A hallmark feature of biological systems is that they are tightly regulated. Whether it is turning genes on and off, controlling cell division, or tuning the activity of enzymes, nature has evolved an intricate array of regulatory measures to ensure that systems can optimally respond to the myriad of environmental queues that determine everything from cell fate to survival. Most often the tuning of an enzyme uses a phenomenon known as allostery, whereby the binding of substrate to one enzyme molecule is coupled to the binding of another molecule. The end result is that binding at one site can influence subsequent binding events at other sites. Thus, the term “allostery,” which is derived from the Greek allos meaning “other” and stereos meaning “structure,” describes the ability of biological molecules to transmit the effects of binding spatially through the protein to other sites. The association of oxygen with tetrameric hemoglobin is the prototypical example (1), and indeed almost every enzyme (2) is allosterically controlled by some ligand. However, is the coupling of spatially distinct events the only way to regulate function? In PNAS, Whittington et al. (3) demonstrate how regulation can arise not only by transmitting binding information spatially but also temporally. This mode of regulation forces a reconsideration of the strategies nature has at its disposal to tune biological systems.
For an enzyme to be tunable, it is well appreciated that there must exist at least two forms of the molecule, one with a high affinity for substrate and the other with low affinity (Fig. 1A). If the relative fraction of molecules occupying the high- and low-affinity states can be adjusted, the enzyme can be mostly unbound under one set of conditions (i.e., more molecules in the low-affinity state), and mostly bound under another (i.e., more molecules in the high-affinity state), thus making the activity tunable. The question is, What strategies will endow an enzyme with such tunability?
Fig. 1.
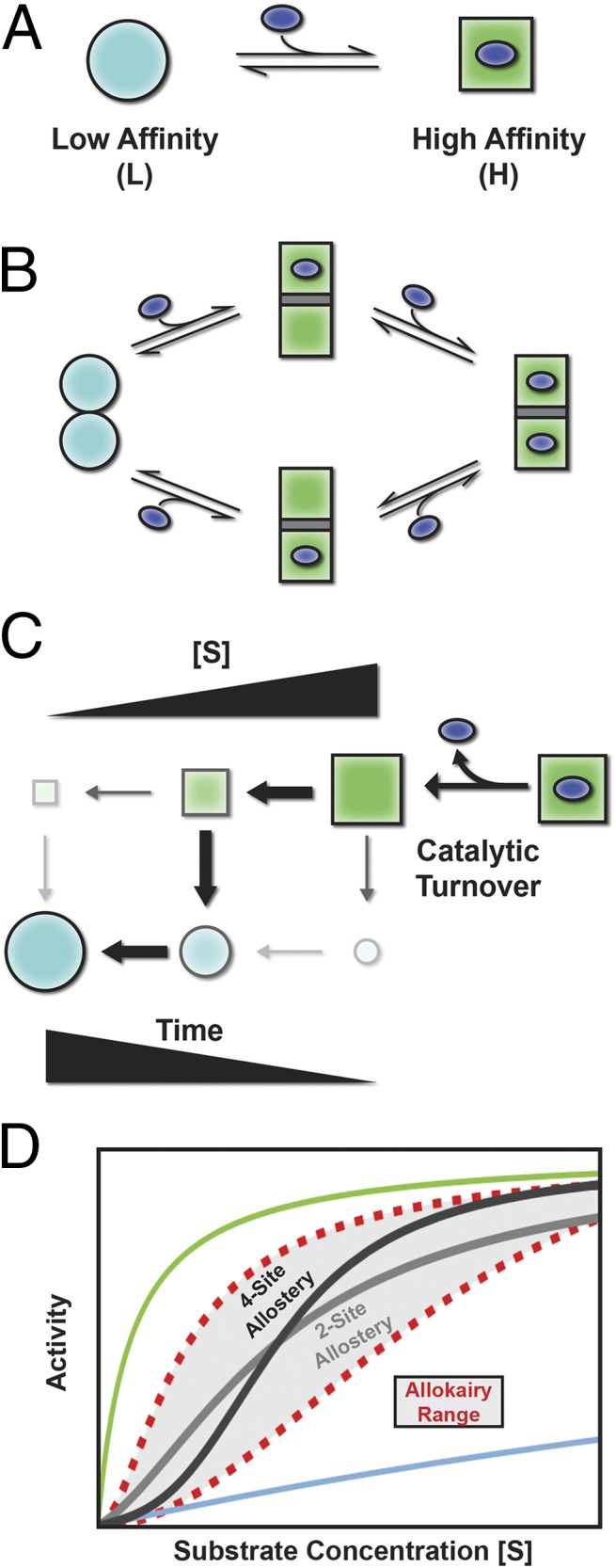
Allosteric and allokairic regulation. (A) Tunability requires that an enzyme can populate at least two states, depicted here as low-affinity (L) and high-affinity (H) with the ligand represented as the blue oval. (B) In allostery, binding and conformational change are coupled. Binding substrate to one molecule of a dimer, for example, converts both to the high-affinity H state, increasing the binding affinity. (C) In allokairy, the enzyme is in the H state after turnover (upper right) and relaxes back to the L state (lower left) determined by the time between turnover and binding, which depends on substrate concentration ([S]). (D) Allostery (two-site dimer, light gray; four-site tetramer, dark gray) and allokairy (red dashed lines) both lead to S-shaped curves when activity is plotted against substrate concentration. The curves for a fully L state or H state are shown in blue and green (top and bottom). Although both regulation mechanisms produce a sigmoidal change in activity, transitioning from the L state at low [S] to the H state at high [S], cooperativity in allokairy is more tunable (gray shading).
For decades, since the existence of regulation was first uncovered (4), the vast majority of regulatory mechanisms relied on the coupling between different sites. This phenomenon is rooted in the very simple principle that if there is a difference in binding affinity between two states, the addition of substrate will stabilize (i.e., make more probable) the state that binds with higher affinity. So, how did nature use this principle to regulate function? By evolving so that the functional unit is an oligomer (i.e., a dimer, trimer, or tetramer, etc.) with all of the copies of the functional units being forced to convert from the low- to high-affinity states together, any stabilization of the high-affinity state caused by substrate binding will be propagated to all other copies. Thus, under low-substrate conditions, all of the oligomers would be in the low-affinity state (Fig. 1B, Left). Addition of substrate would have the dual effect of binding to some of the sites (Fig. 1B, Middle) and also modulating the relative amounts of molecules in the high- and low-affinity states, transforming the remaining empty sites into high-affinity sites. The more substrate that is added, the more sites that become bound and the higher the affinity becomes (Fig. 1B, Right). This principle, the coupling of binding at two spatially separated sites, is at the heart of allostery, and up until now was believed to underlie almost all regulation.
In the late 1960s, however, it was becoming increasingly apparent that the rate of isomerization of an enzyme could also influence regulation, with some enzymes showing a slow response to changes in the concentrations of substrate (5). It was proposed that such “hysteretic” enzymes should display properties that resemble the classic behavior of oligomeric allosteric systems (5–10). However, unlike those allosteric systems, the origin of the effect should not originate from the through-space coupling of different binding sites. The effect should be entirely kinetic, depending instead on a competition between two processes, the binding of the enzyme to more substrate and the relaxation of the enzyme back to its original low-affinity state. For such a system, when substrate concentration is low (Fig. 1C, Left), product release is followed by nearly all of the enzymes relaxing back into the low-affinity state before encountering another substrate molecule. As substrate concentration increases, however, the probability of encountering another substrate molecule after turnover also increases, preventing the enzyme from relaxing back to the low-affinity state (Fig. 1C, Right). The end result is an allosteric-like shift to the high-affinity state with increasing concentration. Aside from establishing a firm theoretical basis, these original studies were essential for having established experimental expectations for hysteretic enzymes.
Whittington et al. (3) provide elegant proof that such hysteretic enzymes do indeed exist. Aside from confirming the original prediction, that even monomeric proteins can display apparent allosteric-like behavior (5, 7), the observation of allosteric-like behavior in a system where allostery is unequivocally absent calls into question how much regulation that seems allosteric is instead kinetic or a combination of allosteric and kinetic. However, the results of Whittington et al. (3) have potentially more far-reaching implications. The fact that disease-causing mutations in glucokinase are associated with disruption of the hysteretic behavior suggests that the effect is functionally relevant and important for regulating glucokinase activity. If this is the case, and the hysteretic behavior is not an artifact but a bona fide regulatory mechanism, optimized through selection, it may be that this new dimension to regulation requires formal distinction. We note that contrary to allostery, which is the transmission of information spatially from one site to another, the glucokinase system reveals that information can also be transmitted temporally (9) to the same, or even a different, site. Indeed, in direct analogy to allostery, this kinetically derived form of regulation is more appropriately termed “allokairy,” which is derived from the Greek allos meaning “other” and kairos meaning “time/event.” In effect, when considering both types of regulation, it would seem that enzymes have evolved strategies to propagate regulatory
The fact that the allokairy and allostery manifest similarly suggests the exciting possibility that allokairy, or regulation through time, may be more prevalent than previously believed.
information through both space and time and, importantly, the effects are manifested in a very similar manner.
So, if nature can use either, what are the functional differences between allostery and allokairy? Which types of systems would use one or the other, or both, mechanisms? In addressing these questions, we note one immediately apparent difference between the two regulatory mechanisms. In allostery, the experimentally observed cooperativity (i.e., the sharpness of the transition between low- and high-affinity states) will depend on how many molecules are involved in the oligomer; the higher the degree of oligomerization, the sharper the transition (Fig. 1D). A limitation of allostery, however, is that the degree of oligomerization is usually fixed (e.g., hemoglobin is a tetramer) and thus the relative sharpness does not vary significantly with conditions. The sharpness of the transition in allokairic systems, however, can be modulated considerably (Fig. 1D) by simply changing the catalytic rate or the rate of interconversion between the high- and low-affinity states. Does nature use allokairy to regulate systems where the degree of cooperativity must be tunable? Do these differences tell us why allokairy, and not allostery, evolved as the regulatory mechanism in glucokinase? The answer to these questions awaits further study. However, the fact that the allokairy and allostery manifest similarly suggests the exciting possibility that allokairy, or regulation through time, may be more prevalent than previously believed.
Acknowledgments
This work was supported by NIH Grant R01-GM63747 and National Science Foundation Grant MCB0446050.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11553.
References
- 1.Perutz MF, et al. Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 2.Changeux JP. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 3.Whittington AC, et al. Dual allosteric activation mechanisms in monomeric human glucokinase. Proc Natl Acad Sci USA. 2015;112:11553–11558. doi: 10.1073/pnas.1506664112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 5.Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970;245(21):5788–5799. [PubMed] [Google Scholar]
- 6.Rabin BR. Co-operative effects in enzyme catalysis: a possible kinetic model based on substrate-induced conformation isomerization. Biochem J. 1967;102(2):22C–23C. doi: 10.1042/bj1020022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ainslie GR, Jr, Shill JP, Neet KE. Transients and cooperativity. A slow transition model for relating transients and cooperative kinetics of enzymes. J Biol Chem. 1972;247(21):7088–7096. [PubMed] [Google Scholar]
- 8.Ricard J, Meunier JC, Buc J. Regulatory behavior of monomeric enzymes. 1. The mnemonical enzyme concept. Eur J Biochem. 1974;49(1):195–208. doi: 10.1111/j.1432-1033.1974.tb03825.x. [DOI] [PubMed] [Google Scholar]
- 9.Neet KE. Cooperativity in enzyme function: Equilibrium and kinetic aspects. Methods Enzymol. 1980;64:139–192. doi: 10.1016/s0076-6879(80)64009-9. [DOI] [PubMed] [Google Scholar]
- 10.Neet KE, Ainslie GR., Jr Hysteretic enzymes. Methods Enzymol. 1980;64:192–226. doi: 10.1016/s0076-6879(80)64010-5. [DOI] [PubMed] [Google Scholar]


