Abstract
Cryptococcus neoformans is a fungus that causes the majority of fatal cryptococcal meningitis cases worldwide. This pathogen is capable of assuming different morphotypes: yeast, pseudohypha, and hypha. The yeast form is the most common cell type observed clinically. The hyphal and pseudohyphal forms are rarely observed in the clinical setting and are considered attenuated in virulence. However, as a ubiquitous environmental pathogen, Cryptococcus interacts with various organisms, and it is known to be parasitic to different hosts. Capitalizing on recent discoveries, morphogenesis regulators were manipulated to examine the impact of cell shape on the cryptococcal interaction with three different host systems: the soil amoeba Acanthamoeba castellanii (a protist), the greater wax moth Galleria mellonella (an insect), and the murine macrophage cell line J774A.1 (mammalian cells). The regulation of Ace2 and morphogenesis (RAM) pathway is a highly conserved pathway among eukaryotes that regulates cytokinesis. Disruption of any of five RAM components in Cryptococcus renders cells constitutively in the pseudohyphal form. The transcription factor Znf2 is the master activator of the yeast to hyphal transition. Deletion of ZNF2 locks cells in the yeast form, while overexpression of this regulator drives hyphal growth. Genetic epistasis analyses indicate that the RAM and the Znf2 pathways regulate distinct aspects of cryptococcal morphogenesis and independently of each other. These investigations using the Cryptococcus RAM and ZNF2 mutants indicate that cell shape, cell size, and likely cell surface properties weigh differently on the outcome of cryptococcal interactions with different hosts. Thus, certain traits evolved in Cryptococcus that are beneficial within one host might be detrimental when a different host is encountered.
Keywords: morphogenesis, amoeba, macrophage, greater wax moth, phagocytosis, hyphae, pseudohyphae, mating
Introduction
Morphological changes are fundamental to the ability of fungi to cause disease in plants and animals. For medically relevant species, it is evident that disrupting the ability to change cell shape results in loss of pathogenicity, illustrated by work in dimorphic species in which strains locked into one cell form have impaired ability to cause disease [1,2]. However, in many fungi it is still not clear what advantages some alternative cell morphologies provide or if they come at a fitness cost to the organism.
Cryptococcus neoformans is the major etiological agent of cryptococcal meningitis, a fatal disease that has been estimated to kill more than 600,000 people worldwide each year [3]. However, as a ubiquitous environmental pathogen, the encounter of this fungus with a susceptible mammalian host is accidental. Therefore, many virulence traits in this pathogen are postulated to be selected through its interaction with the environment or its natural predators. These virulence traits include encapsulation, melanization, and thermotolerance [4]. These traits are beneficial to this pathogen under natural conditions and also during infection in a mammalian host and are thus considered “dual use” factors [5]. Besides the aforementioned traits, cell morphotype also plays an important role in cryptococcal virulence (see reviews [6,7] and references therein), similar to what has been demonstrated in many other environmental fungal pathogens [8]. However, most of the information on the impact of cryptococcal morphotype on its interaction with different hosts was obtained with natural isolates [9]. The difference in the genotype of isolates used as well as the underlying mechanisms responsible for the morphological changes complicates the interpretation of these studies. Critical genetic components controlling cryptococcal morphotypes have been identified in the past few years (as detailed below), making it now possible to conduct comparative analyses using cryptococcal strains in the same genetic background, carrying different mutations, and with different morphological forms.
Three morphotypes can occur in C. neoformans: yeast, pseudohypha, or hypha. Enlarged yeast cells have also been characterized [10-12]. The fungus typically grows as a haploid yeast. The morphological transition from the yeast form to the hyphal form takes place during both bisexual mating (a-α mating) and unisexual mating (mostly α-α mating) [13-19]. Non-mating stimuli can also trigger hyphal growth [20-22]. Regardless of the upstream stimuli, the zinc-finger transcription factor Znf2 is a key regulator for hyphal growth (Fig. 1A). Deletion of the ZNF2 gene restricts Cryptococcus in the yeast form [20,23,24], whereas elevated expression of ZNF2 drives hyphal growth irrespective of environment stimuli or mating type [20,24,25]. Consistent with earlier observations of a reversed relationship between filamentation and virulence (see review [6] and references therein), the znf2Δ mutant in H99 background that is locked in the yeast form is more virulent than the wild-type, while the strain with overexpression of the ZNF2 gene is incapable of causing fatal infection in mice [23,26].
Figure 1.
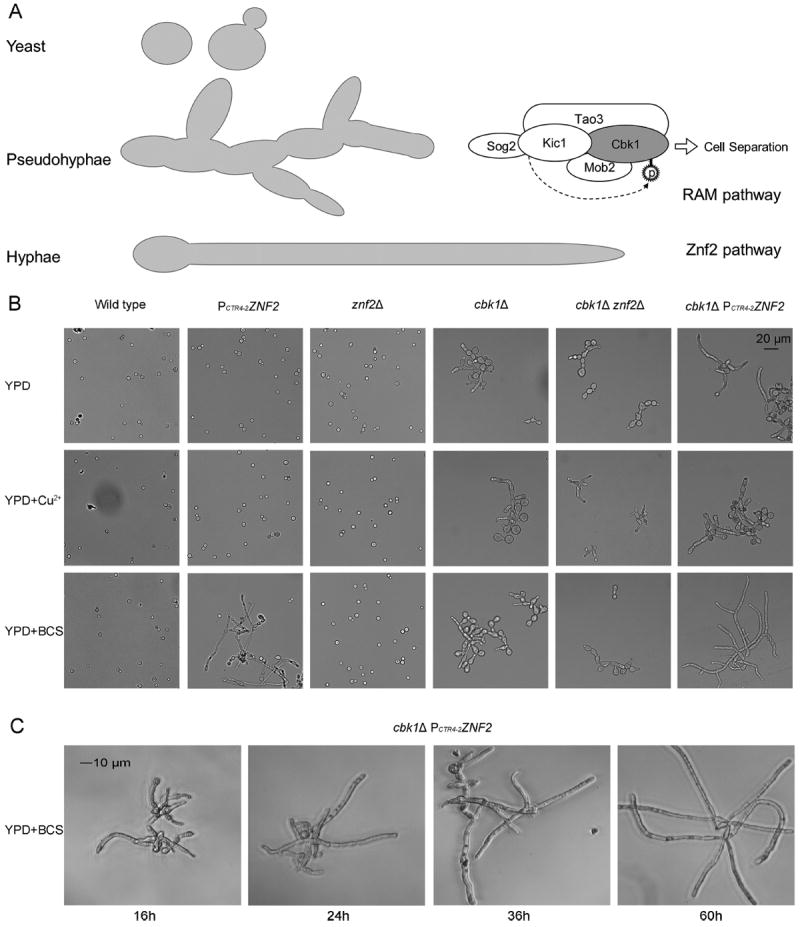
The Znf2 and the RAM pathways control distinct aspects of morphogenesis. (A) Diagram of the three morphotypes of Cryptococcus and the Znf2 and RAM pathways. The five RAM pathway components, Cbk1, Kic1, Mob2, Tao3, and Sog2, are depicted, with Cbk1 being the ultimate kinase of this complex [28]. Znf2 is a master activator of filamentation. (B) The cellular morphology of the wild-type H99 strain, the PCTR4-2ZNF2 strain, the znf2Δ mutant, the cbk1Δ mutant, the cbk1Δ znf2Δ double mutant, and the cbk1Δ PCTR4-2ZNF2 strain was examined from cultures in YPD, YPD+copper, and YPD+BCS media. (C) The cellular morphology of the cbk1Δ PCTR4-2ZNF2 strain was examined microscopically after the induction of ZNF2 expression for the indicated time periods.
Cryptococcus can assume the pseudohyphal growth form in response to nutrient limitation [6,27]. Disruption of the RAM pathway, however, renders this fungus in the pseudohyphal form constitutively [28]. The RAM pathway is a conserved signal transduction network among eukaryotes [29], and it has been shown to impact numerous cellular processes, such as in the best-studied species Saccharomyces cerevisiae to include cell cycle regulation [30,31], cell separation [32-35], mating [32,34], and cell polarization [33,35]. Similar to the RAM pathway in S. cerevisiae, the RAM pathway in C. neoformans is composed of two serine/threonine protein kinases, Cbk1 and Kic1, and three associated proteins, Mob2, Tao3, and Sog2 (Fig. 1A) [28,36]. Tao3 is likely the scaffold protein in the RAM pathway [28,32,33,36], and Mob2 functions as a Cbk1- regulatory subunit [37]. Hym1, a RAM pathway component that physically associates with Kic1 in S. cerevisiae, has not been identified as a RAM component in C. neoformans [28]. The downstream targets of the RAM pathway (i.e., of the Cbk1 kinase) in Cryptococcus remain elusive. There is no cryptococcal homolog of Ace2, one downstream target of the RAM pathway found in S. cerevisiae and S. pombe [28]. Cryptococcal mutants in the RAM pathway (tao3, mob2, kic1, cbk1, and sog2) all have the same pseudohyphal phenotype [28]. In some strain backgrounds they are temperature-sensitive [28,36]. When both mating partners harbor the same mutation, they failed to produce mating filaments or meiotic basidiospores. Mutations in the RAM pathway also drastically attenuate cryptococcal virulence [36].
Thus, both hyphae and pseudohyphae are morphological forms that are attenuated for virulence during Cryptococcus infection in a mammalian host (mouse). This reduction in virulence could be due to the inherent property of these morphological forms where the physical interconnections between cells prevent the fungus from extrapulmonary dissemination and consequently from causing fatal brain infections. Indeed, neither the ZNF2oe strain nor a RAM mutant in the H99 background could be detected in the brain tissue, although both were present in the lungs during infection [36,38]. Given that Cryptococcus likely disseminates to the brain at least in part through a Trojan horse mechanism after being phagocytosed by host cells [39-41], it would be interesting to test if morphotype affects phagocytosis of cryptococcal cells. The pseudohyphal form was isolated as being resistant to phagocytosis by soil amoebae in the 1970s [9]. These natural pseudohyphal isolates were later found to habor mutations in the RAM pathway [28]. It is not known if the hyphal form of Cryptococcus is also resistant to the phagocytosis by soil amoebae and if common factors affect phagocytosis of the two filamentous forms of Cryptococcus by amoebae, wax moth, and mammalian phagocytes.
In this study, we tested if the RAM pathway and the Znf2 pathway regulate independent or common aspects of cryptococcal morphogenesis based on genetic epistasis analyses. To investigate the impact of morphotype on cryptococcal interaction with different hosts, we used the RAM and ZNF2 mutants (single and double mutants) that grow in different morphological forms and tested three different host systems: the soil amoeba Acanthamoeba castellanii (a protist), the wax moth Galleria mellonella (an insect), and the murine macrophage cell line J774A.1 (mammalian cells). Our results indicate that cell shape (polarized form vs yeast form) weigh differently on the outcome of the interaction between Cryptococcus and different hosts. Thus, certain traits evolved in Cryptococcus that are beneficial against one predator might be detrimental when a different host is encountered, and the effect of morphotype is host- and condition-specific.
Methods and materials
Strains, growth conditions, and morphological examination
Strains used in this study are listed in Table S1. The recombinant progeny between RAM pathway gene mutations and ZNF2 alleles were isolated from crosses, and their mating types and genotypes were confirmed by phenotypic assays and polymerase chain reaction (PCR). All C. neoformans cells were maintained on yeast peptone dextrose (YPD) medium or YPD + Cu2+ (20 μM) for the PCTR4–2-ZNF2 strain at 30°C, unless stated otherwise. For crosses to test for amoebic predation, a and α mating pairs with equal number of cells (five microliters each at the cell concentration of 1.5 × 107 cells/ml) were cultured together on V8 agar medium in the dark at 22°C. Hallmarks of successful mating, including the formation of mating hyphae and spores, were examined using an Olympus SZX 16 stereoscope.
For morphological examination, the wild-type and the mutant strains were inoculated into liquid media of YPD, YPD+copper (20 μM), or YPD+ bathocuproinedisulfonic acid (BCS; 200 μM) with the initial OD600 being 0.1. After 60 hours of standing incubation, cellular morphology of these strains was examined under an Olympus CX41 light microscope. Images of these cells were captured using the QCapture software.
Murine macrophage phagocytosis assay
Mouse macrophage cell line J774A.1 (ATCC® TIB-67™) was acquired from the American Type Culture Collection, along with the ATCC-formulated Dulbecco’s Modified Eagle’s Medium (DMEM, catalog no. 30-2002). Fetal bovine serum (FBS) was added into DMEM to a final concentration of 10% immediately prior to the incubation. Three hundred microliters of freshly grown J774A.1 cells were seeded into 24 well microtiter plates, with 2.5 × 105 cells per well. The macrophage cells were cultured at 37°C with 5% CO2 overnight. Old culture media were then replaced by refresh DMEM with 10% FBS. Cryptococcus cells of 2.5 × 106 or more were introduced to each well for the phagocytosis assay. After 30 s of rock mixing, the co-cultures were incubated at 37°C with 5% CO2 for an additional 2 hours. The co-cultures were then washed three times with warm phosphate buffered saline (PBS; 500 μl/well/time) to remove medium and non-adherent cells before fixed with 300 μl 10% formaldehyde made in PBS.
For the prestaining approach, Cryptococcus cells were stained with calcofluor white (20 μg/mL) for 5 minutes before the infection. For the post-stain approach, calcofluor white was used to stain the cells after phagocytosis, and then cells were fixed. The rest of the experiments were performed the same way as described in the previous paragraph. The cells were examined under an inverted microscope (Eclipse Ti, Nikon), and the images were captured using the NIS elements AR 3.0 software.
Amoebic predation assays
Acanthamoeba castellanii (ATCC® 30234™) purchased from ATCC was stored and grown in the PYG medium (ATCC® media 712) as described on the product sheet supplied by ATCC. In brief, 0.25 ml peak density (about 2 weeks growth) culture was transferred into 5 ml fresh PYG liquid media and incubated at 22°C with a 15° horizontal slant.
For the predation assay, the agar plates were cultured for 60 hours with a lawn of cryptococcal cells (original inoculum per plate: 250 μl of a culture with OD600 = 0.5). A 5–10 μl aliquot of a week old amoebic culture (OD600 ≈ 0.8) was dropped onto the center of the cryptococcal lawn. The diameter of the predation halo on each plate was recorded daily. Each strain had three replicates. The amoebic predation zone was defined as Dt–D0 (the diameter on the day of recording subtract the initial diameter). Because the predation zone was not in a perfectly round shape, the diameter of the predation zone was defined as the average of diameters measured with 8 different directions (see Supplemental Fig. 1 for the diagram). The amoebic predation zone was then plotted against the time using the program OriginPro 8.6.
For the amoebic predation assay on mating colonies, the mating pair was first cultured together for a week to allow the mating hyphae to be generated before dropping an aliquot of the amoeba culture on top of the mating colony. Instead of measuring the predation diameter, the amoebic cell number recovered from each cryptococcal mating colony after 2–3 weeks of co-incubation was counted using a hemocytometer.
Galleria mellonella infection
Galleria mellonella at the final instar larval stage were purchased from Vanderhorst Wholesale Inc. The greater wax moth infection experiments were performed as described previously [42]. C. neoformans strains were grown overnight in YPD+BCS (200 μM) liquid medium. Cells were washed three times with PBS and then resuspended in PBS buffer to the final concentration of OD600 = 1. Using a 10μl Hamilton syringe, 5 microliters of Cryptococcus cells (~5 × 104 yeast cells) were injected into G. mellonella hemocoel through the last left proleg, and the control group was injected with 5 microliters of sterile PBS. Before injection, the area was cleaned using an alcohol swab. After 4 hours of incubation at 30°C, the haemolymph of infected larvae (~5 μl) was collected into 45 μl anti-agglutination buffer via gentle squeezing of a cut at the proleg as described previously [43]. The haemolymph material containing haemocytes and fungal cells were then examined microscopically.
Results
The RAM pathway and the Znf2 pathway control distinct aspects of morphogenesis
To investigate the relationship between the RAM pathway and the Znf2 pathway, we crossed the znf2Δ mutant or the conditional ZNF2 overexpression strain (PCTR4-2-ZNF2) with the RAM mutants (tao3Δ, mob2Δ, kic1Δ, cbk1Δ, and sog2Δ). In the PCTR4-2-ZNF2 strain, the expression of ZNF2 is driven by the promoter of the CTR4 gene that encodes a copper transporter [44]. The expression of ZNF2 is thus suppressed by copper and induced by copper starvation in the presence of copper chelator bathocuproinedisulfonic acid (BCS) [20,25]. The isolated double mutants were examined for their morphology and other phenotypes. We found that mutations in the different RAM components in these double mutants conferred the same phenotypes (data not shown), consistent with their established role in one complex [28,36]. Thus, for the following experiments, we chose to use strains with the deletion in the gene encoding the kinase Cbk1 to represent the RAM complex (Fig. 1A).
To compare the morphology of the single and double mutants, we cultured the strains in YPD liquid medium with or without supplementation of copper or BCS. As expected, the wild-type H99 strain and the znf2Δ mutant showed the yeast morphotype under these conditions (Fig. 1B). The cbk1Δ mutant grew in the pseudohyphal form (Fig. 1B). The PCTR4-2-ZNF2 strain grew strictly in the yeast form in the YPD+copper medium, but it became filamentous in the YPD+BCS medium when the expression of ZNF2 was induced (Fig. 1B). Although the majority of the PCTR4-2-ZNF2 cells were in the yeast form in the YPD medium, a few cells became filamentous during prolonged incubation, likely due to the increasingly limited copper level in the medium. It is known that the YPD medium is copper-limiting; for example, growth of the cryptococcal strain defective in the copper regulator Mac1/Cuf1 is poor in this medium [19]. The double mutant cbk1Δ znf2Δ had the same pseudohyphal morphotype as the single cbk1Δ mutant. Interestingly, the cbk1Δ PCTR4-2-ZNF2 strain grew in the hyphal form when the expression of ZNF2 was induced (YPD+BCS). The hyphal production caused by the induction of ZNF2 in the cbk1Δ mutant background was more robust than the expression of ZNF2 alone in the wild-type background (Fig. 1B). We speculate that driving hyphal growth from pseudohyphae might be a more efficient process than switching to hyphal growth from yeast growth. The length of the hyphae increased over time in this cbk1Δ PCTR4-2-ZNF2 strain under the inducing condition (Fig. 1C). In addition, these results show that the mating defect of the RAM mutants is not due to an impediment in their capabilities for filamentous development. These observations also indicate that the mutations in znf2 and cbk1 have somewhat complicated genetic interactions: ZNF2oe is epistatic over cbk1Δ in hyphal formation, while cbk1Δ is epistatic over znf2Δ for pseudohyphal formation. However, they show that the two pathways control distinct morphotype generation and that they act independently based on the phenotypes of the double mutants.
Murine macrophage cells can phagocytose yeasts and hyphae but not pseudohyphae
Although the ZNF2oe strain with constitutive activation of ZNF2 is avirulent in the murine model of cryptococcosis, that strain could persist and amplify in the animal lungs during the first two weeks of infection [23]. A RAM mutant is also significantly attenuated in mouse model, but it can persist in the lungs even after 70 days post infection [36]. This is somewhat surprising given that the RAM mutants used in that experiment grow less well at 37°C in vitro [28,36]. Interestingly, neither the ZNF2oe strain nor the RAM mutant causes infections in the brain, despite their presence in the lungs [23,36]. As a Trojan horse mechanism is important for this facultative intracellular pathogen to spread systemically and to invade the brain [39-41], we hypothesized that the filamentous morphotype (pseudohyphae or hyphae) might be blocked in dissemination from the lungs due to lack of phagocytosis.
To examine the impact of morphotype on the phagocytosis of Cryptococcus by murine macrophages, we infected the J774A.1 cell line with Cryptococcus cells including the wild-type H99 (yeast), the cbk1Δ mutant (pseudohyphae), and the PCTR4-2-ZNF2 strain precultured under the inducing condition (hyphae). After 2 hours of co-incubation, cells were washed with PBS and fixed with formaldehyde. To distinguish extracellular from intracellular cryptococcal cells, we stained the cells with calcofluor white after phagocytosis (post-stain). Calcofluor white is a fluorochrome that binds chitin in the fungal cell wall [45] and it stains extracellular cryptococcal cells. As expected, we found both intracellular yeast cells and adherent extracellular yeast cells of the wild type (Fig. 2B), consistent with Cryptococcus being a facultative intracellular pathogen [46]. To our surprise, some hyphae produced by the PCTR4-2-ZNF2 strain were also phagocytosed (Fig. 2D), although the efficiency of hyphal phagocytosis was only half of that for yeasts (Fig. 2G). By contrast, no pseudohyphae of the cbk1Δ mutant were found either intracellularly or extracellularly (Fig. 2F). To confirm that we did not miss any hidden cryptococcal cells, we cultured Cryptococcus with calcofluor white prior to the co-culture with J774A.1 cells (pre-stain). After 2 hours of co-culturing, cells were again washed with PBS and fixed with formaldehyde. This approach allows the clear visualization of cryptococcal cells, either intracellular or extracellular. Again, both intracellular and adherent extracellular yeast cells (H99) and hyphae (PCTR4-2-ZNF2) were observed (Fig. 2A, C). Consistent with our post-stain approach, we did not find any cbk1Δ mutant pseudohyphae cells (Fig. 2E, 2F). Taken together, the results indicate that the murine macrophage can engulf yeast and hyphae, but not pseudohyphae.
Figure 2.
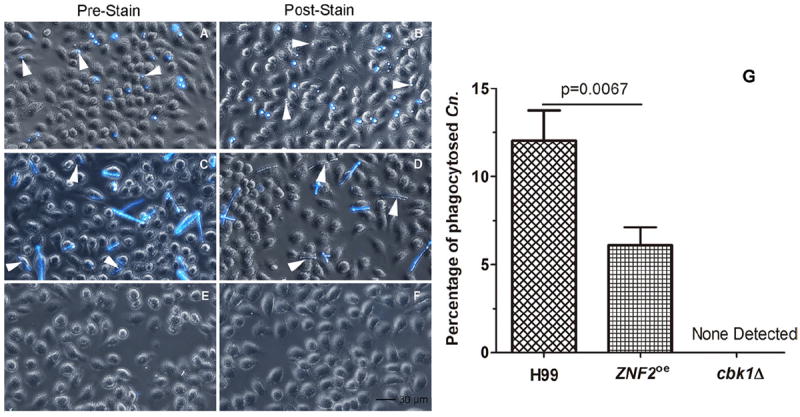
Phagocytosis of cryptococcal cells of different morphology by murine macrophage cells. Murine J774A.1 cells were infected by C. neoformans wild-type strain H99 (A, B), the PCTR4-2ZNF2 strain (C, D), and the cbk1Δ mutant (E, F) for two hours and then fixed. The left panel images were taken with cryptococcal cells stained with calcofluor white prior to the infection (A, C, E). The right panel images were taken with the co-culture stained with calcofluor white after phagocytosis (B, D, F). Statistical analysis was performed on the percentage of phagocytosed C. neoformans (Cn) cells (G). A p value lower than .05 is considered statistically significant. No cbk1Δ mutant cells were phagocytosed and thus no fluorescent signal was detected.
The hyphal and pseudohyphal, but not the yeast, forms of Cryptococcus are resistant to predation by amoebae
The soil amoeba is proposed to act as one of the selective pressures for the evolution of virulence traits in C. neoformans [36,47-49]. Prior observations indicate that the pseudohyphal RAM mutants are resistant to amoebic predation [9,28]. Given the differences that we observed for the hyphae and pseudohyphae in the murine macrophage assays, we decided to examine the impact of morphotype as controlled by Znf2 expression on cryptococcal resistance to amoebic predation.
We designed the following approach to assay amoebic predation. We first cultured a lawn of cryptococcal cells on an agar plate and then dropped the amoebic culture onto the center of the plate. The impact of the predation by amoebae is reflected by the clear zone expanded from the center (Supplemental Fig. 1A). Conditions that favored rapid amplification of Cryptococcus (YPD and PYG media) were not suitable for the detection of amoebic predation (Supplemental Fig. 1B). In comparison, amoebic predation of Cryptococcus on nutrient-limiting V8 juice agar was easily detectable (Supplemental Fig. 1B).
We performed the amoebic predation assay with the wild-type H99 strain, the cbk1Δ mutant, the znf2Δ mutant, and the PCTR4-2-ZNF2 strain on V8 medium with or without the addition of copper or BCS. Consistent with previous results [9,36], the cbk1Δ RAM mutant was highly resistant to the predation by amoebae, and no expansion of the predation zone was detected even after 3 weeks of incubation (data not shown). We found cryptococcal pseudohyphal cells and amoebic cysts at the original inoculation sites, indicating that the pseudohyphal cells can indeed survive the amoebic predation under the tested conditions (data not shown). Amoebae predated the wild-type H99 strain and the znf2Δ mutant similarly well on these media (Fig. 3A), both strains were in the yeast form under these tested conditions. Likewise, amoebae predated the PCTR4-2-ZNF2 strain as well as the wild-type H99 on V8 medium or V8+copper medium (Fig. 3A and Supplemental Fig. 1C). By contrast, the expansion of the amoebic predation zone on this PCTR4-2-ZNF2 strain on the V8+BCS medium (with ZNF2 induction) was drastically slower (Fig. 3A and Supplemental Fig. 1C). We then used microscopy to examine closely three areas on this plate: the area outside of the predation zone, the border of the predation zone, and the area inside the predation zone. The PCTR4-2-ZNF2 strain formed yeast colonies mixed with hyphae extending in all directions on V8+BCS medium (Fig. 3B). At the border and inside the predation zone, yeast cells were being dismantled by amoebae, with hyphae being the “leftovers” (Fig. 3B). Further examination of the PCTR4-2-ZNF2 strain under a compound microscope revealed that amoebae engulfed and digested the yeast cells (Fig. 3C). By contract, amoebae only wrapped around hyphae and the hyphae being attacked were still viable (Fig. 3C). Thus, the cells of the same genetic makeup displayed difference in their resistance to amoebic predation due to their morphological difference.
Figure 3.
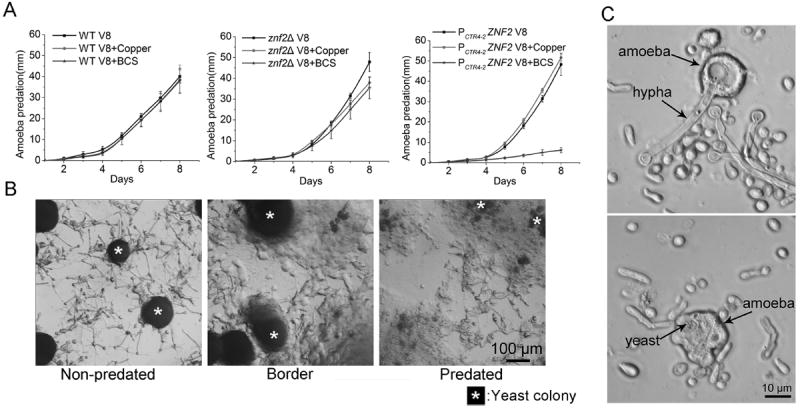
Amoebic predation of Cryptococcus. (A) The dynamics of amoebic predation of Cryptococcus wild-type strain H99, the znf2Δ mutant, and the PCTR4-2ZNF2 strain on V8 juice agar, V8+copper, and V8+BCS was recorded for 8 days. No expansion of predation zone on the cbk1Δ mutant was observed even after three weeks (not shown). (B) The area outside of the predation zone, at the front of the predation, and inside the predation zone of the PCTR4-2ZNF2 strain on V8+BCS agar medium. * indicate the yeast colonies. (C) From the predation zone of the PCTR4-2ZNF2 strain on V8+BCS agar medium, amoebae with attempts to phagocytose hyphae were observed (upper image) and amoebae with completely phagocytosed yeast cells were observed (lower image).
The aforementioned results suggest that amoebae preferentially engulf yeast cells and leave behind hyphae or pseudohyphae. It is known that spontaneous mutations in the RAM pathway genes can occur relatively frequently under natural conditions and the reversion of these mutations also occur under conditions that select against the pseudohyphal form [36,38]. Thus resorting to loss of RAM pathway signaling is a reasonable approach for Cryptococcus to resist the predation by amoebae. Cryptococcus is also known to naturally upregulate ZNF2 and undergoes morphological transition from the yeast growth to hyphal growth during mating [14,15].
The experiments described thus far used transgenic Cryptococcus strains to investigate the role of morphologywith different host organisms. Naturally formed mating hyphae of wild-type cells were then tested in amoebic predation assays. Mating colonies were formed by two pairs of wild-type strains: the serotype A (var. grubii) wild-type congenic pairs H99α and KN99a [50] and the serotype D (var. neoformans) congenic pairs XL280α and XL280a [24]. We included their corresponding znf2Δ a and α strains as a control. It is known that znf2Δ mutants are capable of mating but they are specifically blocked in the hyphal morphogenesis [32]. As shown in Figure 4, the serotype D XL280α-XL280a pair mated robustly and produced abundant hyphae that gave the white fluffy appearance to the mating colony. The serotype A H99α-KN99a pair also mated and produced spotty hyphae at the periphery of the colony. As expected, the znf2Δ mutant mating colonies remained in the yeast form (Fig. 4A). After the addition of amoebae, the yeast cells in the center of the H99α-KN99a mating colony were cleared by amoebae. The corresponding non-filamenting mating colony of the H99 znf2Δ mutants was completely destroyed. Accordingly, more amoebic cells were recovered from the znf2Δ mutant mating colony than those from the wild-type mating colony (Fig. 4B), most likely due to access to more digestible food source (yeast cells). Very minimal impact was observed on the XL280α-XL280a mating colony, where filamentation was much more robust compared to the serotype A (Fig. 4A). Again, increased predation on the XL280 znf2Δ mutant mating colony was observed compared to the wild-type control (Fig. 4A, B). Interestingly, amoebae made a much more modest impact on the XL280 background compared to the H99 background, even for the znf2Δ mutant (Fig. 4B). This suggests that other genetic factors, in addition to morphotype, also influence the efficiency of amoebic predation.
Figure 4.
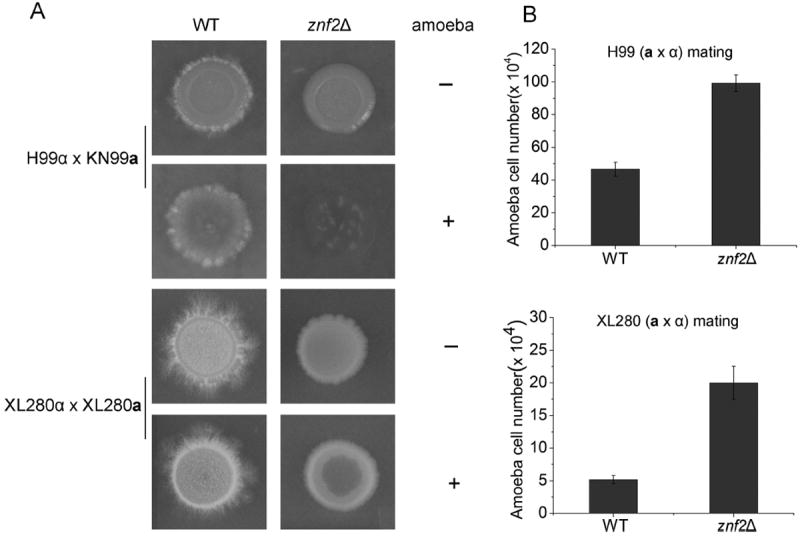
Amoebic predation of mating colonies. (A) The wild-type strain pairs (H99α × KN99a and XL280α × XL280a) and their corresponding znf2Δ mutant pairs were mated on V8 medium. The images of intact colonies and colonies with amoebae predation are shown. (B) Amoebae cells recovered from the mating colonies were quantified.
Collectively, the findings presented here indicate that hyphae and pseudohyphae are resistant to the amoeba A. castellanii while yeast cells, even genetically identical to the filamentous cells, are susceptible to this soil microbial hunter.
Hyphae and pseudohyphae elicit haemocyte nodulation during Galleria mellonella infection
Galleria mellonella (the greater wax moth) has been used as an alternative host system to assess C. neoformans virulence traits. For example, the classic factors important for cryptococcal virulence in mammalian models, like melanin and capsule synthesis, were shown to be important also in the G. mellonella model [42]. Although insects do not have adaptive immune systems as mammals do, they do have sophisticated innate immune responses, including for pathogen recognition, production of antimicrobial peptides, and the formation of a multicellular complex to encapsulate invading pathogens [51-53]. Haemocytes are the major cellular defense mediator in the G. mellonella haemolymph and they are functionally equivalent to macrophages and neutrophils in mammals [52]. Haemocyte aggregation (haemocyte nodulation) is one of the most important defense systems in insects to contain foreign invaders [54,55]. Previous studies demonstrate that the pseudohyphal RAM mutants are attenuated in virulence in the G. mellonella model [36]. Here, we decided to analyze the impact of morphotype on the cellular responses of G. mellonella against Cryptococcus.
We infected G. mellonella with the wild-typeH99 strain, the PCTR4-2-ZNF2 strain, the cbk1Δ mutant, the cbk1Δ PCTR4-2-ZNF2 strain, or the PBS control. All cryptococcal strains were precultured overnight in YPD+BCS medium to induce hypha production in strains that contain the PCTR4-2-ZNF2 construct (Fig. 1C). Haemolymph samples were collected from infected larvae four hours after the injection, and they were immediately examined microscopically for haemocyte nodulation [43,47,56]. All strains elicited Galleria cellular immune responses, as haemocyte nodulation was detected for all of them (Fig. 5). However, pseudohyphae and hyphae induced much stronger responses than yeast cells (Fig. 5). At higher magnification, we found that yeast cells (H99) were phagocytosed by haemocytes, and there was a minute level of clustering of haemocytes compared to the PBS control. By contrast, hyphae and pseudohyphae caused large clustering of haemocytes, with many layers of haemocytes wrapped around the filamentous cryptococcal cells (Fig. 5). This effective containment of hyphae and pseudohyphae by haemocyte nodulation likely contribute to this insect’s resistance to the nonyeast forms.
Figure 5.

Galleria mellonella haemocyte nodulation induced by cryptococcal infections. Larvae were infected with cryptococcal cells (wildtype H99, the znf2Δ mutant, and the PCTR4-2ZNF2 strain) precultured overnight in YPD+BCS liquid medium. Haemolymph from infected larvae was collected at 4 hours post inoculation and was immediately observed microscopically.
Discussion
The results presented in this study demonstrate the importance of morphotype in shaping the interaction between Cryptococcus and various hosts. Both hyphae and pseudohyphae confer resistance to the predation by soil amoebae (Table 1). Thus, being in a filamentous form is beneficial to Cryptococcus in defending itself against this single-celled protist. Similarly, both hyphae and pseudohyphae are resistant to phagocytosis by haemocytes during infection in the greater wax moth. In the wax moth, however, haemocyte nodulation could effectively encase these filamentous cells and disable their infection. On the other hand, yeast cells, although vulnerable for phagocytosis, can effectively kill the infected larvae [36,42]. Surprisingly, yeast and hyphae can both be phagocytosed by murine macrophages, whereas no phagocytosis was observed for pseudohyphae (Fig. 2). In this case, cell surface molecules differentially presented on hyphae versus pseudohyphae might be more critical than the physical shape. Nonetheless, strains in both filamentous forms are incapable of disseminating from the lungs inmouse models and are consequently unable to cause fatal brain infections [20,36]. Thus, being in the yeast form is clearly advantageous for cryptococcal systemic spread in a mammalian host. Hence, as a facultative intracellular pathogen, resistance to phagocytosis is only part of the equation in determining the overall level of virulence of Cryptococcus in a multicellular host.
Table 1.
Morphotype affects the interaction between C. neoformans and its hosts.
| Yeast | Pseudohyphae | Hyphae | |
|---|---|---|---|
| Acanthamoeba castellanii | Phagocytosed | Survived | Survived |
| Galleria mellonella | Phagocytosed | Contained by aggregated haemocytes | Contained by aggregated haemocytes |
| Murine macrophage cell J774A.1 | Phagocytosed | No adherence or phagocytosis | Phagocytosed |
It is reasonable to assume that these different morphotypes differ in their cell surface composition or structure (protein and complex carbohydrates). For instance, Znf2 is known to control multiple cell surface factors in Cryptococcus, including proteins for cellular adhesion [20,21,55]. The effect of mutations in the RAM pathway on cryptococcal cell surface is unknown; however, these cells were not detected as adhered to macrophages. While grown on media with cell wall perturbing agents, the ZNF2oe strain (hyphae) showed the highest sensitivity to Caspofungin and Congo red, followed by the RAM mutant (pseudohyphae) and then the wild type (yeast) (Supplemental Fig. 2A). As Caspofungin inhibits (1–3)-β-D-glucan synthase and Congo red binds to β-1,4-glucans [57,58], this result suggests that ZNF2 overexpression and RAM pathway mutantions affect the β-glucan to a different extent. Moreover, both hyphae and pseudohyphae were more sensitive to the cell wall and membrane disturbing detergent SDS than yeasts (Supplemental Fig. 2A). By constract, no gross difference in the production of chitin/chitosan or capsule between the yeast, hyphae, or pseudohyphae was detected through Calcofluor white and India ink staining (Supplemental Fig. 2B). However, these assays could not detect subtle changes in their composition or structure. Further detailed analyses of the cell wall of these morphotypes are warranted.
The physical size of the pathogen and of the phagocytes appears to be a critical factor for phagocytosis. For example, a previous study showed that haemocytes of G. mellonella are effective in phagocytosis of Aspergillus fumigatus conidia that are smaller than 3 micrometers [38]. Although we could easily detect phagocytosis of cryptococcal yeast cells of 3–5 micrometers by G. mellonella haemocytes (Fig. 5), it is relatively rare to observe phagocytosis of filaments [59]. Attempts to engulf and destroy filaments are often likely to be futile as we observed for amoebae (Fig. 3C). It is possible that G. mellonella resorts to and relies primarily on haemocyte nodulation for particles of larger sizes, like hyphae or pseudohyphae (Fig. 5). What is surprising to us is that the J774A.1 macrophage, which is of similar size as the amoebae used in this study, can efficiently engulf cryptococcal hyphae (Fig. 2). It is unclear why macrophages failed to phagocytose pseudohyphae. Clustering of cells and/or lower adherence of the RAM mutant might be responsible for this phenomenon.
Differences in genotype, in addition to the variations in morphotype, also affect the outcome of the interaction between Cryptococcus and the host. For instance, yeast cells in the XL280 background are more resistant against amoebic predation than yeast cells in the H99 background (Fig. 4). This is contrary to their virulence level in mouse models [50,60]. Moreover, some yeasts might be digested and serve as a food source for amoebae ([61] and this study), while some other cryptococcal yeasts could revert that relationship and replicate inside amoebae after being phagocytosed [47]. In a murine model, alveolar macrophages could rapidly phagocytose yeast cells after the intratracheal infection [46]. However, yeast cells of some strains can adopt different strategies to avoid phagocytosis [62,63].
Often a microbial population grown under natural condition is not homogenous, even if the population is derived from a single parental cell. These cells could be heterogenous in cellular physiology, morphology, or size. The balance of different subpopulations could be shifted in response to different environmental stimuli, maximizing the community fitness or enhancing microbial survival under disparate conditions. Pigeon guano, a natural niche of C. neoformans, provides a stimulating environment for cryptocccal mating [64]. A cryptococcal mating colony, for example, contain yeasts, pseudohyphae, germ tubes, and hyphae. A subpopulation of the hyphae could further differentiate into fruiting bodies and generate recombinant progeny that are then distributed to new locations in the environment [43]. The stochasticity in gene expression in cells of the same morphotype and the dynamics in morphotype transition are part of the bet-hedging strategy of this fungus to survive under changing and unpredictable selective pressures. Thus, although it is important to understand the impact of a pure morphotype on the cellular interaction between cryptococcus and the host, it is equally important to extrapolate such findings to the population level with caution. Similarly, the interpretation of the interaction of Cryptococcus with multicellular hosts at the cellular level should be placed in the context of the host organism as a whole.
Supplementary Material
Acknowledgments
We thank Drs. Bing Zhai and Qingming Qin for their assistance with the macrophage experiments. We gratefully acknowledge the financial support from the National Institute of Allergy and Infectious Diseases (grants R21 AI094364 to A. I., R01 AI097599 and R21 AI107138 to X. L.). A. I. is supported on an Australian Research Council Future Fellowship. X. L. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material
Supplementary material is available at Medical Mycology online (http://www.mmy.oxfordjournals.org/).
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1.Nemecek JC, Wuthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312(5773):583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 2.Holbrook ED, Rappleye CA. Histoplasma capsulatum pathogenesis: making a lifestyle switch. Curr Opin Microbiol. 2008;11(4):318–324. doi: 10.1016/j.mib.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Park BJ, Wannemuehler KA, Marston BJ, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 4.Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: ASM Press; 1998. [Google Scholar]
- 5.Casadevall A, Steenbergen JN, Nosanchuk JD. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr Opin Microbiol. 2003;6(4):332–337. doi: 10.1016/s1369-5274(03)00082-1. [DOI] [PubMed] [Google Scholar]
- 6.Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol. 2009;9(4):401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Lin X. Morphogenesis in fungal pathogenicity: shape, size, and surface. PLoS Pathogens. 2012;8(12):e1003027. doi: 10.1371/journal.ppat.1003027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sil A, Andrianopoulos A. Thermally dimorphic human fungal pathogens-polyphyletic pathogens with a convergent pathogenicity trait. Cold Spring Harb Perspect Med. 2014 doi: 10.1101/cshperspect.a019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neilson JB, Ivey MH, Bulmer GS. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Iimmun. 1978;20(1):262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaragoza O, Nielsen K. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr Op in Microbiol. 2013;16(4):409–413. doi: 10.1016/j.mib.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaragoza O, Garcia-Rodas R, Nosanchuk JD, et al. Fungal cell gigantism during mammalian infection. PLoS Pathogens. 2010;6(6):e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okagaki LH, Strain AK, Nielsen JN, et al. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathogens. 2010;6(6):e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon-Chung KJ. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67(6):1197–1200. [PubMed] [Google Scholar]
- 14.Alspaugh JA, Davidson RC, Heitman J. Morphogenesis of Cryptococcus neoformans. Contrib Microbiol. 2000;5:217–238. doi: 10.1159/000060352. [DOI] [PubMed] [Google Scholar]
- 15.Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host & Microbe. 2010;8(1):86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68(4):821–833. [PubMed] [Google Scholar]
- 17.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434(7036):1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 18.Wickes BL, Mayorga ME, Edman U, et al. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. Proc Natl Acad Sci U S A. 1996;93(14):7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin X, Huang JC, Mitchell TG, et al. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genetics. 2006;2(11):e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Zhai B, Lin X. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathogens. 2012;8(6):e1002765. doi: 10.1371/journal.ppat.1002765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Tian X, Gyawali R, et al. Fungal adhesion protein guides community behaviors and autoinduction in a paracrine manner. Proc Natl Acad Sci U S A. 2013;110(28):11571–11576. doi: 10.1073/pnas.1308173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu J, Morris IR, Wickes BL. The production of monokaryotic hyphae by Cryptococcus neoformans can be induced by high temperature arrest of the cell cycle and is independent of samesex mating. PLoS Pathogens. 2013;9(5):e1003335. doi: 10.1371/journal.ppat.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X, Jackson JC, Feretzaki M, et al. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genetics. 2010;6(5):e1000953. doi: 10.1371/journal.pgen.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai B, Zhu P, Foyle D, et al. Congenic strains of the filamentous form of Cryptococcus neoformans for studies of fungal morphogenesis and virulence. Infect Immun. 2013;81(7):2626–2637. doi: 10.1128/IAI.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Tian X, Gyawali R, et al. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathogens. 2014;10(6):e1004185. doi: 10.1371/journal.ppat.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Del Poeta M. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryotic Cell. 2004;3(5):1067–1075. doi: 10.1128/EC.3.5.1067-1075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SC, Phadke S, Sun S, Heitman J. Pseudohyphal growth of Cryptococcus neoformans is a reversible dimorphic transition in response to ammonium that requires Amt1 and Amt2 ammonium permeases. Eukaryotic Cell. 2012;11(11):1391–1398. doi: 10.1128/EC.00242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell. 2006;17(9):3768–3780. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maerz S, Seiler S. Tales of RAM and MOR: NDR kinase signaling in fungal morphogenesis. Curr Op inMicrobiol. 2010;13(6):663–671. doi: 10.1016/j.mib.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Bogomolnaya LM, Pathak R, Guo J, et al. Roles of the RAM signaling network in cell cycle progression in Saccharomyces cerevisiae. Current Genetics. 2006;49(6):384–392. doi: 10.1007/s00294-006-0069-y. [DOI] [PubMed] [Google Scholar]
- 31.Brace J, Hsu J, Weiss EL. Mitotic exit control of the Saccharomyces cerevisiae Ndr/LATS kinase Cbk1 regulates daughter cell separation after cytokinesis. Mol Cell Biol. 2011;31(4):721–735. doi: 10.1128/MCB.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson B, Kurischko C, Horecka J, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14(9):3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du LL, Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(2):503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidlingmaier S, Weiss EL, Seidel C, et al. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(7):2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Racki WJ, Becam AM, Nasr F, et al. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 2000;19(17):4524–4532. doi: 10.1093/emboj/19.17.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magditch DA, Liu TB, Xue C, et al. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans. PLoS Pathogens. 2012;8(10):e1002936. doi: 10.1371/journal.ppat.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 andMob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107(6):739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 38.Renwick J, Daly P, Reeves EP, et al. Susceptibility of larvae of Galleria mellonella to infection by Aspergillus fumigatus is dependent upon stage of conidial germination. Mycopathologia. 2006;161(6):377–384. doi: 10.1007/s11046-006-0021-1. [DOI] [PubMed] [Google Scholar]
- 39.Kechichian TB, Shea J, Del Poeta M. Depletion of alveolar macrophages decreases the dissemination of a glucosylceramidedeficient mutant of Cryptococcus neoformans in immunodeficient mice. Infect Immun. 2007;75(10):4792–4798. doi: 10.1128/IAI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlier C, Nielsen K, Daou S, et al. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77(1):120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabiiti W, Robertson E, Beale MA, et al. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Investigat. 2014;124(5):2000–2008. doi: 10.1172/JCI72950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mylonakis E, Moreno R, ElKhoury JB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Iimmun. 2005;73(7):3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upadhyay S, Torres G, Lin X. Laccases involved in 1,8-dihydroxynaphthalene melanin biosynthesis in Aspergillus fumigatus are regulated by developmental factors and copper homeostasis. Eukaryotic Cell. 2013;12(12):1641–1652. doi: 10.1128/EC.00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ory JJ, Griffith CL, Doering TL. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 2004;21(11):919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- 45.Herth W, Schnepf E. The fluorochrome, calcofluor white, binds oriented to structural polysaccharide fibrils. Protoplasma. 1980;105(1–2):129–133. [Google Scholar]
- 46.Feldmesser M, Kress Y, Novikoff P, et al. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68(7):4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A. 2001;98(26):15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon-Chung KJ, Fraser JA, Doering TL, et al. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4(7):a019760. doi: 10.1101/cshperspect.a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Casadevall A. Amoeba provide insight into the origin of virulence in pathogenic fungi. Adv Exp Med Biol. 2012;710:1–10. doi: 10.1007/978-1-4419-5638-5_1. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen K, Cox GM, Wang P, et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71(9):4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mak P, Zdybicka-Barabas A, Cytrynska M. A different repertoire of Galleria mellonella antimicrobial peptides in larvae challenged with bacteria and fungi. Dev Comp Immunol. 2010;34(10):1129–1136. doi: 10.1016/j.dci.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32(10):1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 53.Cytrynska M, Mak P, Zdybicka-Barabas A, et al. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides. 2007;28(3):533–546. doi: 10.1016/j.peptides.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32(10):1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 55.Bogus MI, Kedra E, Bania J, et al. Different defense strategies of Dendrolimus pini, Galleria mellonella, and Calliphora vicina against fungal infection. J Insect Physiol. 2007;53(9):909–922. doi: 10.1016/j.jinsphys.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 56.Strand MR, Pech LL. Immunological basis for compatibility in parasitoid-host relationships. Ann Rev Entomol. 1995;40:31–56. doi: 10.1146/annurev.en.40.010195.000335. [DOI] [PubMed] [Google Scholar]
- 57.Wood PJ, Fulcher RG. Dye interactions. A basis for specific detection and histochemistry of polysaccharides. J Histochem Cytochem. 1983;31(6):823–826. doi: 10.1177/31.6.6841974. [DOI] [PubMed] [Google Scholar]
- 58.Deresinski SC, Stevens DA. Caspofungin. Clin Infect Dis. 2003;36(11):1445–1457. doi: 10.1086/375080. [DOI] [PubMed] [Google Scholar]
- 59.Jackson JC, Higgins LA, Lin X. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PloS One. 2009;4(1):e4224. doi: 10.1371/journal.pone.0004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101(1):177–194. [PMC free article] [PubMed] [Google Scholar]
- 61.Bunting LA, Neilson JB, Bulmer GS. Cryptococcus neoformans: gastronomic delight of a soil ameba. Sabouraudia. 1979;17(3):225–232. doi: 10.1080/00362177985380341. [DOI] [PubMed] [Google Scholar]
- 62.Luberto C, Martinez-Marino B, Taraskiewicz D, et al. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J Clin Invest. 2003;112(7):1080–1094. doi: 10.1172/JCI18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozel TR, Gotschlich EC. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J Immunol. 1982;129(4):1675–1680. [PubMed] [Google Scholar]
- 64.Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryotic Cell. 2007;6(6):949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


