Abstract
Indole, a derivative of the amino acid tryptophan, is a toxic signaling molecule, which can inhibit bacterial growth. To overcome indole-induced toxicity, many bacteria have developed enzymatic defense systems to convert indole to non-toxic, water-insoluble indigo. We previously demonstrated that, like other aromatic compound-degrading bacteria, Acinetobacter baumannii can also convert indole to indigo. However, no work has been published investigating this mechanism. Here, we have shown that the growth of wild-type A. baumannii is severely inhibited in the presence of 3.5 mM indole. However, at lower concentrations, growth is stable, implying that the bacteria may be utilizing a survival mechanism to oxidize indole. To this end, we have identified a flavoprotein oxygenase encoded by the iifC gene of A. baumannii. Further, our results suggest that expressing this recombinant oxygenase protein in Escherichia coli can drive indole oxidation to indigo in vitro. Genome analysis shows that the iif operon is exclusively present in the genomes of A. baumannii and Pseudomonas syringae pv. actinidiae. Quantitative PCR and Western blot analysis also indicate that the iif operon is activated by indole through the AraC-like transcriptional regulator IifR. Taken together, these data suggest that this species of bacteria utilizes a novel indole-detoxification mechanism that is modulated by IifC, a protein that appears to be, at least to some extent, regulated by IifR.
Introduction
More than 85 species of bacteria produce indole, an aromatic organic compound that is known to act as an extracellular signaling and regulatory molecule in a variety of physiological processes [1, 2]. Tryptophanase, encoded by the tnaA gene, is a tryptophan indole-lyase that produces indole, ammonium, and pyruvate from the amino acid tryptophan in Escherichia coli [3, 4]. Notably, the tryptophanase operon, encoding both tryptophanase and tryptophan permease, is regulated by glucose and tryptophan [5, 6]. For example, E. coli cultured in lysogeny broth (LB) media can secrete up to 0.5 mM of indole in the stationary phase, but when cultured in medium containing excess tryptophan, the indole concentration can reach up to 5 mM in the media [7]. Further, this increased concentration of indole is even higher inside the cell compared to the extracellular media as indole has a high affinity for lipids and will transverse the hydrophobic membrane of the cell [8] allowing this diffusible signaling molecule to regulate gene expression and numerous downstream processes, including fungal and bacterial growth.
During cellular growth, indole appears to exhibit both oxidant toxicity [9] and proton ionophoric activity [8, 10], which modulate the inhibition of cell division. Further, this inhibition can be induced by very low concentrations of indole. For example, in Aspergillus niger, just 0.43 mM of indole is sufficient to inhibit growth [11]. Similarly, a low level of indole (20 μg/ml; 0.17 mM) also inhibits the growth of four lactic acid bacterial strains known to function in the human intestine [12]. In fact, some bacteria in the intestinal tract are known to secrete indole or indole derivatives in order to inhibit the growth of other bacteria [2], thereby eliminating competition in their environment. Thus, it is not surprising that these compounds can greatly affect the host’s immune system as small changes in concentration can result in major fluctuations in the composition of gut microbiota [2, 13].
Notably, several types of bacterial, plant, and animal species have developed enzymatic defense systems to interfere with indole signaling and overcome indole-induced oxidant toxicity [2]. For instance, E. coli expressing wild-type and mutant cytochrome P450 enzymes from mammalian, human, and bacterial sources can convert indole into non-toxic indigo and indigoids [14–20]. In a similar manner, many bacterial species, including Acinetobacter spp., have been shown to transform indole to indigo in the presence of an aromatic inducer, such as phenol and aromatic hydrocarbons [21, 22]. Furthermore, recombinant E. coli expressing exogenous genes from this species also convert indole to various indigoids [21, 23]. While research to identify the species specific genes responsible for this enzymatic defense against indole toxicity is ongoing, the full function and regulation of these genes is largely unknown.
A. baumannii is a gram-negative bacterial pathogen found in human feces [24] and the rhizosphere [25]. In these environments, A. baumannii may encounter indole produced by other living organisms as well as industrial sources [26]. However, it is unclear whether the growth of A. baumannii is affected by indole. In the present study, we have shown that high concentrations of indole disrupt A. baumannii growth. However, at low indole concentrations, this bacterial species appears to utilize an enzymatic defense against this toxic compound. Furthermore, we have also characterized the function and regulation of an oxygenase encoded by the iif (iif stands for indole induced flavoprotein) operon, which appears to be involved in the oxidation and detoxification of indole in this species of bacteria.
Materials and Methods
Bacterial growth conditions and primers
A. baumannii and E. coli strains were routinely grown in LB medium or LB agar [27] at 37°C with vigorous shaking. M9 minimal medium (Amresco, Solon, OH) was used for the indole toxicity test. Ampicillin, chloramphenicol, and kanamycin were added to the medium when needed at 100 μg/ml, 12.5 μg/ml, and 50 μg/ml, respectively. Primers used in this study are listed in Table 1.
Table 1. Primers used in this study.
| Primer | Sequence a | Gene/direction |
|---|---|---|
| Cloning primers | ||
| OxyF | 5′-GCAGCCGATTATCACTTACTAGGCCG-3′ | |
| OxyR | 5′-CTCTTGGGTTTTCGGCATTAATCGC-3′ | |
| IifCF | 5′-GCATGGATCCATGCGCCGTATTGCAATTGTTG-3′ | |
| IifCR | 5′-GCATCTGCAGGACCTTAGGCCACTTTTGCTGTG-3′ | |
| BslF | 5′-GCCTCACTGATTAAGCATTGG-3′ | |
| BslR | 5′-CATCAGAGCAGCCGATTGTCTG-3′ | |
| IifRF | 5′-CACCGCCAATGATTGAAGCCG-3′ | |
| IifRR | 5′-CCTACTGGCGAATACGACCAG-3′ | |
| cIifRF | 5′-GGATCCAGAGCAATACCTCACGCTAAAAG-3′ | |
| cIifRR | 5′-GGATCCTGCAAGTTCGCTATGTTCACC-3′ | |
| cIifPF | 5′-GGATCCTTGGTAATTCCCCAATCATCC-3′ | |
| cIifPR | 5′-CCTAGTAAGTGATAATCGGCTGCCCTGACATTGTTATGGCTTTATCTTCTATC-3′ | |
| cIifCF | 5′- GATAGAAGATAAAGCCATAACAATGTCAGGGCAGCCGATTATCACTTACTAGG -3′ | |
| cIifCR | 5′-GGATCCGACCTTAGGCCACTTTTGCTGTG-3′ | |
| RT-PCR primers | ||
| IifF1 | 5′-TTAGACCCAACAGGCTTACCAGTTG-3′ | iifA/forward |
| IifR1 | 5′-CAATCGCCTTACCTTTGGCATC-3′ | iifB/reverse |
| IifF2 | 5′-TCATCCCGAAGAAGTCGCCAATG-3′ | iifB/forward |
| IifR2 | 5′-GATTTCATCGGCAGTACGGTTG-3′ | iifC/reverse |
| IifF3 | 5′-GCCAATAACTTTGATGACCCGC-3′ | iifC/forward |
| IifR3 | 5′-AATGGCGGATCTAAAGACAGAGATG-3′ | iifD/reverse |
| IifF4 | 5′-AGTATGAAGGCGGAGATCACCTG-3′ | iifD/forward |
| IifR4 | 5′-CGGCAAACCTAATAATGTCCCTGC-3′ | iifE/reverse |
| RT-qPCR primers | ||
| qF911_02007F | 5′-TAGGTTTAGGCGCAGCTATGCC-3′ | F911-02007 b /forward |
| qF911_02007R | 5′-ATCACTTTGGCCCAGCCATAGC-3′ | F911-02007 b /reverse |
| qIifAF | 5′-GCACGACCAAAAAGCAACCATTAC-3′ | iifA/forward |
| qIifAR | 5′-CATGACGGATATGCTCTTCCCAG-3′ | iifA/reverse |
| qIifBF | 5′-CTGAGCCGTGCTTTATACAATGACC-3′ | iifB/forward |
| qIifBR | 5′-GAAACCGAATTGACGCGAATACC-3′ | iifB/reverse |
| qIifCF | 5′-CAACCGTACTGCCGATGAAATC-3′ | iifC/forward |
| qIifCR | 5′-AGCCCAATGCCTTCTACGGC-3′ | iifC/reverse |
| qIifDF | 5′-GGCCGGAAAATAGGCATGAC-3′ | iifD/forward |
| qIifDR | 5′-AAAATGCCCCGAAAGCTGATG-3′ | iifD/reverse |
| qIifEF | 5′-GTATTGGGCTGCCACATATTGG-3′ | iifE/forward |
| qIifER | 5′-GCCCAGCTTGCATATCATTTGC-3′ | iifE/reverse |
| qIifRF | 5′-TGAACCAAGCCAACTCCCAC-3′ | iifR/forward |
| qIifRR | 5′-GCCTACATTTCGGCAGGTTTC-3′ | iifR/reverse |
| qGyrBF | 5′-GGCGGCTTATCTGAGTTTGT-3′ | gyrB/forward |
| qGyrBR | 5′-TTTGTGGAATGTTATTTGTG-3′ | gyrB/reverse |
aRestriction recognition sites are underlined.
bF911-02007 is the locus_tag for the gene located immediately upstream iifA.
Indole and indigo toxicity test
To test the effects of different concentrations of indole on the growth of wild-type A. baumannii, 1 ml of overnight LB culture media was collected and centrifuged. The supernatant was discarded and the pellet was resuspended in 500 μl of M9. This aliquot was then transferred to a flask containing 50 ml of M9 and incubated. Thus, after adding varying amounts of indole (dissolved in absolute ethanol) (0.5–5 mM), the culture was incubated at 37°C, 200 rpm for 16 h. The final concentration of ethanol was 0.1% and ethanol acted as the sole carbon source in this environment. The optical density at 600 nm (OD600) was used to determine the growth of the bacteria under the various conditions. Experiments were performed in triplicate.
To test the toxicity of indigo, the experimental culture condition was conducted as described above. After adding indole or indigo (5 mM), the culture was incubated at 37°C, 200 rpm for 16 h. The drop plate counting method was used to count the total number of bacteria [28]. The experiment was carried out three times.
Next, we compared the effects of 3 mM indole on the growth of the iifC and iifR mutants, described in the following sections, to the wild-type A. baumannii using the methods described above. Notably, the growth rate was measured every 2 h using the OD600. Experiments were performed three times.
Screening of indigo-forming E. coli colonies and identification of functional genes
We previously constructed a fosmid library representing 10-fold coverage of the A. baumannii ATCC19606 genome [23]. To isolate blue indigo-forming fosmid colonies, individual clones from the fosmid library were cultured on LB plates containing chloramphenicol overnight at 37°C, followed by incubation at 4°C for 7 days. In doing so, the indigo-producing fosmid, pOXY, was identified. pOXY was then purified and the insert end sequences were determined by sequencing as previously described [23].
Sequence analysis
BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi), MacVector software [29], and Pfam [30] were used to search the GenBank database, align sequences, and search the protein domains, respectively. The helical regions of proteins were predicated by YASPIN [31].
Quantitative reverse transcription PCR (RT-qPCR) and reverse transcription PCR (RT-PCR)
Gene expression was investigated using RT-qPCR. For RNA purification, we collected 1 ml of A. baumannii overnight LB culture media for centrifugation. The supernatant was then discarded and cells were resuspended in 500 μl M9. The resuspended cells were transferred into 50 ml of M9 containing 1.5 mM indole and 0.1% ethanol and incubated at 37°C, 200 rpm for 6 h. RNA was extracted during the exponential-phase of the cultures using TRIzol reagent (Invitrogen). After resuspension in TRIzol, chloroform was added, and the mixture was centrifuged at 4°C, 12,000 × g for 15 min. An equal volume of 70% ethanol was then added to the supernatant and RNA was extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The concentration of extracted RNA was determined using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc.; Wilmington, DE). Aliquots of the samples were analyzed on an RNA 6000 Nano chip in order to measure the RNA integrity using an Agilent 2100 bioanalyzer (Agilent Technologies; Santa Clara, CA). RNA was reverse transcribed using M-MLV reverse transcriptase (Promega) and qPCR was performed in triplicate with a LightCycler 1.5 instrument (Roche Diagnostics; Mannheim, Germany) using the LightCycler FastStart DNA MasterSYBRE Green I kit (Roche). Relative gene expression was calculated as previously described [32], using gyrase subunit B gene (gyrB) expression as an internal control. Fold change was determined by the 2(-ΔΔ C T) method and the graphs represent means ± SD. Primer pairs used for RT-qPCR are listed in Table 1. The following PCR cycling conditions were used: 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 10 s, followed by 60°C for 15 s.
For RT-PCR, Total RNA was purified from A. baumannii cultured in M9 medium containing 1.5 mM indole using TRIzol and reverse transcribed as described above. The cDNA was amplified by PCR with Herculase II Fusion DNA polymerase (Agilent Technologies) and specific primer pairs for iifA-iifB, iifB-iifC, iifC-iifD, and iifD-iifE intergenic regions (Table 1).
Transposon insertion
To identify the indigo-producing gene in the pOXY clone, the fosmid was purified and pOXY insertion mutants were generated by transposition in vitro with the EZ-Tn5 <KAN-2> transposon (Epicentre) following the manufacturer’s instructions. Transposon-tagged pOXY (pOXY::KAN-2) fosmids were then transformed into E. coli EPI300 (Epicentre) and selected on LB agar containing chloramphenicol and kanamycin. The transposon-tagged fosmids, which did not produce blue pigment when expressed in E. coli, were isolated, and the transposon insertion sites were determined with sequencing as previously described [23]. Of these fosmids, we selected nine, all of which appear to insert the KAN-2 transposon into the iifC gene (designated pOXY-iifC::KAN-2 plasmids).
iifC gene amplification and protein expression vector construction
A DNA fragment containing the indigo-producing gene iifC and the 223-bp region upstream of the putative translation initiation site was amplified by PCR using primers (OxyF and OxyR). The amplified DNA fragment was cloned using StrataClone Blunt PCR Cloning Kit (Agilent Technologies, Santa Clara, CA). The PCR products were ligated with Topoisomerase I-charged pSC-B vector arms and transformed into the provided StrataClone SoloPack competent cells according to the manufacturer’s protocol. Transformants were selected on LB agar containing ampicillin. The resulting plasmid was named pSCB-OXY.
To construct the IifC protein expression plasmid, the functional iifC gene was first amplified via PCR using the IifCF and IifCR primers. The PCR product was then digested with BamHI and PstI restriction enzymes and ligated into the BamHI/PstI digested pQE-80L expression plasmid (Qiagen) to create the pQE80L-IifC plasmid. This expression plasmid was then transformed into two E. coli strains: CY15000, a bacterial strain that harbors a mutation in the tnaA gene [33] obtained from the E. coli Genetic Stock Center (CGSC7682); and DH5α competent cells.
Isolation and characterization of pigments
Thin-layer chromatography (TLC) analysis was used to examine the pigments produced in the E. coli DH5α(pQE80L-IifC) and E. coli CY15000(pQE80L-IifC). E. coli DH5α(pQE80L-IifC) and E. coli CY15000(pQE80L-IifC) were cultured at 37°C overnight in LB broth. For pigments purification, 500 μl overnight culture was extracted using an equal volume of ethyl acetate. The ethyl acetate layer was separated by centrifugation and then evaporated. The blue pigment was resuspended in ethyl acetate, spotted on a silica gel plate (Silica gel 60 F254; catalogue number 05554; Merck), and developed with chloroform-methanol/50-1 (v/v).
Mutation of the iifC and iifR genes and complementation testing
Mutation of the iifC and iifR genes (designed ΔiifC and ΔiifR strains) was conducted using conjugation-mediated allelic exchange and confirmed with PCR as described in the S1 Methods section. Further, plasmids were also constructed for each mutant to perform complementation testing to determine their functional role in indigo production.
Indoline oxidization test
Further, in order to determine if indoline can be oxidized by IifC, E. coli CY15000(pQE80L-IifC) cultures were grown at 37°C overnight in LB broth supplemented with ampicillin and indoline. Following overnight cultivation, changes in the color of the media were monitored.
Recombinant protein purification and enzymatic activity assay
To produce recombinant IifC protein, E. coli CY15000(pQE80L-IifC) was cultured in LB at 37°C until an OD600 of 0.4 was reached. The recombinant protein was expressed by adding 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to the culture. The culture was then incubated with agitation at 20°C for 4 h, after which the cells were pelleted by centrifugation and stored at -20°C until use. Protein expression was examined using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) [34], followed by Coomassie blue G250 staining. The recombinant IifC protein was then purified and enzyme activity was analyzed using previously optimized assay procedures [23]. The flavin adenine dinucleotide (FAD) concentration in purified IifC was determined following the procedure described elsewhere [35].
Preparation of IifC and IifR polyclonal antibodies and Western blot analysis
Rabbit polyclonal antibodies against the purified recombinant IifC and IifR proteins were generated using a previously documented antiserum purification procedure [23]. Western blot analysis was then performed as previously described [23] with some modifications specific to these novel antibodies. Briefly, bacterial proteins were separated by 8% SDS-PAGE, blotted onto a polyvinylidene fluoride (PVDF) membrane, and blocked with Tris buffered saline (TBS; 50 mM Tris-HCl [pH 7.6], 0.9% NaCl) containing 5% milk for 1 h. The PVDF membrane was then incubated with either anti-IifC (dilution, 1:20,000) or anti-IifR (dilution, 1:25,000), along with a mouse monoclonal antibody against the alpha-subunit of RNA polymerase (dilution, 1:1,000; Neoclone, Madison, WI) for 1 h. After washing the membrane 6 times with TTBS buffer (TBS containing 0.25% Tween 20) for 10 min each, it was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (dilution, 1:2,000; Santa Cruz Biotechnology, Dallas, TX) or horseradish peroxidase-conjugated goat anti-mouse IgG (dilution, 1:2,000; Santa Cruz Biotechnology) secondary antibody for 1 h and washed with TTBS buffer 5 times, 10 min each. Protein expression was visualized with enhanced chemiluminescence reagent (Pierce ECL Western Blotting Substrate; Thermo Scientific).
Results
Indole toxicity assay
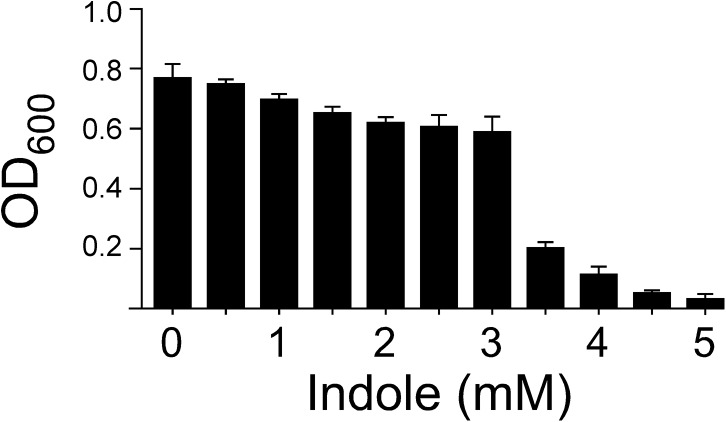
To determine whether indole inhibits the growth of A. baumannii, A. baumannii were cultured in M9 media containing varying concentration of indole (0.5–5 mM). The OD600 readings gradually decreased with the increasing indole concentration, with a severe decline in cell density at 3.5 mM (Fig 1). The toxicity assay was used to compare the effects of indole and indigo on bacteria growth. Unlike indole, 5 mM indigo does not inhibit the growth of A. baumannii (S1 Fig). These data indicate a possible defense against indole toxicity in A. baumannii when the indole concentration is low. To this end, A. baumannii ATCC19606 cultured in M9 medium with indole were observed to produce indigo (S2 Fig), suggesting the involvement of an enzymatic indole-conversion pathway.
Fig 1. Effect of indole on the growth of wild-type A. baumannii ATCC 19606.
Wild type A. baumannii ATCC 19606 was grown in M9 medium supplemented with 0–6 mM indole for 16 h.
Identification of indigo producing genes in A. baumannii
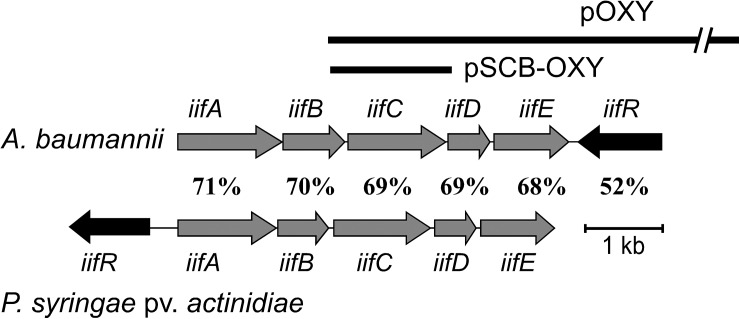
A fosmid library of A. baumannii ATCC19606 was previously constructed [23] and 960 fosmid clones were screened on LB plates in this study. One fosmid clone, pOXY, was consistently observed to produce blue pigments, leading us to isolate this colony. Sequence analysis and paired-end sequence mapping of the clone revealed that pOXY contains a 35,647 bp region of A. baumannii ATCC19606 genomic DNA, starting at the 130,733th bp and ending at the 166,379th bp of the 13th contig (Accession number APRG01000013) (Fig 2).
Fig 2. The gene organization of the iif operon in A. baumannii ATCC 19606 and P. syringae pv. actinidiae ICMP 19070.
Gray arrows represent the five-gene operon in A. baumannii ATCC 19606 and the dark-gray arrows represent the AraC-like regulator IifR. Thick lines represent the inserts of pOXY and pSCB-OXY. In A. baumannii ATCC 19606, the locus_tag for iifA, iifB, iifC, iifD, iifE, and iifR is F911_02006, F911_02005, F911_02004, F911_02003, F911_02002, and F911_02001, respectively. In P. syringae pv. actinidiae ICMP 19070, the locus_tag for iifA, iifB, iifC, iifD, iifE, and iifR is A259_30100, A259_30105, A259_30110, A259_30115, A259_30120, and A259_30095, respectively.
Sequence analysis of the iif operon of A. baumannii
Using RT-PCR to amplify the four putative intergenic regions of the iif operon found in A. baumannii, it appears that these four regions are amplified as a single iifABCDE operon in the presence of indole (S3 Fig). Further, this iif organization is prevalent in multiple A. baumannii strains, including ATCC19606, AB0057, AB307-0294, AB900, ACICU, AYCC 17978, and AYE. Aside from these A. baumannii strains, this iif operon is also found in Pseudomonas syringae pv. actinidiae and the arrangement of the operon in these two species is identical (Fig 2). Further, sequence analysis of the regulatory iifR gene indicates that the transcription direction of the gene is opposite to that of those in the main operon in both species; however, the iifR gene in A. baumannii is located downstream of the operon, while it is located upstream of the operon in P. syringae pv. actinidiae. The protein identity between each iif gene in A. baumannii ATCC19606 and P. syringae pv. actinidiae is also shown in Fig 2.
The G+C content of each iif gene present in A. baumannii ATCC19606 and the domains of the protein products encoded by iifABCDER are listed in Table 2. Using a Pfam database search, we found that IifA belongs to the dienelactone hydrolase family and shows strong homology (72% identity) to the putative dienelactone hydrolase of Pseudomonas sp. GM102 (accession no. EJL95871). Further, IifB appears to be a putative short-chain dehydrogenase. Sequence analysis of the iifC gene indicates that it is 1,239 bp long and the ATG start codon is preceded by a TGGAG ribosome-binding site. Although our sequence analysis did not reveal any significant Pfam domains, IifC showed 64.8% (268/413), 59.8% (241/403) and 47.4% (196/413) identity to MoxY (uncultured bacterium; GenBanK accession No. ABQ12175), StyA1 of Rhodococcus opacus 1CP (GenBanK accession Nos. ACR43973 and AFO70154), and StyA2B of R. opacus 1CP (GenBanK accession Nos. ACR43974 and AFO70155), respectively (S4 Fig). IifD showed 67.9% (114/168) identity to the flavin reductase of Pseudomonas fuscovaginae (accession No. WP_029379307), while IifE showed 73.6% (217/295) identity to the protein involved in the MetA-pathway of phenol degradation-like protein of Pseudomonas fuscovaginae (accession No. WP_019363187). Moreover, sequence analysis of IifR revealed that this protein contains two helix-turn-helix motifs (residues 255 to 332; C-terminal DNA binding domain) and belongs to the AraC/XylS family (S5 Fig). Multi sequence alignment of the DNA binding domain of this family revealed that, like AraC and XylS, IifR of A. baumannii ATCC19606 and P. syringae pv. actinidiae also contains the consensus binding sequence utilized by the AraC/XylS family [36].
Table 2. The iif genes of A. baumannii ATCC19606.
| Genes | Locus_tag | G+C content (%) | Product size (aa) | Protein Accession No. | Pfam family (Description) a | Pfam number (E-Value) a |
|---|---|---|---|---|---|---|
| iifA | F911-02006 | 44.04 | 416 | ENW75089 | DLH/Dienelactone hydrolase family | PF01738 (1.1e-49) |
| iifB | F911-02005 | 45.67 | 261 | ENW75088 | Adh_short_C2 (Enoyl-Acyl carrier protein reductase) | PF13561 (3.3e-31) |
| iifC | F911-02004 | 45.50 | 412 | ENW75087 | No significant matches | |
| iifD | F911-02003 | 42.94 | 176 | ENW75086 | Flavin_Reduct (Flavin reductase like domain) | PF01613 (2.2e-33) |
| iifE | F911-02002 | 40.72 | 315 | ENW75085 | Phenol_MetA_deg (Putative) MetA-pathway of phenol degradation | PF13557 (2.2e-63) |
| iifR | F911-02001 | 37.77 | 352 | ENW75084 | AraC_binding_2 (AraC-binding-like domain), HTH_18 (Helix-turn-helix domain) | PF14525 (6.1e-39), PF12833 (2.6e-28) |
aDetermined by searching the Pfam database.
Identification of iifC as an important player in indigo production
In order to determine the genes functioning in the conversion of indole to indigo in A. baumannii, RT-qPCR was performed to investigated the differential gene expression of iifABCDE, iifR, and the gene located immediately upstream of iifA (locus_tag F911_02007) in the presence and absence of indole. The expression of iifA, iifB, iifC, and iifD was increased in the presence of indole (18.6 ± 1.6 fold, 24.7 ± 1.8 fold, 24.2 ± 1.5 fold, and 14.7 ± 0.6, respectively) (Fig 3). F911_02007, iifE, and iifR, on the other hand, showed only slight changes in expression (0.8 ± 0.1 fold, 1.5 ± 0.1 fold, and 1.7 ± 0.3 fold, respectively). Therefore, it is likely that one or more of the genes located in the iifABCD region of the operon is responsible for indigo production in this bacterial species.
Fig 3. Expression of iif genes A. baumannii ATCC 19606 following the addition of indole.
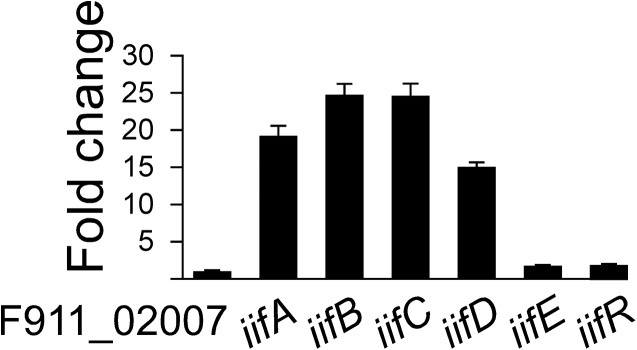
Expression of the iif genes was examined by RT-qPCR. Graph shows the fold change for each gene following the addition of 1.5 mM indole after normalization to gyrB. Reactions were done in triplicate.
To further investigate the pigment-producing genes located in the fosmid pOXY, transposon insertion analysis was performed with the EZ-Tn5 <KAN-2> transposon. Transposon tagged fosmids were transformed into E. coli Epi300. Nine transposon-tagged fosmids, which could not produce blue pigment when expressed in E. coli were isolated. Sequence analysis of these fosmids revealed that the EZ-Tn5 <KAN-2> insertion sites of all of these clones were located in the iifC gene, indicating that this gene is likely responsible for producing blue pigment when expressed in E. coli. The EZ-Tn5 <KAN-2> target sites are listed in S1 Table.
Additionally, a DNA fragment containing the intact iifC gene as well as the 223 bp upstream sequence was amplified and subcloned to generate the pSCB-OXY plasmid (also featured in Fig 2). Both the pOXY and pSCB-OXY plasmids were observed to form blue colonies when expressed in E. coli on LB agar plates.
An IifC expression plasmid (pQE80L-IifC) was constructed for protein overexpression and introduced into two E. coli strains: DH5α and CY15000 (a tnaA mutant). Following expression plasmid transformation, but prior to the addition of IPTG and indoline, overnight cultures of both strains were extracted with ethyl acetate, concentrated, and analyzed by TLC. Unlike E. coli DH5α(pQE80L-IifC), which produced indigo, the tnaA mutant E. coli CY15000(pQE80L-IifC) was not able to produce indigo or indirubin (S6 Fig). Taken together, these data demonstrate that in E. coli strains with fully functioning tryptophanase proteins, which will naturally be producing low levels of indole, the expression of iifC is sufficient for indigo production. Moreover, in the E. coli CY15000(pQE80L-IifC) cultures, when the non-functional tryptophanase was bypassed by adding indoline, the culture medium turned a deep pink color when were cultured overnight (S7 Fig).
Purification of IifC protein and enzyme assay
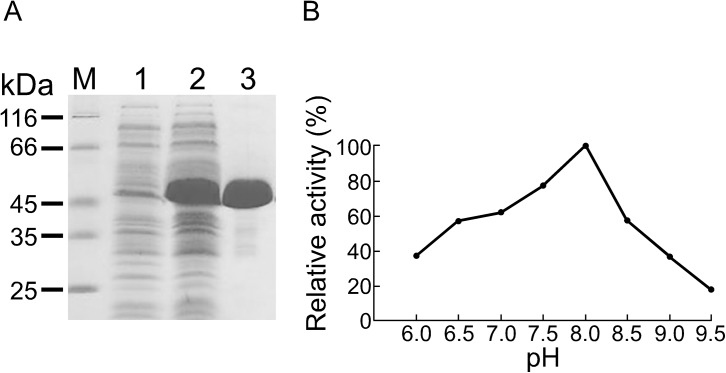
IifC protein expression was initially carried out at 37°C with vigorous shaking, resulting in the proteins being well-expressed in soluble form. After sonication to disrupt the cells and centrifugation to remove cell debris, the clarified protein solution (crude extract) was loaded directly onto a 2.6 × 40 cm column of anion-exchange matrix equilibrated with 10 mM phosphate buffer, pH 7.0, containing 5 mM EDTA. Protein was then eluted with a linear, ascending gradient of potassium chloride. The protein was concentrated and observed to be nearly homogeneous (Fig 4A). Further, the color of the purified protein solution was light yellow. FAD content assay indicated that 1 mol of recombinant IifC contained 0.48 mol of FAD. Purification of the IifC protein was 17.6-fold with a yield of 45% (S2 Table), and the optimum pH for the enzyme was observed in the alkaline range (varying between 6.0 and 9.5). The maximum activity rate was achieved at pH 8.0 (Fig 4B). Therefore, all further substrate specificity and kinetic experiments were conducted at pH 8.0.
Fig 4. IifC protein purification and kinetics.
(A) Overexpression and purification of recombinant IifC. Cell extracts were prepared before (lane 1) and after (lane 2) IPTG induction. Recombinant IifC protein (lane 3) was further purified using Q-Sepharose High Performance anion-exchange column. (B) Enzymatic activity of IifC in the presence of indole in various pH buffers. pH dependent activity assay was carried out in 100 mM potassium phosphate buffer between pH 6.0 and 7.5, 100 mM Tris-HCl buffer between pH 8.0 and 9.5.
Next, we investigated the enzyme kinetics of IifC. The conversion of indole into indigo was dependent upon the presence of IifC, NADH, FAD, and indole in the assay. The IifC K m values for indole and NADH were 0.20 ± 0.05 mM and 0.27 ± 0.03 mM, respectively, while the k cat was 0.38 ± 0.03 min-1.
Indole toxicity and expression of IifC in the iifC and iifR mutants
In contrast to wild-type A. baumannii ATCC19606 (S2 Fig), neither the ΔiifC or ΔiifR strain could produce indigo when cultured in M9 medium with 3 mM indole. To verify the functions of the iifC and iifR genes, we performed complementation experiments. Using electroporation, we transformed pComIifC and pComIifR plasmids into ΔiifC and ΔiifR strains, respectively. ΔiifC(pComIifC) and ΔiifR(pComIifR) could both produce indigo when cultured in M9 medium with indole (S2 Fig).
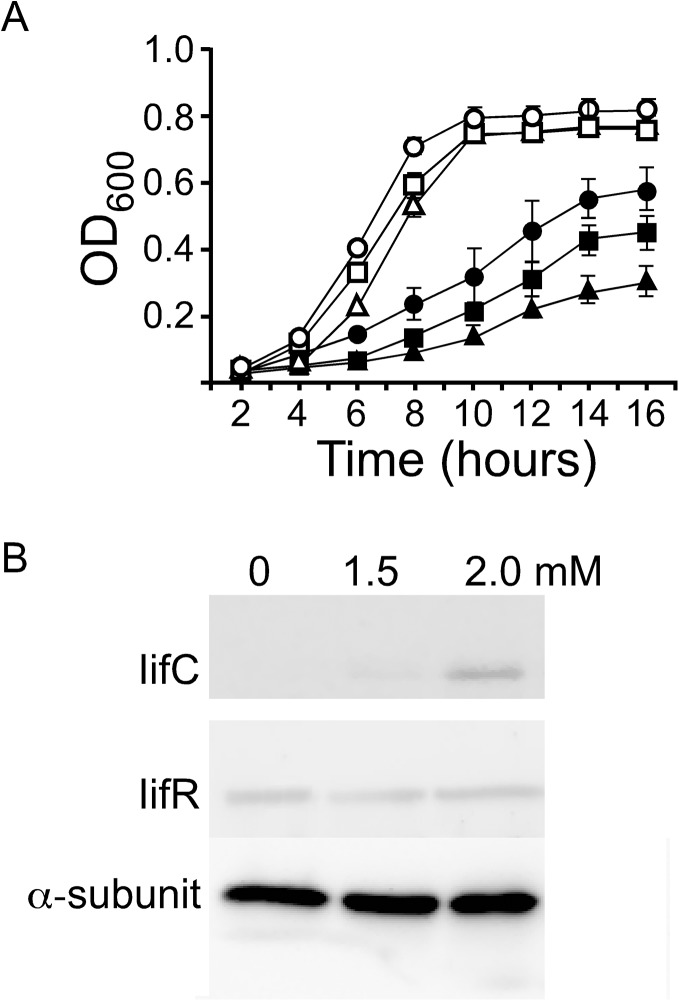
We also compared the growth of wild-type A. baumannii ATCC19606, ΔiifC, ΔiifR, ΔiifC(pComIifC), and ΔiifR(pComIifR) strains cultured in M9 medium with 3 mM indole. From the OD curves, it is evident that, in the absence of indole, the growth of these strains is similar (Fig 5A). However, in the presence of 3 mM indole, the wild-type, ΔiifC(pComIifC) and ΔiifR(pComIifR) strains grew much more slowly. Further, the growth of the ΔiifC and ΔiifR strains was further inhibited compared to the wild-type, with the ΔiifR strain being more severely affected by indole than the ΔiifC strain (Fig 5A).
Fig 5. Effect of indole on the growth of wild-type A. baumannii ATCC 19606 and mutant strains.
(A) Wild type (●), iifC (■), and iifR (▲) mutants were grown in M9 medium containing 0 or 3 mM indole (highlighted by the empty shapes or solid shapes, respectively). The results present the mean ± SD of three independent cultures. The growth of wild-type, ΔiifC(pComIifC) and ΔiifR(pComIifR) strains is similar. (B) Western blot of the total proteins isolated from A. baumannii ATCC 19606 cultured in M9 medium with 0, 1.5, and 2.0 mM indole. The alpha-subunit of RNA polymerase (α-subunit) was used as a loading control.
To further determine the function of IifC in these mutant strains, bacteria were cultured in M9 medium with and without indole, and IifC protein was detected by Western blotting. Our data indicate that IifC protein expression is induced by indole in a dose-dependent manner (Fig 5B). While the levels of IifC increase in A. baumannii ATCC19606, this strain expressed the same amount of IifR protein when cultured in M9 medium, regardless of the level of indole. Moreover, the iifR mutant does not appear to express IifC, irrespective of the presence of indole. These data indicate that while IifC appears to be the main enzyme functioning in converting indole to indigo, IifR likely plays an important regulatory role in this process.
Discussion
In this study, we have shown that A. baumannii utilizes the indole-converting protein IifC to protect itself from the effects of this compound at low concentrations, while at higher (3.5 mM) concentrations the indole toxicity overcomes these defenses and inhibits growth. Further, it appears that IifR also plays a regulatory role in this pathway. To our knowledge, this is the first time IifC has been documented to function as an enzymatic defense against indole toxicity in this bacterial strain.
Protection against indole-induced cytotoxicity has been observed in a number of bacterial species. While some species degrade indole or use it as a carbon source [26, 37–39], many species utilize oxidative enzymes that convert indole to non-toxic indigo. Further, recombinant E. coli expressing exogenous genes from several species, including Acinetobacter spp. [21, 23], Bacillus subtilis WU-S2B [40], Burkholderia cepacia [41], Pseudomonas spp. [42–44], Methylophaga sp. [45], Ralstonia eutropha [46], Rhodococcus spp. [47–49], or the bacterial metagenome [50–52] also convert indole to various indigoids. Moreover, in the presence of an aromatic inducer, Acinetobacter spp. [21, 22], Comamonas sp. [53], and Pseudomonase spp. [54–59] also transform indole into indigoids.
A. baumannii is a notable opportunistic pathogen of the genus Acinetobacter [60] that is known to survive in diverse environments, including soil [25]. Further, different aromatic compounds released from plant and crude oil are also present in the soil [61] and Acinetobacter spp. can degrade and utilize a variety of these molecules as carbon and energy sources [61]. For example, previous studies have shown that A. baumannii can use the aromatic compounds benzoate, p-hydroxybenzoate and indole-3-acetic acid (IAA) [23, 62]. In our previous report [23] and the present study, we have demonstrated that, like other aromatic compound-degrading bacteria, A. baumannii can also convert toxic indole to non-toxic indigo. Although these bacteria can oxidize indole, no work has been published investigating the mechanism of this phenomenon.
In the search to understand indole conversion, a number of genes have been discovered that can oxidize this compound to indigo. For example, the iacA gene of A. baumannii, which encodes an indole oxygenase, can be induced by the plant hormone IAA. However, bacteria harboring an iacA mutation can still convert indole into indigo, indicating that there is likely another indole oxygenase gene in A. baumannii that is the primary enzyme in this reaction.
Thus, to investigate other genes involved in indigo production, we isolated a fosmid clone, pOXY, from our A. baumannii genomic library. Sequence analysis revealed that A. baumannii harbors the iif operon, a sequence of five structural genes that is regulated by an AraC/XylS family member. Interestingly, it appears that the iif operon is only present in two bacterial species, A. baumannii and P. syringae pv. actinidiae. While these genes in both species are organized identically and display strong similarity, the average G + C content of the iif operon is quite different, indicating that it has existed in these two organisms for a long time. Unfortunately, aside from iifC, which was characterized in this study, the functions of the rest of the iif genes are still unclear. For example, sequence analysis showed that the iifA gene encodes a dienelactone hydrolase-like protein. Dienelactone hydrolases are involved in the degradation of 2, 4-dichlorophenoxyacetic acid (2, 4-D) and other chloroaromatics [63], but the function of IifA, and how A. baumannii degrades 2, 4-D is still unknown.
Notably, the third gene of the iif operon, iifC, appears to be the primary gene involved in indigo production and indole resistance. Indigo and indigoids are both important dye and drug compounds that are produced by chemical synthesis, isolated from plant sources, and synthesized by bacteria. Interestingly, because E. coli naturally produces indole, recombinant E. coli engineered to also express exogenous indigo-producing genes (IPGs) will be blue in color because the IPG will convert the endogenous indole to indigo. Therefore, recombinant E. coli can be exploited as a host for gene cloning and IPG isolation and analysis.
Since their initial discovery in 1983, many IPGs have been identified, including the cytochrome P450 enzymes and a variety of flavoproteins. Aside from the genes encoding the cytochrome P450 enzymes, these IPG encoded flavoproteins can be roughly divided into two types: Type I, which contain an FAD-binding domain (FBD); and Type II, which do not contain this domain. Table 3 lists the Type I flavoproteins that can oxidize indole in E. coli. We have previously showed that the Type II IPGs belong to the acyl-CoA dehydrogenase family [23]. Notably, the acyl-CoA dehydrogenase-like flavoproteins encoded by these IPGs do not have a conserved FBD, but contain the Pfam domains Acyl-CoA-dh-N and Acyl-CoA-dh-2 or Acyl-CoA-dh-N and Acyl-CoA-dh-M. Further, these enzymes are all similar in size, ranging from 387 to 416 amino acids [23]. On the other hand, the sizes of the flavoproteins encoded by the Type I IPGs are larger than 400 amino acids (Table 3). Moreover, the protein domains of these flavoproteins are much more diverse that those of Type II IPGs and can contain flavoprotein monooxygenase (FMO)-like, FAD_binding_3, or Pyr_redox_2 Pfam domains. iifC is a Type I IPG, but after a Pfam database search, no significant domains were identified. However, similar to styrene monooxygenase, IifC also appears to contain the FAD-binding fingerprints GxGxxG, GG, and Dx6G at amino acid positions 8, 119, and 140, respectively.
Table 3. Indigo producing genes encoding FAD containing proteins.
| Gene/Source | Length (bp) | Product size (aa) | Protein Accession No. | FAD-binding sequence (GxGxxG-GG-DxxxxxxG) | FAD/NAD(P) interaction motif (GDxxxxxxP) | Pfam family a | Reference | Pfam family |
|---|---|---|---|---|---|---|---|---|
| iifC/A. baumannii | 1239 | 412 | ENW75087 | GAGQSG-GG-ELVLLAAG | ADALVVNDP | No significant matches | This study | No significant matches |
| fmo/Methylophaga sp. | 1371 | 456 | AAM18566 | GAGPSG-//-DYVVCCTG | RDVIMGRLP | FMO-like | [45] | FMO-like |
| smo/metagenome | 1251 | 416 | ABV24041 | GAGIAG-GG-DLLVVSSG | GDVHSVVDP | FAD_binding_3 | [52] | FAD_binding_3 |
| styA2B/R. opacus 1CP | 1221 | 406 | ACR43973 | GAGQAG-GG-DLVILAAG | ADVVVLNDP | Pyr_redox_2 | [49] | Pyr_redox_2 |
aDetermined by searching the Pfam database.
In addition to flavins, the cofactor NAD(P)H is also involved in FMO reactions. Flavins are reduced by NAD(P)H via electron transfer and then sequentially converted to hyroperoxyflavins, which are an unstable intermediate. FMOs can bind and stabilize these hyroperoxyflavins, and subsequently oxygenate a variety of substrates [64]. In most two-component FMOs, a reductase gene is located immediately downstream of a monooxygenase gene [52]. For example, C2-hpah and C1-hpah encode a p-hydroxyphenylacetate hydroxylase, a two-component FMO of A. baumannii isolated from Thai soil [65]. Further, in P. fluorescens ST styA and styB genes encode a two-component styrene monooxygenase (SMO) [66]. Notably, the Gdx6P motif, important for the FAD/NAD(P) interaction [49], is also found at amino acid position 292 of IifC. In addition, our results also shown that 1 mol of purified IifC contained 0.48 mol of FAD. Due to the dissociation of non-covalently-bound FAD from proteins during purification, both FMO [45] and IifC purified from E. coli contained approximately 0.5 mol of FAD per mol of these two proteins. Although StyA2B contains the FAD-binding fingerprints, recombinant His10-StyA2B purified from E. coli with Ni-chelate affinity chromatography contained trace amounts of FAD [49]. Therefore, the binding of FAD to FAD-binding protein in the buffer for purification may have influenced the dissociation of FAD from enzymes. Further, we also discovered that iifD, located directly downstream of iifC, is a putative flavin oxidoreductase, which could be functioning as one component of the two-component FMO. Like C2-hpah and StyA, IifC is also larger than IifD. Taken together, these data suggest that the proteins encoded by the iifC and iifD genes could constitute a putative two-component FMO.
Like Fmo protein of Methylophage sp. [45], recombinant IifC protein purified from E. coli binds to FAD. As a flavoprotein oxygenase, the purified IifC contains FAD, preventing the determination of the K m value for this cofactor. However, for the other cofactors, indole and NADH, IifC protein has a K m value of 0.20 ± 0.05 mM and 0.27 ± 0.03 mM, respectively, and a k cat of 0.38 ± 0.03 min-1. Notably, the K m for indole is much lower than the concentration required to significantly inhibit the growth of cultured A. baumannii. Further, the indole K m of IifC was four times lower than that previously found for IacA (0.80 ± 0.04 mM) and the catalytic efficiency of the enzyme (k cat/K m) of IifC (1.9 min-1mM-1) was higher than that of IacA (1.1 min-1mM-1) [23]. Thus, it would appear that IifC is more efficient at converting indole to indigo. This was not surprising as we suspect indole may be the natural substrate of IifC, while the natural substrate of IacA is IAA. However, because IAA and indole are structurally similar, IacA could also oxidize indole [23], just at a lower efficiency. In fact, when examining current research it appears that although other enzymes encoded by IPGs can oxidize indole, the natural substrate of these enzymes is not indole. For example, cytochrome P450 enzymes, which have wide substrate specificity [67], can oxidize indole. In addition, O’Connor et al. [56] reported that since indole and styrene are very similar structurally, SMOs can also use indole even though this is not their natural substrate. However, while these other IPGs may have the ability to convert indole, IifC is induced by indole in vivo, supporting our hypothesis that indole is the natural substrate of this enzyme, making it the key player in indole resistance. Not surprisingly, indoline, a compound that is structurally similar to indole, was also converted to an uncharacterized water-soluble pigment in a similar manner by IifC.
While IifC is likely the primary enzyme involved in indole conversion, mutagenesis and complementation analysis showed that IifR is required for IifC expression. Sequence analysis indicates that IifR may be a member of the AraC/XylS family, all of which, with the exception of CelD, are positive transcriptional activators [36, 68]. Notably, AraC/XylS family members are known to regulate bacterial genes involved in carbon metabolism, stress response, and manifestation of pathogenesis [36, 69]. Therefore, in the presence of the appropriate inducer, AraC/XylS-like proteins could likely activate cognate operon expression. In fact, the expression of several operons involved in the degradation of aromatic compounds has been shown to vary depending on the concentration of the AraC/XylS inducer [70]. Interestingly, in the present study, we observed that the expression of IifC was activated by indole in a dose-dependent manner. Taken together, these results indicate that IifR is an AraC-like protein that functions to control iif operon expression. However, while the N-terminal non-conserved domain of AraC and XylS can bind arabinose and benzoate [71, 72], respectively, further study is needed to determine where IifR can bind indole. Alternatively, another activator/cascade could be modulating this regulatory protein, such as that observed for other AraC/XylS family members [70, 72]. Western blot analysis also revealed that the expression of IifR was constitutive irrespective of indole concentrations, indicating that this protein may not be regulating the indole-induced stress response alone.
Notably, a number of other indole-inducible proteins involved in indole resistance have been characterized previously, in addition to IifC and IifR. For example, AhpC, found in E. coli, and a novel 36-kDa protein found in Brevibacterium flavum have both been reported to be induced by indole [9]. We can also not rule out the possibility that IifR could be an activator for any number of other indole-inducible genes in this pathway. Further, we suspect that these various other genes and regulatory proteins may also be contributing synergistically with IifC and IifR to help protect the bacteria from the toxic effects of indole and, therefore, warrant further investigation.
In conclusion, our work suggests that indole oxidization (detoxification) in A. baumannii is performed by IifC, which is in turn controlled by a member of the AraC/XylS family, IifR. While these data help advance our understanding of how bacteria counteract the toxicity of the aromatic compound, indole, additional work is necessary to elucidate the full signaling cascade and functional mechanism underlying this phenomenon.
Supporting Information
A. baumannii ATCC 19606 was grown in M9 medium containing 0.1% ethanol as carbon source (control). To test the toxicity of indole and indgo, A. baumannii ATCC 19606 was cultivated in M9 medium supplemented with 5 mM indole (indole) or 5 mM indigo (indigo). After incubation for 16 h at 37°C, the cell cultures were serially diluted and 10 μl-drops of selected dilutions were plated onto LB agar plates for viable counts.
(DOCX)
Indigo was produced by wild-type A. baumannii ATCC 19606 (A), ΔiifC(pComIifC) (B), and ΔiifR(pComIifR) (C). In contrast, ΔiifC (D) and ΔiifR (E) cannot produce indigo. These strains were grown in LB broth overnight. Bacteria from 0.5 ml of the overnight cultures was collected by centrifugation, washed with M9 medium, and transferred to fresh M9 medium (50 ml) supplemented with 3.0 mM indole and 0.1% ethanol. Indigo was observed in the medium after 16 h.
(DOCX)
(A) Diagram of the intergenic regions of iifA-iifB, iifB-iifC, iifC-iifD, and iifD-iifE (represented by thick black lines) that were amplified by RT-PCR. (B) The RT-PCR amplification products were analyzed by agarose gel electrophoresis. RT-PCR amplifications were conducted with reverse transcriptase (lanes 1–4) or without reverse transcriptase (negative control; lanes 5–8) using total RNA as the template. Lanes 1, 2, 3, and 4 are the RT-PCR products of the intergenic region of iifAB, iifBC, iifCD, and iifDE, respectively. These cDNA were not amplified in the negative control lanes. Lane M: 100-bp ladder marker.
(DOCX)
Asterisks indicate identical amino acids in IifC, StyA, and StyA2B. The FAD-binding fingerprints GxGxxG, GG, and Dx6G are also shown. IifC showed 64.8% (268/413), 59.8% (241/403), and 47.4% (196/413) identity to MoxY (uncultured bacterium; GenBanK accession No. ABQ12175), StyA1 of Rhodococcus opacus 1CP (GenBanK accession Nos. ACR43973 and AFO70154) and StyA2B of R. opacus 1CP (GenBanK accession Nos. ACR43974 and AFO70155), respectively.
(PDF)
Multi sequence alignment of the DNA binding domains of the IifR of A. baumannii (IifR/Ab), IifR of P. syringae pv. actinidiae (IifR/Ps), AraC of E. coli (AraC/Ec; SWISSPROT accession no. P03021), and XylS of P. putida (XylS/Pp; SWISSPROT accession no. P07859) were carried out manually. This region contains two potential helix-turn-helix (HTH) DNA binding motifs (first and second HTH motifs). Underlined sequences indicate the helical regions.
(PDF)
Ethyl acetate extracts of E. coli DH5α(pQE80L-OXY) (lane 2) and E. coli CY15000(pQE80L-OXY) (lane 3) culture broth were analyzed by TLC. Indirubin (lane 1) and indigo (lane 4) were used as marker.
(DOCX)
E. coli CY15000(pQE80L) (tubes 1–3) and E. coli CY15000(pQE80L-OXY) (tubes 4–6) were inoculated into 3 ml of LB broth containing 1 μl (tubes 1 and 4), 2 μl (tubes 2 and 5), or 3 μl (tubes 3 and 6) of indoline. After incubating for an additional 16 h, the medium turned a deep pink color in the cultures of E. coli CY15000(pQE80L-OXY) (tubes 4–6).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors acknowledge the High-throughput Genome Analysis Core Facility of National Core Facility Program for Biotechnology, Taiwan, for sequencing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Work was supported by grants from the 496 Ministry of Science and Technology, Taiwan (grant NSC 102-2320-B-309 -003 and MOST 103-2320-B-309- 001-). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hu M, Zhang C, Mu Y, Shen Q, Feng Y. Indole affects biofilm formation in bacteria. Indian J Microbiol. 2010; 50: 362–368. 10.1007/s12088-011-0142-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010; 34: 426–444. 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- 3. Newton WA, Morino Y, Snell EE. Properties of crystalline tryptophanase. J Biol Chem. 1965; 240: 1211–1218. [PubMed] [Google Scholar]
- 4. Snell EE. Tryptophanase: structure, catalytic activities, and mechanism of action. Adv Enzymol Relat Areas Mol Biol. 1975; 42: 287–333. [DOI] [PubMed] [Google Scholar]
- 5. Isaacs H Jr, Chao D, Yanofsky C, Saier MH Jr. Mechanism of catabolite repression of tryptophanase synthesis in Escherichia coli . Microbiology. 1994; 140: 2125–2134. [DOI] [PubMed] [Google Scholar]
- 6. Yanofsky C, Horn V, Nakamura Y. Loss of overproduction of polypeptide release factor 3 influences expression of the tryptophanase operon of Escherichia coli . J Bacteriol. 1996; 178: 3755–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology. 2013; 159: 402–410. 10.1099/mic.0.064139-0 [DOI] [PubMed] [Google Scholar]
- 8. Gaimster H, Cama J, Hernandez-Ainsa S, Keyser UF, Summers DK. The indole pulse: a new perspective on indole signalling in Escherichia coli . PLoS One. 2014; 9: e93168 10.1371/journal.pone.0093168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garbe TR, Kobayashi M, Yukawa H. Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch Microbiol. 2000; 173: 78–82. [DOI] [PubMed] [Google Scholar]
- 10. Chimerel C, Field CM, Pinero-Fernandez S, Keyser UF, Summers DK. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim Biophys Acta. 2012; 1818: 1590–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kamath AV, Vaidyanathan CS. New pathway for the biodegradation of indole in Aspergillus niger . Appl Environ Microbiol. 1990; 56: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nowak A, Libudzisz Z. Influence of phenol, p-cresol and indole on growth and survival of intestinal lactic acid bacteria. Anaerobe. 2006; 12: 80–84. [DOI] [PubMed] [Google Scholar]
- 13. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009; 106: 3698–3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Celik A, Speight RE, Turner NJ. Identification of broad specificity P450CAM variants by primary screening against indole as substrate. Chem Commun. 2005; 29: 3652–3654. [DOI] [PubMed] [Google Scholar]
- 15. Gillam EM, Aguinaldo AM, Notley LM, Kim D, Mundkowski RG, Volkov AA, et al. Formation of indigo by recombinant mammalian cytochrome P450. Biochem Biophys Res Commun. 1999; 265: 469–472. [DOI] [PubMed] [Google Scholar]
- 16. Gillam EM, Wunsch RM, Ueng YF, Shimada T, Reilly PE, Kamataki T, et al. Expression of cytochrome P450 3A7 in Escherichia coli: effects of 5' modification and catalytic characterization of recombinant enzyme expressed in bicistronic format with NADPH-cytochrome P450 reductase. Arch Biochem Biophys. 1997; 346: 81–90. [DOI] [PubMed] [Google Scholar]
- 17. Li QS, Schwaneberg U, Fischer P, Schmid RD. Directed evolution of the fatty-acid hydroxylase P450 BM-3 into an indole-hydroxylating catalyst. Chemistry. 2000; 6: 1531–1536. [DOI] [PubMed] [Google Scholar]
- 18. Lu Y, Mei L. Co-expression of P450 BM3 and glucose dehydrogenase by recombinant Escherichia coli and its application in an NADPH-dependent indigo production system. J Ind Microbiol Biotechnol. 2007; 34: 247–253. [DOI] [PubMed] [Google Scholar]
- 19. Rosic NN. Versatile capacity of shuffled cytochrome P450s for dye production. Appl Microbiol Biotechnol. 2009; 82: 203–210. 10.1007/s00253-008-1812-8 [DOI] [PubMed] [Google Scholar]
- 20. Zhang ZG, Liu Y, Guengerich FP, Matse JH, Chen J, Wu ZL. Identification of amino acid residues involved in 4-chloroindole 3-hydroxylation by cytochrome P450 2A6 using screening of random libraries. J Biotechnol. 2009; 139: 12–18. 10.1016/j.jbiotec.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doukyu N, Toyoda K, Aono R. Indigo production by Escherichia coli carrying the phenol hydroxylase gene from Acinetobacter sp. strain ST-550 in a water-organic solvent two-phase system. Appl Microbiol Biotechnol. 2003; 60: 720–725. [DOI] [PubMed] [Google Scholar]
- 22. Qu Y, Pi W, Ma F, Zhou J, Zhang X. Influence and optimization of growth substrates on indigo formation by a novel isolate Acinetobacter sp. PP-2. Bioresour Technol. 2010; 101: 4527–4532. 10.1016/j.biortech.2010.01.033 [DOI] [PubMed] [Google Scholar]
- 23. Lin GH, Chen HP, Huang JH, Liu TT, Lin TK, Wang SJ, et al. Identification and characterization of an indigo-producing oxygenase involved in indole 3-acetic acid utilization by Acinetobacter baumannii . Antonie Van Leeuwenhoek. 2012; 101: 881–890. 10.1007/s10482-012-9704-4 [DOI] [PubMed] [Google Scholar]
- 24. Dijkshoorn L, van Aken E, Shunburne L, van der Reijden TJ, Bernards AT, Nemec A, et al. Prevalence of Acinetobacter baumannii and other Acinetobacter spp. in faecal samples from non-hospitalised individuals. Clin Microbiol Infect. 2005; 11: 329–332. [DOI] [PubMed] [Google Scholar]
- 25. Sachdev D, Nema P, Dhakephalkar P, Zinjarde S, Chopade B. Assessment of 16S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol Res. 2010; 165: 627–638. 10.1016/j.micres.2009.12.002 [DOI] [PubMed] [Google Scholar]
- 26. Yin B, Gu JD, Wan N. Degradation of indole by enrichment culture and Pseudomonas aeruginosa Gs isolated from mangrove sediment. Int Biodeter Biodegr. 2005; 56: 243–248. [Google Scholar]
- 27. Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli . J Bacteriol. 1951; 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen CY, Nace GW, Irwin PL. A 6 x 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli . J Microbiol Methods. 2003; 55: 475–479. [DOI] [PubMed] [Google Scholar]
- 29. Olson SA. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol Biol. 1994; 25: 195–201. [DOI] [PubMed] [Google Scholar]
- 30. Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014; 42: D222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin K, Simossis VA, Taylor WR, Heringa J. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics. 2005; 21: 152–159. [DOI] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 33. Yanofsky C, Horn V. Rifampin resistance mutations that alter the efficiency of transcription termination at the tryptophan operon attenuator. J Bacteriol. 1981; 145: 1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 35. Zhang M, Robertus JD. Molecular cloning and characterization of a full-length flavin-dependent monooxygenase from yeast. Arch Biochem Biophys. 2002; 403: 277–283. [DOI] [PubMed] [Google Scholar]
- 36. Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997; 61: 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bak F, Widdel F. Anaerobic degradation of indolic compounds by sulfate-reducing enrichment cultures, and description of Desulfobacterium indolicum gen. nov., sp. nov. Arch Microbiol. 1986; 146: 170–176. [Google Scholar]
- 38. Claus G, Kutzner HJ. Degradation of indole by Alcaligenes spec. Syst Appl Microbiol. 1983; 4: 169–180. 10.1016/S0723-2020(83)80046-0 [DOI] [PubMed] [Google Scholar]
- 39. Doukyu N, Aono R. Biodegradation of indole at high concentration by persolvent fermentation with Pseudomonas sp. ST-200. Extremophiles. 1997; 1: 100–105. [DOI] [PubMed] [Google Scholar]
- 40. Furuya T, Takahashi S, Ishii Y, Kino K, Kirimura K. Cloning of a gene encoding flavin reductase coupling with dibenzothiophene monooxygenase through coexpression screening using indigo production as selective indication. Biochem Biophys Res Commun. 2004; 313: 570–575. [DOI] [PubMed] [Google Scholar]
- 41. Rui L, Reardon KF, Wood TK. Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl Microbiol Biotechnol. 2005; 66: 422–429. [DOI] [PubMed] [Google Scholar]
- 42. Ensley BD, Ratzkin BJ, Osslund TD, Simon MJ, Wackett LP, Gibson DT. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983; 222: 167–169. [DOI] [PubMed] [Google Scholar]
- 43. Leveau JH, Gerards S. Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol Ecol. 2008; 65: 238–250. 10.1111/j.1574-6941.2008.00436.x [DOI] [PubMed] [Google Scholar]
- 44. McClay K, Boss C, Keresztes I, Steffan RJ. Mutations of toluene-4-monooxygenase that alter regiospecificity of indole oxidation and lead to production of novel indigoid pigments. Appl Environ Microbiol. 2005; 71: 5476–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi HS, Kim JK, Cho EH, Kim YC, Kim JI, Kim SW. A novel flavin-containing monooxygenase from Methylophaga sp strain SK1 and its indigo synthesis in Escherichia coli . Biochem Biophys Res Commun. 2003; 306: 930–936. [DOI] [PubMed] [Google Scholar]
- 46. Drewlo S, Bramer CO, Madkour M, Mayer F, Steinbuchel A. Cloning and expression of a Ralstonia eutropha HF39 gene mediating indigo formation in Escherichia coli . Appl Environ Microbiol. 2001; 67: 1964–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hart S, Koch KR, Woods DR. Identification of indigo-related pigments produced by Escherichia coli containing a cloned Rhodococcus gene. J Gen Microbiol. 1992; 138: 211–216. [DOI] [PubMed] [Google Scholar]
- 48. Kwon NR, Chae JC, Choi KY, Yoo M, Zylstra GJ, Kim YM, et al. Identification of functionally important amino acids in a novel indigo-producing oxygenase from Rhodococcus sp. strain T104. Appl Microbiol Biotechnol. 2008; 79: 417–422. 10.1007/s00253-008-1445-y [DOI] [PubMed] [Google Scholar]
- 49. Tischler D, Eulberg D, Lakner S, Kaschabek SR, van Berkel WJ, Schlomann M. Identification of a novel self-sufficient styrene monooxygenase from Rhodococcus opacus 1CP. J Bacteriol. 2009; 191: 4996–5009. 10.1128/JB.00307-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, et al. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli . Appl Environ Microbiol. 2005; 71: 7768–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singh A, Singh Chauhan N, Thulasiram HV, Taneja V, Sharma R. Identification of two flavin monooxygenases from an effluent treatment plant sludge metagenomic library. Bioresour Technol. 2010; 101: 8481–8484. 10.1016/j.biortech.2010.06.025 [DOI] [PubMed] [Google Scholar]
- 52. van Hellemond EW, Janssen DB, Fraaije MW. Discovery of a novel styrene monooxygenase originating from the metagenome. Appl Environ Microbiol. 2007; 73: 5832–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qu Y, Zhang X, Ma Q, Ma F, Zhang Q, Li X, et al. Indigo biosynthesis by Comamonas sp. MQ. Biotechnol Lett. 2012; 34: 353–357. 10.1007/s10529-011-0778-2 [DOI] [PubMed] [Google Scholar]
- 54. Alemayehu D, Gordo LM, O'Mahony MM, O'Leary ND, Dobson AD. Cloning and functional analysis by gene disruption of a novel gene involved in indigo production and fluoranthene metabolism in Pseudomonas alcaligenes PA-10. FEMS Microbiol Lett. 2004; 239: 285–293. [DOI] [PubMed] [Google Scholar]
- 55. Doukyu N, Arai T, Aono R. Effects of organic solvents on indigo formation by Pseudomonas sp. strain ST-200 grown with high levels of indole. Biosci Biotechnol Biochem. 1998; 62: 1075–1080. [DOI] [PubMed] [Google Scholar]
- 56. O'Connor KE, Dobson AD, Hartmans S. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl Environ Microbiol. 1997; 63: 4287–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Oshima T, Kawai S, Egami F. Oxidation of indole to indigotin by Pseudomonas indoloxidans . J Biochem. 1965; 58: 259–263. [DOI] [PubMed] [Google Scholar]
- 58. Pathak H, Madamwar D. Biosynthesis of indigo dye by newly isolated naphthalene-degrading strain Pseudomonas sp. HOB1 and its application in dyeing cotton fabric. Appl Biochem Biotechnol. 2010; 160: 1616–1626. 10.1007/s12010-009-8638-4 [DOI] [PubMed] [Google Scholar]
- 59. Qu Y, Ma Q, Zhang X, Zhou H, Li X, Zhou J. Optimization of indigo production by a newly isolated Pseudomonas sp. QM. J Basic Microbiol. 2012; 52: 687–694. 10.1002/jobm.201100516 [DOI] [PubMed] [Google Scholar]
- 60. Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008; 46: 1254–1263. 10.1086/529198 [DOI] [PubMed] [Google Scholar]
- 61.Williams P, Kay CM. The catabolism of aromatic compounds by Acinetobacter In: Gerischer U, editor. Acinetobacter molecular biology; 2008. pp. 99–118.
- 62. Park SH, Kim JW, Yun SH, Leem SH, Kahng HY, Kim SI. Characterization of beta-ketoadipate pathway from multi-drug resistance bacterium, Acinetobacter baumannii DU202 by proteomic approach. J Microbiol. 2006; 44: 632–640. [PubMed] [Google Scholar]
- 63. Kumar A, Trefault N, Olaniran AO. Microbial degradation of 2,4-dichlorophenoxyacetic acid: Insight into the enzymes and catabolic genes involved, their regulation and biotechnological implications. Crit Rev Microbiol. 2014; 1–15. 10.3109/1040841X.2014.917068 [DOI] [PubMed] [Google Scholar]
- 64. van Berkel WJ, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol. 2006; 124: 670–689. [DOI] [PubMed] [Google Scholar]
- 65. Thotsaporn K, Sucharitakul J, Wongratana J, Suadee C, Chaiyen P. Cloning and expression of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii: evidence of the divergence of enzymes in the class of two-protein component aromatic hydroxylases. Biochim Biophys Acta. 2004; 1680: 60–66. [DOI] [PubMed] [Google Scholar]
- 66. Beltrametti F, Marconi AM, Bestetti G, Colombo C, Galli E, Ruzzi M, et al. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1997; 63: 2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rosic NN, Huang W, Johnston WA, DeVoss JJ, Gillam EM. Extending the diversity of cytochrome P450 enzymes by DNA family shuffling. Gene. 2007; 395: 40–48. [DOI] [PubMed] [Google Scholar]
- 68. Brautaset T, Lale R, Valla S. Positively regulated bacterial expression systems. Microb Biotechnol. 2009; 2: 15–30. 10.1111/j.1751-7915.2008.00048.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Egan SM. Growing repertoire of AraC/XylS activators. J Bacteriol. 2002; 184: 5529–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tropel D, van der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev. 2004; 68: 474–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bustos SA, Schleif RF. Functional domains of the AraC protein. Proc Natl Acad Sci U S A. 1993; 90: 5638–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gallegos MT, Marques S, Ramos JL. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996; 178: 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. baumannii ATCC 19606 was grown in M9 medium containing 0.1% ethanol as carbon source (control). To test the toxicity of indole and indgo, A. baumannii ATCC 19606 was cultivated in M9 medium supplemented with 5 mM indole (indole) or 5 mM indigo (indigo). After incubation for 16 h at 37°C, the cell cultures were serially diluted and 10 μl-drops of selected dilutions were plated onto LB agar plates for viable counts.
(DOCX)
Indigo was produced by wild-type A. baumannii ATCC 19606 (A), ΔiifC(pComIifC) (B), and ΔiifR(pComIifR) (C). In contrast, ΔiifC (D) and ΔiifR (E) cannot produce indigo. These strains were grown in LB broth overnight. Bacteria from 0.5 ml of the overnight cultures was collected by centrifugation, washed with M9 medium, and transferred to fresh M9 medium (50 ml) supplemented with 3.0 mM indole and 0.1% ethanol. Indigo was observed in the medium after 16 h.
(DOCX)
(A) Diagram of the intergenic regions of iifA-iifB, iifB-iifC, iifC-iifD, and iifD-iifE (represented by thick black lines) that were amplified by RT-PCR. (B) The RT-PCR amplification products were analyzed by agarose gel electrophoresis. RT-PCR amplifications were conducted with reverse transcriptase (lanes 1–4) or without reverse transcriptase (negative control; lanes 5–8) using total RNA as the template. Lanes 1, 2, 3, and 4 are the RT-PCR products of the intergenic region of iifAB, iifBC, iifCD, and iifDE, respectively. These cDNA were not amplified in the negative control lanes. Lane M: 100-bp ladder marker.
(DOCX)
Asterisks indicate identical amino acids in IifC, StyA, and StyA2B. The FAD-binding fingerprints GxGxxG, GG, and Dx6G are also shown. IifC showed 64.8% (268/413), 59.8% (241/403), and 47.4% (196/413) identity to MoxY (uncultured bacterium; GenBanK accession No. ABQ12175), StyA1 of Rhodococcus opacus 1CP (GenBanK accession Nos. ACR43973 and AFO70154) and StyA2B of R. opacus 1CP (GenBanK accession Nos. ACR43974 and AFO70155), respectively.
(PDF)
Multi sequence alignment of the DNA binding domains of the IifR of A. baumannii (IifR/Ab), IifR of P. syringae pv. actinidiae (IifR/Ps), AraC of E. coli (AraC/Ec; SWISSPROT accession no. P03021), and XylS of P. putida (XylS/Pp; SWISSPROT accession no. P07859) were carried out manually. This region contains two potential helix-turn-helix (HTH) DNA binding motifs (first and second HTH motifs). Underlined sequences indicate the helical regions.
(PDF)
Ethyl acetate extracts of E. coli DH5α(pQE80L-OXY) (lane 2) and E. coli CY15000(pQE80L-OXY) (lane 3) culture broth were analyzed by TLC. Indirubin (lane 1) and indigo (lane 4) were used as marker.
(DOCX)
E. coli CY15000(pQE80L) (tubes 1–3) and E. coli CY15000(pQE80L-OXY) (tubes 4–6) were inoculated into 3 ml of LB broth containing 1 μl (tubes 1 and 4), 2 μl (tubes 2 and 5), or 3 μl (tubes 3 and 6) of indoline. After incubating for an additional 16 h, the medium turned a deep pink color in the cultures of E. coli CY15000(pQE80L-OXY) (tubes 4–6).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.