelles (both were loaded with DiI dye, 0.4 mg/ml), respectively. Cells were incubated at 37°C for 15 min, and washed three times with PBS buffer (pH 7.4). These dishes were then directly examined with the DeltaVision imaging system.
Targeting efficacy of clinical specimens
Peripheral blood specimens from AML patients were drawn for this study after informed consent was obtained. Leukemic cells together with some normal PBMC were isolated using Ficoll gradient. These isolated cells then passed through CD34+ MACS columns (Miltenyi, Bergisch Gladbach, Germany) to enrich CD34+ leukemic cells. Over 96% of the isolated cells were CD34+ cells (data not shown). Normal hematopoietic stem cells were obtained from leftover allogeneic stem cell transplantation specimens. To determine the targeting efficiency of nanomicelles coated with CLL1-targeting peptide, enriched CD34+ cells were incubated with different concentrations of nontargeting or targeting nanomicelles at 37°C for 30 min. These cells were washed three times with PBS buffer (pH 7.4, 0.5% BSA), and the fluorescence density was detected by flow cytometry.
RESULTS
Development of ligands targeting CLL1
The strategy of panning the PhD C7C phage peptide display library is shown in Suppl. Information S1, Figure A). After panning, a total of 36 phage clones were sequenced. After alignment, two consensus sequences were identified: CXLR(S/T)AAVC and CXLRSSGPC (Table 1), in which “X” represents several amino acids that can be replaceable. Serine and threonine belong to the same hydrophilic class of amino acids with only one methyl group difference. Of these two major sequences, one common motif exists: “LR(S/T)”.
To confirm that these ligands indeed bind to CLL1, A549 cells were transfected with a CCL1-RFP expression vector or RFP (SI, Figure B). Expression of CLL1-RFP could be visualized by the appearance of red fluorescence. Since all these peptides contain the LR(S/T) motif, we randomly select the peptide CDLRSAAVC for subsequent in-depth analysis. The A549 cells expressing CCL1-RFP or RFP were incubated with phage displaying the CDLRSAAVC peptide, and probed with anti-M13 monoclonal antibody-FITC conjugate. Phage expressing the CDLRSAAVC peptide were detected on the surface of A549 cells expressing CLL1-RFP, but not on cells expressing RFP. This peptide is named CLL1-L1 and was used for all subsequent experiments.
We also performed flow cytometry studies to determine cell binding. CLL1-L1 was conjugated to biotin through the polyethylene glycol (PEG) linker. Cells were incubated with biotinylated CLL1-L1 and probed with streptavidin-PE. No significant binding of CLL1-L1 to PBMC and A549 control cells was observed while significant binding of CLL1-L1 to A549 cells expressing CLL1-GFP was detected (Figure 1A).
To further determine the binding specificity, cell lines of various origins were probed with CLL1-L1 or a bladder cancer–specific ligand PLZ4 as a negative control. Specifically, CLL1-L1 bound to A549 and K562 cells that expressed recombinant CLL1, but not to the parental A549, K562, or other cell lines (Figure 1, B).
Development and characterization of nanoparticles targeting CLL1
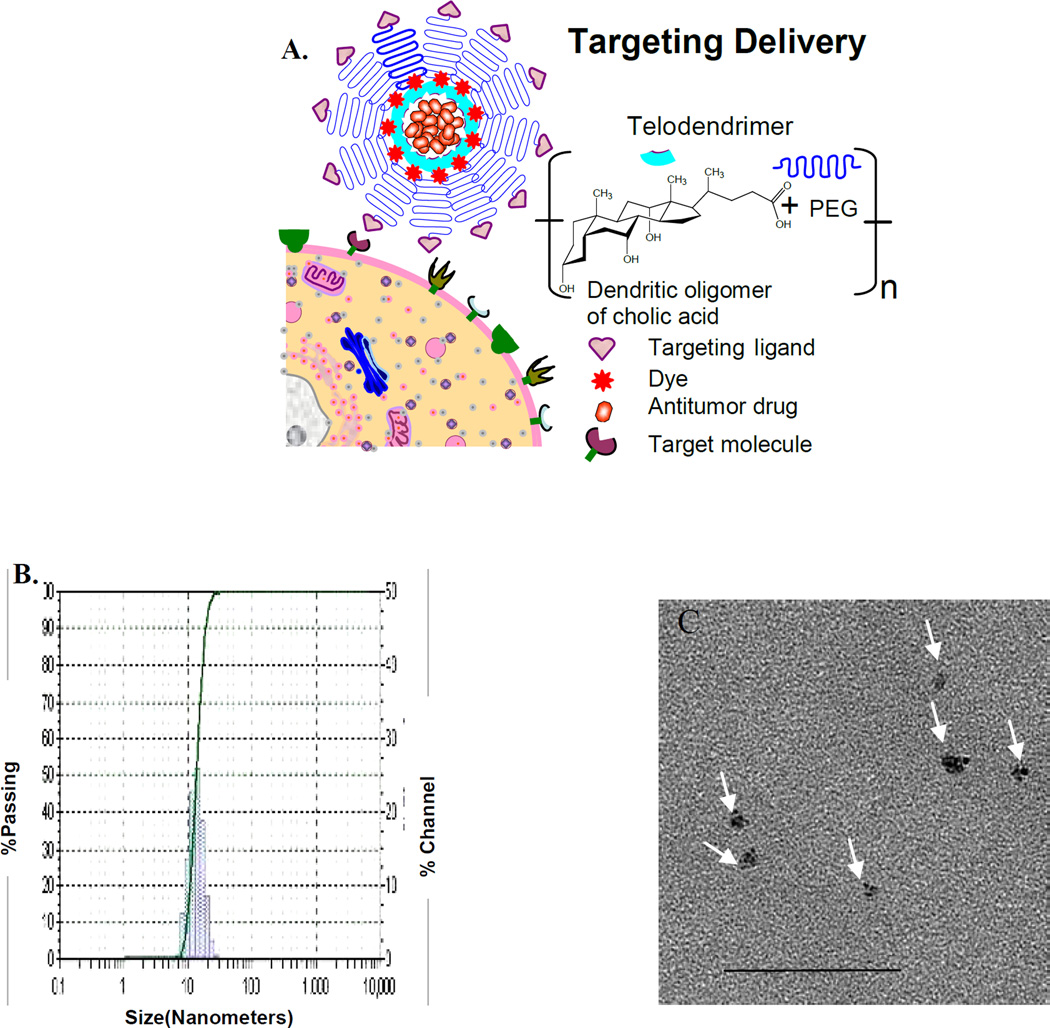
Next, we determined if CLL-L1 could be used to develop targeting nanotherapeutics by displaying CLL-L1 on the surface of our recently developed nanomicelles (Figure 2A). The average size of the targeting nanomicelle was approximately 13.5 nm with a narrow size distribution (Figure 2B, left panel). This size distribution was confirmed with transmission electron microscopy (Figure 2B, right panel). The size of the nanomicelles is attractive for treating LSC since the particles are small enough to easily distribute though the vasculature and bone marrow, but are large enough to be highly loaded with daunorubicin (DNR). DNR is one of the only two first-line chemotherapeutic drugs (together with cytarabine) for AML. We determined the drug loading capacity, and found that up to 5 mg of DNR could be loaded in 20 mg of telodendrimer. Considering a typical clinical dose of DNR is 45 mg/m2, this is equivalent to a telodendrimer dose of 225 mg/m2. This dose of telodendrimer is well below the nontoxic level of telodendrimer at 1 mg/mL in cell culture,8 and less than one fourth of the telodendrimer dose used in our dog studies when the nanomicelles were loaded with paclitaxel, another chemotherapeutic drug (data not shown)‥ This suggests that the dose of telodendrimer of our targeting nanomicelle is within the tolerable range and that toxicity will be limited by the total dose of DNR. Further preclinical and clinical trials will be performed to determine the toxicity of DNR in the nanomicelle formulation.
Figure 2.
Characterization of targeting nanomicelles. (A) Schematic. (B) Size determination of nanomicelles with dynamic light scattering (left panel) and TEM (right panel). The median size of nanomicelles loaded with DNR and coated with CLL1-targeting peptide was about 13.5 nm with narrow size distribution. Scale bar, 50 nm.
Cellular uptake of CLL1-targetting nanomiceles
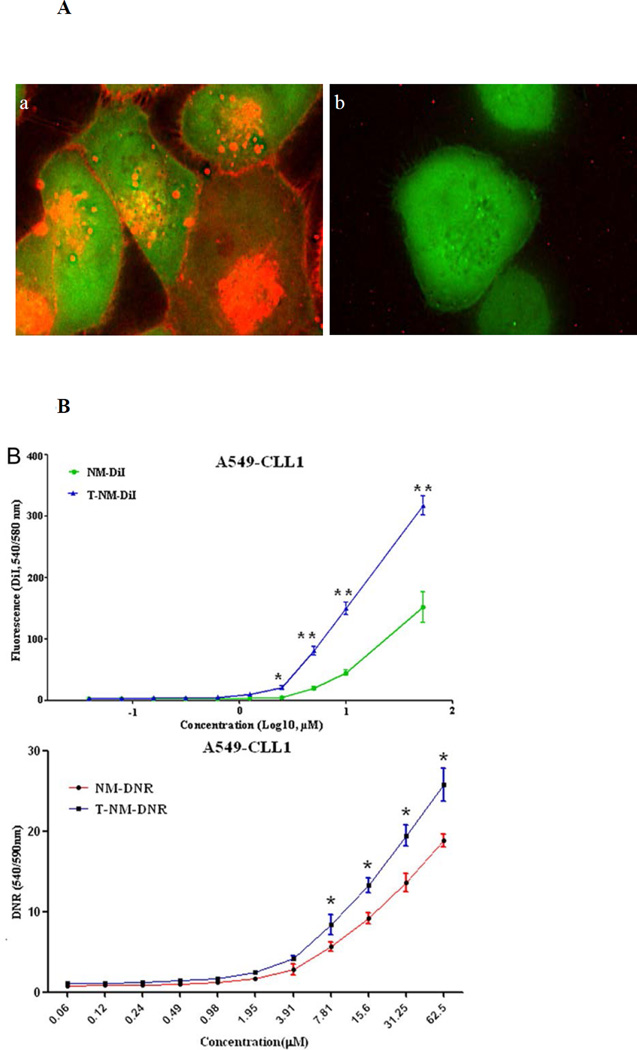
Next, we determined if CLL1-L1 could enable the drug-loaded nanomicelles to be taken up into cells expressing CLL1 and deliver the cytotoxic cargo across the cell membrane. This is a critical requirement in order for targeting nanomicelles to overcome the relatively high drug resistance of LSC. Since visualizing the binding and drug delivery of nanomicelles to LSC is technically demanding in suspension cells with little cytoplasm, we chose to work with adherent A549 cells transfected with GFP-CLL1 as a model system (SI, S4). A549 cells transfected with GFP vector alone were used as the negative control. Nanomicelles were loaded with DiI, which exhibits intense red fluorescence, as a model for DNR for monitoring delivery and intracellular distribution. Within ten minutes of incubation, strong DiI fluorescence was visible in A549 cells that expressed GFP-CLL1, but not in control A549 cells expressing GFP. However, these experiments lacked sufficient resolution to differentiate whether the DiI was on the surface or inside the cells.
To determine if the DiI-loaded targeting nanomicelles could penetrate into cells, high-resolution 3-dimensional microscopy was performed with the DeltaVision system. The images were then deconvolved using a known optical transfer function and DeltaVision software algorithms per manufacturer’s protocol. To mimic the in vivo metabolism and clearance of nanomicelles, cells were incubated with the DiI-loaded targeting nanomicelles for 15 minutes and washed with PBS to remove any unbound nanomicelles. Red fluorescence (nanomicelle loaded with DiI) was not only localized on the cell membrane, but, strikingly, the DiI was transported to the cell interior, including nucleus, of the cells expressing GFP-CLL1 (Figure 3A, panel a), while no significant red fluorescence was observed in the control cells transfected with GFP vector only (Figure 3A, panel b). To quantify the DiI delivery, the same number of A549 cells, expressing GFP or CLL1-GFP, were treated with the same concentration of DiI-loaded nanomicelles, washed and lysed, and DiI was measured with the excitation wavelength of 549 nm and emission wavelength of 565 nm. There was a dose-dependent and statistically significant difference in DiI delivery between the targeting and non-targeting nanomicelles (Figure 3B, B, upper panel, nanomicelles loaded with DiI; lower panel, nanomicelles loaded with DNR). These data suggest that drugs loaded into targeting nanomicelles can be delivered preferentially into cells that express CLL1.
Figure 3.
Penetration of targeting nanomicelles to A549 cells expressing CLL1. (A) Tomography analysis to determine intracellular distribution of targeting nanomicelles. A549 cells expressing GFP-CLL1 (panel a) or GFP (panel b) were incubated with targeting nanomicelles loaded with DiI for 15 minutes before washing. (B) Preferential delivery of targeting nanomicelles to A549 cells expressing CLL1. A549 cells expressing CLL1 were incubated with nontargeting (NM) or targeting nanomicelles (T-NM) loaded with DiI (upper panel) or DNR (lower panel) for 30 minutes before washing. Because of nonspecific uptake of nanomicelles by cells, there was no plateau of fluorescence intensity as the nanomicelle concentration increased. *P < 0.05; **P < 0.001.
Targeting nanomicelles coated with CLL1-L1 could target LSC from clinical specimens
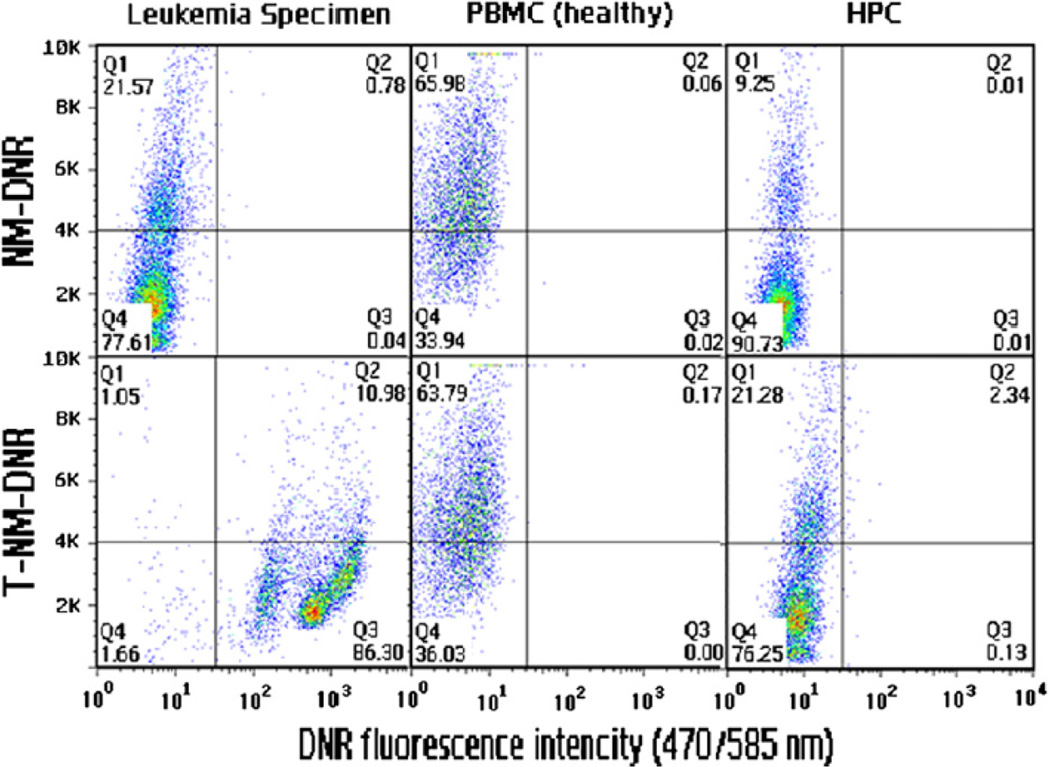
Next, we determined if targeting nanomicelles decorated with CLL1-L1 could target clinical leukemia specimens from AML patients. First, we compared the drug delivery between leukemic cells, PBMC and normal hematopoietic stem cells. Significant DNR delivery was observed for leukemic cells while little delivery was observed with PBMC and normal hematopoietic stem cells (Figure 4).
Figure 4.
Targeting leukemic cells with nanomicelles decorated with CLL1-targeting peptides. PMBCs, normal hematopoietic stem cells (HPC), and leukemia cells were incubated with nontargeting (NM, upper panels) or targeting (T-NM, lower panels) nanomicelles loaded with DNR for 30 minutes, and analyzed with flow cytometry after washing. x-axis, DNR fluorescence intensity; y-axis, side gate for cell size.
Next, we determined if there existed a dose-dependent DiI delivery difference between non-targeting and targeting nanomicelles to CD34+ leukemic cells (SI, S5). We first isolated CD34+ leukemic cells using affinity separation columns (Mitenyi, Bergisch Gladbach, Germany). After incubation with non-targeting or targeting nanomicelles at different concentration for 30 minutes, cells were washed with PBS and analyzed with flow cytometry. There was a dose-dependent difference in drug delivery of DiI between these two types of nanomicelles (SI S5, Figure A, nanomicelles loaded with DNR; Figure B, nanomicelles loaded with DiI). We tested leukemia specimens from four patients. The targeting nanomicelles could target three out of the four specimens, which is consistent with a previous report that not all LSCs express CLL1 [5].
Discussion
To our knowledge, this is the first report demonstrating the screening and synthesis of a peptide that binds with high affinity to cells expressing CLL1 and with low nonspecific binding to other cell types. The CLL1 molecule is expressed on LSC, but not on normal hematopoietic stem cells, which makes it an attractive cell surface target for LSC-seeking nanotherapeutics. We also showed that nanomicelles decorated with CLL1-targeting peptides bind to cells expressing CLL1, but more importantly, deliver the drug load directly into the target cells.
The targeting nanomicelles developed in this project can potentially improve the treatment outcomes of AML through the following three mechanisms: (1) targeting LSC through direct drug delivery into the interior of LSC; (2) killing of leukemia cells throughout the body with chemotherapeutic drugs released from nanomicelles into blood circulation; and (3) formulation of chemotherapeutic drugs inside the nanoparticles allowing administration of high-dose chemotherapy without increasing the toxicity.
One major concern in targeting LSC is whether the targeting nanomicelles can deliver sufficient drug concentration to LSC to overcome the chemoresistance and improve the treatment outcomes of AML. Based on our calculation and available data described below, drug concentration should be sufficient to overcome the resistance. It has been shown that high-dose chemotherapy itself can improve treatment outcomes as demonstrated in the Eastern Cooperative Oncology Group (ECOG)1900 trial (DNR at 90 mg/m2 versus the standard dose of 45 mg/m2) [13–15]. Higher doses (135 mg/m2) of DNR in the liposomal formulation together with cytarabine could be delivered and induce complete remission in refractory or recurrent AML [16]. Paclitaxel could be loaded in our nanomicelle platform and administrated three times the free parental drug that significantly improved the treatment outcomes without increasing the toxicity [7]. This suggests that the nanomicelle itself can potentially improve the treatment outcomes of AML just by re-formulating daunorubicin in nanomicelles. The nanomicelles are covered with PEG rendering them “stealthy” with low non-specific uptake. Furthermore, we have recently demonstrated that by incorporating slightly negative charge residues to the surface of the nanoparticles, we can further lower the nonspecific particle uptake by normal organs such as liver and lung that may decrease toxicity (data not shown). This decreased toxicity itself will improve treatment outcomes since the 4-week mortality rate during the induction chemotherapy ranges from 5% to 57% [17].
Our data support the feasibility of using targeting nanomicelles to deliver DNR or other drugs into LSCs as a means to overcome resistance mediated by cancer stem cells. When nanomicelles were decorated with CLL1-L1, consistently high concentrations of DNR were observed in cells expressing CLL1-L1 and in clinical leukemia specimens (Figures 3 and 4). Furthermore, the drug load was observed at the nucleus (Figure 3, B), suggesting the targeting nanomicelles were able to deliver the alkylating agent DNR proximally to its DNA target. In addition, the nanomicelle platform can allow us to simultaneously load different drugs into the same nanomicelles. For example, DNR can be loaded along with drugs targeting apoptosis, or combined with small interfering RNA to target antiapoptosis or other drug-resistant mechanisms, making them more efficacious. However, the in vivo antileukemia efficacy and toxicity of DNR-loaded targeting nanomicelles must be determined by clinical trials.
Targeting and killing LSC alone may not be sufficient in the treatment of AML. LSC account for a small population of all leukemia cells. Even if all LSC are eliminated, the remaining leukemia cells and their progenitor cells can continue to cause symptoms. Furthermore, the progenitor cells may still have limited proliferation capacity and generate more leukemia cells, or may even de-differentiate back into LSC. This is supported by a clinical trial demonstrating that the anti-CD20 antibody rituximab as a single agent is not effective in multiple myeloma, even though myeloma stem cells express the CD20 cell surface molecules [18,19]. The advantage of our targeting nanomicelles is that not only are LSC targeted, but the loaded drugs will be released into systemic circulation and kill leukemia cells throughout the body.
So far, no molecule has been identified that is specific to cancer stem cells. CLL1 and CD123 are two molecules expressed on LSC, but not on normal hematopoietic stem cells. However, they are also expressed on some normal hematopoietic cells. Anti-CD123 therapy, either with neutralizing antibodies or immunotoxins, preferentially kills AML LSC with little effect on normal stem cells, and decreases the effectiveness of AML engraftment in NOD-SCID mice [20–22]. Similar anti-leukemia activity was also observed with an anti-CLL1 antibody [23]. Nevertheless, the long half-life of antibody (weeks to months) will kill the newly regenerated hematopoietic cells and may affect the hematological recovery because both CD123 and CLL1 are also expressed on normal hematopoietic cells. The advantage of our targeting nanomicelles over antibody-based therapy is that its in vivo half-life of less than 24 hours is relatively short (data not shown). Our targeting nanomicelles are expected to kill some normal hematopoietic cells as any other chemotherapeutic agents do, but he short half-life means that the subsequently newly regenerated hematopoietic cells will not be affected.
In conclusion, we have identified several peptides that target CLL1, a molecule that is expressed on LSC surface, but not on normal hematopoietic stem cells. Nanomicelles decorated with CLL1-targeting peptides allow delivery of the small molecule content directly into LSC.
Supplementary Material
References
- 1.Pan CX, Zhu W, Cheng L. Implications of cancer stem cells in the treatment of cancer. Future Oncol. 2006;2:723–731. doi: 10.2217/14796694.2.6.723. [DOI] [PubMed] [Google Scholar]
- 2.Misaghian N, Ligresti G, Steelman LS, Bertrand FE, Basecke J, Libra M, Nicoletti F, Stivala F, Milella M, Tafuri A, Cervello M, Martelli AM, McCubrey JA. Targeting the leukemic stem cell: the Holy Grail of leukemia therapy. Leukemia. 2009;23:25–42. doi: 10.1038/leu.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terpstra W, Ploemacher RE, Prins A, van Lom K, Pouwels K, Wognum AW, Wagemaker G, Lowenberg B, Wielenga JJ. Fluorouracil selectively spares acute myeloid leukemia cells with long-term growth abilities in immunodeficient mice and in culture. Blood. 1996;88:1944–1950. [PubMed] [Google Scholar]
- 4.Copland M, Hamilton A, Elrick LJ, Baird JW, Allan EK, Jordanides N, Barow M, Mountford JC, Holyoake TL. Dasatinib (BMS-354825) targets an earlier progenitor population than imatinib in primary CML but does not eliminate the quiescent fraction. Blood. 2006;107:4532–4539. doi: 10.1182/blood-2005-07-2947. [DOI] [PubMed] [Google Scholar]
- 5.Bakker AB, van den Oudenrijn S, Bakker AQ, Feller N, van Meijer M, Bia JA, Jongeneelen MA, Visser TJ, Bijl N, Geuijen CA, Marissen WE, Radosevic K, Throsby M, Schuurhuis GJ, Ossenkoppele GJ, de Kruif J, Goudsmit J, Kruisbeek AM. C-type lectin-like molecule-1: a novel myeloid cell surface marker associated with acute myeloid leukemia. Cancer Res. 2004;64:8443–8450. doi: 10.1158/0008-5472.CAN-04-1659. [DOI] [PubMed] [Google Scholar]
- 6.van Rhenen A, van Dongen GA, Kelder A, Rombouts EJ, Feller N, Moshaver B, Stigter-van Walsum M, Zweegman S, Ossenkoppele GJ, Jan Schuurhuis G. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007;110:2659–2666. doi: 10.1182/blood-2007-03-083048. [DOI] [PubMed] [Google Scholar]
- 7.Xiao K, Luo J, Fowler WL, Li Y, Lee JS, Xing L, Cheng RH, Wang L, Lam KS. A self-assembling nanoparticle for paclitaxel delivery in ovarian cancer. Biomaterials. 2009;30:6006–6016. doi: 10.1016/j.biomaterials.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 9.Lam KS, Lebl M, Krchnak V. The "One-Bead-One-Compound" Combinatorial Library Method. Chem Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Aina OH, Lam KS, de Vere White R, Evans C, Henderson P, Lara PN, Wang X, Bassuk JA, Pan CX. Identification of a bladder cancer-specific ligand using a combinatorial chemistry approach. Urol Oncol. 2011 doi: 10.1016/j.urolonc.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 12.Kaiser E, Colescott RL, Bossinger CD, Cook PI. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal Biochem. 1970;34:595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]
- 13.Information NaPR. High Dose Chemotherapy Significantly Prolongs Survival for Patients with Acute Myeloid Leukemia. Cancer News on the Net®. 2008 [Google Scholar]
- 14.Fernandez HF, Sun Z, Yao X, Litzow MR, Luger SM, Paietta EM, Racevskis J, Dewald GW, Ketterling RP, Bennett JM, Rowe JM, Lazarus HM, Tallman MS. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361:1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, Sonneveld P, Maertens J, Jongen-Lavrencic M, von Lilienfeld-Toal M, Biemond BJ, Vellenga E, van Marwijk Kooy M, Verdonck LF, Beck J, Dohner H, Gratwohl A, Pabst T, Verhoef G. High-dose daunorubicin in older patients with acute myeloid leukemia. N Engl J Med. 2009;361:1235–1248. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- 16.Cortes J, Estey E, O'Brien S, Giles F, Shen Y, Koller C, Beran M, Thomas D, Keating M, Kantarjian H. High-dose liposomal daunorubicin and high-dose cytarabine combination in patients with refractory or relapsed acute myelogenous leukemia. Cancer. 2001;92:7–14. doi: 10.1002/1097-0142(20010701)92:1<7::aid-cncr1285>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 17.Estey EH. Therapeutic options for acute myelogenous leukemia. Cancer. 2001;92:1059–1073. doi: 10.1002/1097-0142(20010901)92:5<1059::aid-cncr1421>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 18.Zojer N, Kirchbacher K, Vesely M, Hubl W, Ludwig H. Rituximab treatment provides no clinical benefit in patients with pretreated advanced multiple myeloma. Leuk Lymphoma. 2006;47:1103–1109. doi: 10.1080/10428190600564803. [DOI] [PubMed] [Google Scholar]
- 19.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, Smith BD, Civin CI, Jones RJ. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feuring-Buske M, Frankel AE, Alexander RL, Gerhard B, Hogge DE. A diphtheria toxin-interleukin 3 fusion protein is cytotoxic to primitive acute myeloid leukemia progenitors but spares normal progenitors. Cancer Res. 2002;62:1730–1736. [PubMed] [Google Scholar]
- 21.Hogge DE, Yalcintepe L, Wong SH, Gerhard B, Frankel AE. Variant diphtheria toxin-interleukin-3 fusion proteins with increased receptor affinity have enhanced cytotoxicity against acute myeloid leukemia progenitors. Clin Cancer Res. 2006;12:1284–1291. doi: 10.1158/1078-0432.CCR-05-2070. [DOI] [PubMed] [Google Scholar]
- 22.Jin L, Lee EM, Ramshaw HS, Busfield SJ, Peoppl AG, Wilkinson L, Guthridge MA, Thomas D, Barry EF, Boyd A, Gearing DP, Vairo G, Lopez AF, Dick JE, Lock RB. Monoclonal antibody-mediated targeting of CD123, IL-3 receptor alpha chain, eliminates human acute myeloid leukemic stem cells. Cell Stem Cell. 2009;5:31–42. doi: 10.1016/j.stem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Singh S, Pardoux C, Zhao J, Hsi ED, Abo A, Korver W. Targeting C-type lectin-like molecule-1 for antibody-mediated immunotherapy in acute myeloid leukemia. Haematologica. 2010;95:71–78. doi: 10.3324/haematol.2009.009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.