Significance
Seed weight is a complex trait controlled by polygenes, and its underlying regulatory mechanisms, especially those involving polyploidy crops, remain elusive. Brassica napus L., which is the second leading crop source of vegetable oil around the world, is an important tetraploid (4×) crop. Our results have generated three significant findings. (i) By combining the linkage and associated analysis, this study revealed the first (to our knowledge) quantitative trait locus (QTL) in rapeseed, which will provide insights for QTL cloning in polyploidy crops. (ii) The functional gene and marker could be useful in rapeseed breeding. (iii) We revealed a maternal regulatory pathway affecting seed weight that differs from the mechanisms described in previous reports.
Keywords: seed weight, silique length, ARF18, cell growth, maternal effect
Abstract
Seed weight (SW), which is one of the three major factors influencing grain yield, has been widely accepted as a complex trait that is controlled by polygenes, particularly in polyploid crops. Brassica napus L., which is the second leading crop source for vegetable oil around the world, is a tetraploid (4×) species. In the present study, we identified a major quantitative trait locus (QTL) on chromosome A9 of rapeseed in which the genes for SW and silique length (SL) were colocated. By fine mapping and association analysis, we uncovered a 165-bp deletion in the auxin-response factor 18 (ARF18) gene associated with increased SW and SL. ARF18 encodes an auxin-response factor and shows inhibitory activity on downstream auxin genes. This 55-aa deletion prevents ARF18 from forming homodimers, in turn resulting in the loss of binding activity. Furthermore, reciprocal crossing has shown that this QTL affects SW by maternal effects. Transcription analysis has shown that ARF18 regulates cell growth in the silique wall by acting via an auxin-response pathway. Together, our results suggest that ARF18 regulates silique wall development and determines SW via maternal regulation. In addition, our study reveals the first (to our knowledge) QTL in rapeseed and may provide insights into gene cloning involving polyploid crops.
The rapid growth of the world population has increased the global requirement for food, which in turn warrants significant improvement in crop grain yield. As one of the three direct factors influencing crop grain yield, seed weight (SW) has been widely accepted as a complex trait that is controlled by polygenes. Therefore understanding the genetic and molecular basis of SW is extremely important for crop-improvement programs.
The size of seeds is influenced by a variety of cellular processes (1). In Arabidopsis, some mutants such as ap2, arf2, da1, eod3, ttg2, and klu control seed size mainly by regulating cell elongation in the integument surrounding the seed (1–5). In mini3, iku1, iku2, and shb1 mutants, premature cellularization or proliferation of the endosperm in the early phase of seed development affects seed mass (6–10). The met1 gene has been determined to have parent-of-origin effects on seed size because of the loss of methylation in cytosine residues in CG islands (11). In rice, a total of 47 quantitative trait loci (QTLs) for grain length and 48 for grain width have been identified (12). Recent studies have shown that certain genes such as GW2, GIF1, qSW5, GS3, GS5, GW8, and qGL3 regulate grain size (13–20). Among these, GW2 and qSW5 were determined to regulate grain weight by increasing cell number in the outer glume, whereas the others affected grain weight by directly regulating cell division and/or cell expansion of grain. Despite this progress, no genes responsible for SW have been identified in polyploid crops.
Polyploidy is produced by the multiplication of a single genome (autopolyploid) or the combination of two or more divergent genomes (allopolyploid), which commonly occurs in flowering plants, including many important agricultural crops (21). The complexity of polyploid genomes results in difficulties in QTL localization such as the inaccuracies caused by homologous sequences from different chromosomes and the interactions between homolog genes. For example, Brassica napus L. (AACC), the world’s second leading crop source of vegetable oil following soybean, is a tetraploid (4×) species containing two ancestors, namely, Brassica rapa and Brassica oleracea, both of which underwent whole-genome triplication (22). B. rapa harbors the Brassica A genome and is closely related to B. oleracea, which contains the Brassica C genome (23). Rapeseed is an important global agricultural crop that has recently gained attention in the field of plant genomics. Despite the identification of more than 80 QTLs for SW across the 19 chromosomes of the B. napus genome (24–27), no genes associated with these QTLs have been identified.
We previously examined the F2 populations derived from a cross between the rapeseed lines zy72360 and R1 and detected a major QTL for SW. This QTL was localized to chromosome A09 and explained ∼30% of the phenotypic variation observed in SW. Interestingly, other rapeseed lines also harbored this QTL, as detected by Yang et al. (26) and Li et al. (27). In the present study, we used map-based cloning and targeted-region association to clone and characterize the gene related to the major QTL for SW. We further elucidated the possible mechanism regulating SW to provide critical information for breeding high-yield crops.
Results
Fine Mapping of the Major SW-Regulating Gene on Chromosome A9.
In the present study we used B. napus lines zy72360 and R1, which showed significant differences in SW (zy72360: 5.53 ± 0.43 g; R1: 3.86 ± 0.25 g) (Fig. 1A) and silique length (SL) (zy72360: 85 ± 5.4 mm; R1: 60 ± 6.5 mm) (Fig. 1B). First, we mapped the SW QTL between markers TSNP08 and TSNP149 within the F2 population. With newly developed markers based on insertions and deletions (indels) and SNPs in the region of A09 between zy72360 and R1 (Fig. 1C and Table S1), we localized the SW QTL to a high-resolution linkage map by progeny testing of homozygous recombinant plants (BC4F2) and narrowed the SW locus to a 147-kb region between markers TSNP13 and TSNP16 (Fig. 1D). Based on the SNP information on the 147-kb region between zy72360 and R1, we further narrowed the distance to 120 kb by PCR confirmation of the recombinant lines (Fig. 1E).
Fig. 1.
Fine mapping of the SW QTL in rapeseed using NILs and association populations. (A) SW in parents of zy72360 and R1. (Scale bar, 0.5 cm.) **P = 0.01. (B) SL in parents of zy72360 and R1. (Scale bar, 0.5 cm.) **P = 0.01. (C) The SW locus was detected on chromosome A09 in the F2 population. Positional cloning narrowed the SW locus to a 1.1-Mb region between P534 and BrBAC248. (D) Testing of six recombinant plants (BC3F4) narrowed the SW locus to the region between markers TSNP13 and TSNP16 (147 kb). The seeds from the main inflorescence were used to determine the SW for each plant. Open bars indicate the S1 homozygous regions, gray bars indicate the S2 homozygous regions, and black bars indicate the heterozygous regions. The SWs of the progenies were significantly different. (E) PCR identification narrowed the SW locus to the region between SNP3 and SNP5 (120 kb). (F) Scanning of the association of SW with 11 marker loci on the A09 linkage group in a rapeseed association population. Eleven maker loci are arranged on the horizontal axis according to their physical positions on chromosome A9.
Table S1.
Molecular marker primers used in fine mapping and association analysis
| Primer name | Forward primer (5′→3′) | Reverse primer (5′→3′) |
| Fine mapping | ||
| niab018 | ACAATTGTTCTAGGCGTTCAAA | ATCAGTATCCTGGTTCTGG |
| EST-45 | CGATTCTTGATTCACGGGTTT | TATGAGGCTCGCTTCACGTCG |
| SF32915 | TCCTCAGCCACCAAGGTTAC | CGATGAAAAGCTTTAAACGGA |
| P515(Br231) | TTCGATTGTGGTTCTTATTGG | CCATATTTATCTTAGTTCTCGCC |
| BrBAC256a | ATCATTGCAACGGTAGCCTC | GCGGGTGTTAGTTTATCCGA |
| EST013 | ACGAGCAAGAGTGCTGCC | TCGTAGCTTCTGTCTCTTCGC |
| In/del201 | CGTTGAGTTCTTGGACTTTAC | CAGACCAAGTAAGTCAACTGA |
| SF0145 | TGAGCTTGTCTCGTCAATTAGG | TGGATATGAATTTTCCACGCT |
| SF1402 | GATCAACCAATCAATCGCCTT | TTGCTCATGTCCAAATCTGTCT |
| BrBAC275 | CAGATGGGGCCAAGTTACAT | CATTCTTCACCAACACACCG |
| P530 | TGCACTAAAATAGCTGCAAAGA | GGTTTACAAGACGTCGAAGAAG |
| M42189(Br0188) | GAGCAGCAGAACCAAAGGAC | ATCATGACGATGACAAGCCA |
| P534 | TCGGGATCCTGAACAGTG | ATGCAGCAGAGTAGCGGA |
| ns380 | TTGGACCATTTTGCTAGTTTAGTAAACAGTGGAAT | GATGATGATGATATTGGGGACTAACCAGGATT |
| BrBAC248 | AAGTGGCGTCAATGTGTCAA | TCTACACAAAGCACAATGCCA |
| BrBAC229 | AAAAACATAAAGACAACTTTGATGAC | GAGATTTTGACAAACGACTTGAAA |
| SF12680 | AGCAGGAACAGCTCGAACAT | CTGGTGTCCCAACTGTTGAA |
| M18(SF023) | CACAAGCATTCTACCATAGCAAAGTC | TGCACATATGGCATGTTGTTTG |
| SNP011 | AGATCGTCAAGAAGAAGAATAAGA | AGATTTTGAGGGTTAGTCCGAAGTT |
| GACAGACCTTTTAGTTACTTTATTTACAC | GCTCCGAGGGGTTTCTTTTCTTT | |
| SNP104 | TTGCATCTTCAAGTGTGCTTGCA | GTCAGGGAAAATCAGTACTTG |
| ATTTGAATTGCT/CAAGCACTTTTAGCGT (Taqman probe) | ||
| SNP073 | GTCAAGTTGGGGAATATCATCTTG | TAACTCAGAGATTTAGTTATCGG |
| ATCGCTGTAAGTTCTCGAAAAACTA | ACAAGCGCCCATGACAATCG | |
| TSNP08 | GAGGAACTGCTTCCTCCAGAAG | GCATTTCTACATCAGTGATCTTGAG |
| CCCTAAATGTG/ATTGAGGCCTCTGG (Taqman probe) | ||
| TSNP10 | CGATCCGAACCGAACTCGAA | CCAGCCTAAAGTAGACCATAG |
| CGAATTATCCG/AGACCGAAACC (Taqman probe) | ||
| NS380 | TTGGACCATTTTGCTAGTTTAGTAAACAGTGGAAT | GATGATGATGATATTGGGGACTAACCAGGATT |
| TSNP149 | AGTCGTAATTCAATCATCTCATG | CCATATATGATCAACTATTTCGAAG |
| CAATCATCTCATGACCTTTTTC/TCCTGC (Taqman probe) | ||
| TSNP18 | GGAACCGGGATTGTCGACTT | ACTCTGCAAATGCAGACGGC |
| GTATGTAAGTGGAC/TTAGATTCACGACA (Taqman probe) | ||
| TSNP09 | GAATCATCGACGATTAACCATGC | AGGAGCGGCCAAGGTACTGAAG |
| GCTCCTTACCGCG/AAACAACCTG (Taqman probe) | ||
| TSNP10 | GGTGACGCACATGCACGATGG | CATTGGAGTATATCTCACCTC |
| TTCAC/TCTCCAAAATAGGGGAACTCTATA (Taqman probe) | ||
| TSNP11 | CCAAGCAAATAATCGTGC | GATGGTGACGAAGAAGAG |
| AATTTTGGGATCTA/TGCTCATCCTTG (Taqman probe) | ||
| TSNP12 | GGATCACTGTTTCGGGCTGTT | TTTCCCTCCTGTGAAGGTGTAGT |
| CCATCTGAGGAGTTCA/GAAAGCAAGTAC (Taqman probe) | ||
| SNP025 | CTTCTTACGGAAGAGGAGTCCT | GCTTAGCTGTTAGACTGTCTGAACT |
| CTCCTTCTTTGAGGCACCAGC | TCCTCCTCCGGAGAAGTATGG | |
| TSNP13 | CAGTTGGTCGGTCTGTGGAA | CGGTTCGCTGCGCTTGTC |
| CTTC/GGGTAACAGCAAACGGAAAGGAG (Taqman probe) | ||
| In/del032 | GAGGTACCACTACTTTTCCTG | ATGTACACACTTACCTAGGCTTG |
| TAGGCAGAGCATGACACAGATGAAG | GTTTGAGAATAGCTAATAGTACCT | |
| TSNP16 | GGAAGTACCATTTATCGTTAA | CACACATGAACGTGTAAG |
| TSNP23 | CGAAGCTCTCGACAACGCCC | CGAAGCTCTCAGCAACGCCC |
| SNP680 | GCCAAGTTGAAAGTGGATACC | CGTTGTGAGTACTACTAAGTC |
| CCATTTACCAAGTGATCCAGACTATC | TGTATAAACTTTCAACTTCGC | |
| SNP1 | CCAATAGAGCCACGAATCCTGA | GCCGTACTTATGTTCTTGTGC |
| SNP2 | TTCAGCTTTGAACCCACCGC | GAAACGACCACTGGCGTCTGC |
| SNP3 | GTGTTCTTGAGGTACCACTAC | CTGAATGGTTAACTGAACCTGC |
| SNP4 | CCATTTAACCCCACATGAGTC | ATGGCTTAGAAGATGACGATG |
| SNP5 | ACCGATTTAAGCGCACGGCTC | AGAGGAACAAGGAGCCGAAG |
| SNP6 | ACACGTTCGTCATTCATTAACT | CCTATGAGTTTCGTAAGCGTT |
| SNP7 | CTAGACTTTTCATCATAGCCAT | TAGCGACGCATCGATCGTCTG |
| SNP8 | TAAGCAGGCCTTCTCATACTC | CTGTTATTGTCATTATTGCTAG |
| Association analysis | ||
| SSR-90 | CTCGGATCACGCTAAGCATT | TTCTCTTTCCCTTTTCAATTCC |
| SSR-72 | ACACCGGATTTCATAGCTCG | TGCAGAAGACGAGTCCAGTG |
| TSNP12 | GGATCACTGTTTCGGGCTGTT | TTTCCCTCCTGTGAAGGTGTAGT |
| CCATCTGAGGAGTTCA/GAAAGCAAGTAC (Taqman probe) | ||
| SNP025 | CTTCTTACGGAAGAGGAGTCCT | GCTTAGCTGTTAGACTGTCTGAACT |
| In/del032 | GAGGTACCACTACTTTTCCTG | ATGTACACACTTACCTAGGCTTG |
| SNP322 | GGGTATCCAAAGAGAATACTCA | GTGAAAGAGAGAAGAGATGCC |
| In/del637 | GTTTAAACATAGTACAACGACCATAT | CTGAATCATTGAATTAACCAACTAATATA |
| SSR-25 | TTGTTCCTAACAAACGGAAAA | AAAAGAAACAGCCCAGCTCA |
| SNP049 | GGAAGAAGAAGAAGTTTAAC | CCTCGCCTCTACGACTACATCC |
| SSR-87 | AAAATAACAATATGAAAGCCAAAAC | GGATGATTTCGTCGTCTGGT |
| SSR-89 | TCAACTTGACATGTTCACTAATAGT | CAATAGACACGGAAATGGGCCATT |
To narrow the distance of the SW QTL further, association analysis was conducted using 380 lines that showed a large range of phenotypic variation for SW. Scanning of the association of SW with 11 QTL-linked markers on the A09 linkage group generally displayed an obvious peak between SSR-72 and SSR-89 (Fig. 1F). Using the results of fine mapping and targeted-region association, we narrowed the distance to 50 kb and identified seven putative ORFs in this region. Irrespective of the near isogenic line (NIL) or association population, the SW and the SL genes were colocated within the QTL region; The phenomenon might be induced by pleiotropism in this QTL.
ARF18 Decreases SW and SL in Rapeseed.
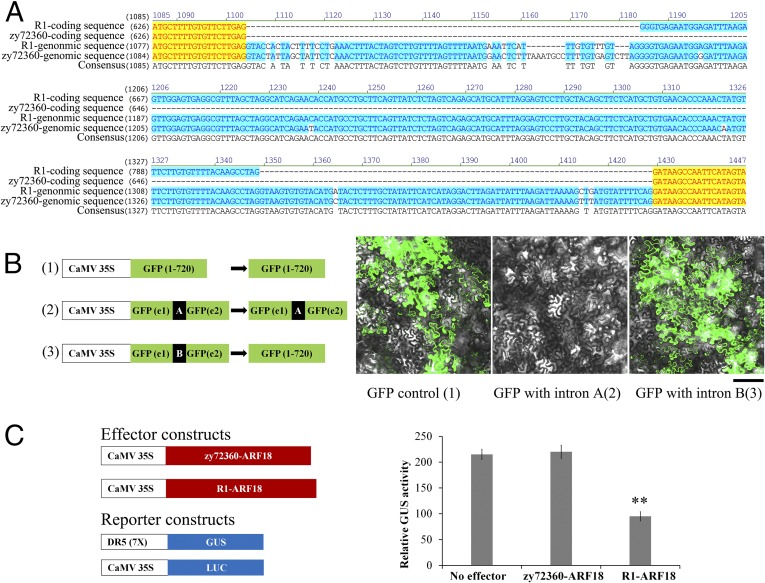
Based on the genome sequence of rapeseed (www.ncbi.nlm.nih.gov/assembly/GCA_000686985.1/), we cloned the seven genes including the upstream regulatory and coding regions from the parents zy72360 and R1 (Table S2). Sequence comparison showed that three genes, namely, BnaA09g55580D, BnaA09g55560D, and BnaA09g55520D, showed differences in amino acids, and the upstream regulation sequence of BnaA09g55570D showed SNPs and indels between the parents. These four genes were chosen for functional identification by overexpression in Arabidopsis under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Except for the BnaA09g55580D-overexpressing transgenic lines, these genes showed no change in SW (Fig. S1). Therefore, BnaA09g55580D was implicated as a candidate gene for SW. Genome and cDNA sequence analysis of R1 showed that the BnaA09g55580D gene comprised 12 exons and 11 introns. BnaA09g55580D homologs from B. rapa and Arabidopsis thaliana were detected in the National Center for Biotechnology Information (NCBI) database and were annotated as auxin-response factor 18 (ARF18) proteins; therefore, we named the BnaA09g55580D gene “BnaA.ARF18.a” based on the standard nomenclature of Østergaard and King (28). Comparison of the ARF18 coding sequences in zy72360 and R1 showed that, in addition to differences in a 6-bp indel site and four SNPs inducing changes in four amino acids, the seventh exon (165 bp) in zy72360 was lost (Fig. 2A and Fig. S2).
Table S2.
Primers used for cloning seven candidate genes including the upstream regulatory and coding regions
| Gene name | Annotation | Primer name | Primer sequence (5′-3′) |
| BnaA09g55580D | Auxin-response factor 18 (ARF18) | 580F1 | GTCTATTTAGGAGAATTGCCCAT |
| 580R1 | CATGCTCTGACTAGAGATAACTG | ||
| 580F2 | GAGGTACTATTAGCTATTCTCAA | ||
| 580R2 | CCTAATGACGATTAATCATCCGCT | ||
| 580cdsF | ATGGCGAATGTAGATGGAGATG | ||
| 580cdsR | CTCTCACACCTAATGACGATTAA | ||
| BnaA09g55570D | Zinc finger transcription factor of the Dof family involved in the control of seed germination | 570F | GGGTATCCAAAGAGAATACTCA |
| 570R | GTGAAAGAGAGAAGAGATGCC | ||
| 570cdsF | ATGATAAACGTAAAACCAATGGA | ||
| 570cdsR | TCACCACGAAGATCCTCCT | ||
| BnaA09g55560D | P-loop containing nucleoside triphosphate hydrolases superfamily protein | 560F1 | AACACGCCTGAGGCTACCA |
| 560R1 | ATCTTCTCCTCGTTTCCGG | ||
| 560F2 | ACAGTCTCGGAGGAGCTGGA | ||
| 560R2 | CACAGCCAATCACAGATCTC | ||
| 560cdsF | ATGGGGATTCTCCGGAACTC | ||
| 560cdsR | CTACGGCTGAGATACCATTG | ||
| BnaA09g55550D | Unknown protein | 550F1 | CTAGTCTATCAAACCTACGTAC |
| 550R1 | GGCGCAACCTCCCAAGATCCCAG | ||
| 550F2 | TCCTCGCACCACCCATCGT | ||
| 550R2 | TCAACAACACTCTTAGACATGTG | ||
| BnaA09g55540D | Unknown protein | 540F | ATCACAATGTATCAATTGTG |
| 540R | TCTACGCTGGTCCATGTATA | ||
| BnaA09g55530D | Cytochrome p450 78a9 (CYP78A9) | 530F1 | TTTTAGGAAGGCGACGAGTAT |
| 530R1 | CTCTGGGTACGTTACGTACACATGT | ||
| 530F2 | CACCGTGTCAGTTCCTCTGAAT | ||
| 530R2 | CACCTGAAGGCAGCAATTTCATCTCA | ||
| BnaA09g55520D | Homeobox 12 (ATHB12) | 520F1 | CGAAACTAGCCAAACTCGTAAC |
| 520R1 | ATTCTCAGAGCCAACTACAATC | ||
| 520F2 | GTTTAAACATAGTACAACGACCATAT | ||
| 520R2 | CTGAATCATTGAATTAACCAACTAATATA | ||
| 520cdsF | ATGGAAGAAGGAGACGTTT | ||
| 520cdsR | TCATGACCAAAAGTCCCACCA |
Fig. S1.
SW analysis in transgenic Arabidopsis lines overexpressing BnaA09g55580D (A), BnaA09g55570D (B), BnaA09g55560D (C), and BnaA09g55520D (D). **P < 0.01.
Fig. 2.
Structural characterization and functional identification of ARF18. (A) ARF18 structure and mutation sites, including nucleotide substitutions and deletions, in zy72360 and R1. (B and C) Comparison of SW (B) and the SL (C) in Arabidopsis (vector control) and ARF18 transgenic lines in Arabidopsis. (Scale bars, 0.5 mm in B and 1 mm in C.) All data are expressed as mean ± SD. (D and E), Comparison of SW (D) and SL (E) in the zy72360 line with a promoter-driven expression of R1-ARF18. (Scale bars, 0.5 cm.) All data are expressed as mean ± SD. **P = 0.01.
Fig. S2.
Comparison of the ARF18 CDS sequences from zy72360 and R1.
A total of 30 independent homogenous T3 transgenic Arabidopsis lines with overexpression of ARF18 from R1 and zy72360, respectively, were analyzed. Among these, 10 ARF18-overexpressing lines from R1 showed phenotypes distinct from that of the WT line. For example, in the two lines with high ARF18 expression levels, R1-ARF18-5 and R1-ARF18-8, seed sizes were smaller (thousand kernel weight: 17.6 ± 0.38 mg and 17 ± 0.4 mg, respectively) than in the WT line (thousand kernel weight: 19.6 ± 0.34 mg) (Fig. 2B and Fig. S3A). On the other hand, overexpression of ARF18 in zy72360 did not affect seed size (thousand kernel weight: 19.3 ± 0.41 mg and 19.7 ± 0.5 mg, respectively), as compared with that of WT plants (Fig. 2B and Fig. S3A). The transgenic R1-ARF18-5 and R1-ARF18-8 plants also showed significantly shorter siliques (13.62 ± 0.4 mm and 13.38 ± 0.34 mm, respectively) compared with the plants containing ARF18 from zy72360 (15.35 ± 0.39 mm and 15.54 ± 0.35 mm, respectively), which yielded siliques similar in length to those of WT plants (15.4 ± 0.41 mm) (Fig. 2C). The Arabidopsis ARF18 mutant was planted also. Compared with WT Arabidopsis, the mutant generated larger seeds (21.2 ± 0.52 mg) and longer siliques (17 ± 0.4 mm) (Fig. S3 C and D).
Fig. S3.
(A) Real-time PCR analysis of ARF18 expression levels in the siliques of transgenic Arabidopsis lines and WT Arabidopsis control. (B) Real-time PCR analysis of ARF18 expression levels in the siliques of transgenic rapeseed lines overexpressing R1-ARF18 gene and zy72360 control. Data presented are mean values of three biological replicates; error bars indicate SDs. (C and D) Comparison of SW (C) and SL (D) in the Arabidopsis control and the arf18 mutant (CS719283). (Scale bars, 0.5 mm in C; 1 mm in D.) All data are shown as mean ± SD. **P < 0.01.
Based on these findings, ARF18 and its upstream regulatory region (the 1.6 kb that included the 5′ noncoding region) that originated from R1 was transfected into rapeseed line zy72360 by a hypocotyl transgenic system of Agrobacterium. A total of 25 independent homogenous T2 transgenic lines were used for analysis of phenotypes, including SW and SL. The SW in more than 30% of these transgenic lines decreased by at least 15% (Fig. S4A). For example, in the two transgenic lines R1-ARF18-BnT5 and R1-ARF18-BnT11, which had high expression levels of R1-ARF18, the SW decreased (4.61 ± 0.3 g and 4.81 ± 0.22 g, respectively) compared with that of the zy72360 control (5.50 ± 0.38 g) (Fig. 2D and Fig. S3B). The SL also shortened in these two lines (70 ± 4 mm and 72 ± 3 mm, respectively) compared with the zy72360 control (86 ± 6 mm) (Fig. 2E).
Fig. S4.
(A) Analysis of SW and SL in rapeseed lines overexpressing R1-ARF18 showing obvious phenotypes. *P < 0.05; **P < 0.01. (B) Analysis of SW and SL in transgenic Arabidopsis overexpressing R1-ARF18 (without exon 7). Values are shown as means ± SD. (C) Prediction of ARF18 protein structure using SWISS-MODEL. ARF18 from zy72360 was predicted to be a monomer, and ARF18 from R1 was predicted to be a homodimer. (D) SW of NIL (SW) × NIL (SW) F1, NIL (SW) × R1 F1, R1 × R1 F1, and R1 × NIL (SW) F1. Values are shown as means ± SD.
Incorrect Splicing of the Sixth Intron Induces Nonfunctional ARF18 That Originated from zy72360.
A search inquiry of the NCBI database identified two BnaA09g55580D homologs in B. rapa and one in Arabidopsis. A hypothetical protein in Capsella rubella was identified also. These proteins have high amino acid sequence identities (B. rapa 1: 86%; B. rapa 2: 99%; A. thaliana: 81%; C. rubella: 80%) with ARF18 of B. napus (rapeseed) (Fig. 3A). Thus far, none of these homologous genes has been characterized functionally. Previous studies have shown that ARFs act as transcription factors and comprise three domains: an N-terminal DNA-binding domain (DBD), a C-terminal dimerization domain (CTD) similar to domains III and IV of the Aux/IAA family of proteins, and the middle region (MR), which determines that the ARF protein functions as an transcriptional activator/repressor of auxin-responsive genes (Fig. 3A). Subcellular localization in tobacco showed that R1-ARF18 localized to the nucleus (Fig. 3B).
Fig. 3.
Comparative analysis of ARF18 proteins from zy72360 and R1. (A) Amino acid alignments of ARF18 proteins from other species with rapeseed ARF18. The black lines indicate the three conserved domains. (B) Nuclear localization of the ARF18-GFP fusion protein in tobacco epidermal cells. Cells transformed with the plasmid were viewed under a fluorescent filter to show GFP (Left), under a bright field for cell morphology (Center), and in combination (Right). (Scale bar, 50 μm.) (C) Putative conserved domains detected in ARF18 proteins from the alleles of zy72360 and R1.
Comparison of the parental ARF18 proteins showed that the 55-aa deletion in ARF18 of zy72360 was located between the DBD and the MR (Fig. 3 A and C). We deleted the 165-bp segment in ARF18 from R1 by fusion PCR and overexpressed it in Arabidopsis. The transgenic lines showed normal phenotypes, suggesting that the 55-aa deletion in ARF18 generated a nonfunctional protein (Fig. S4B). To corroborate that the 165-bp deletion in zy72360 was induced by sequence variation, two GFP reporter vectors driven by a CaMV 35S promoter were constructed. The sixth intron of R1 (69 bp) or the sequence from zy72360 corresponding to the sixth intron site in R1 (80 bp) was inserted into the middle of GFP gene (Fig. 4 A and B). Detection of transient expression in tobacco leaf (Nicotiana tabacum L.) showed that only the vector inserted with the intron of R1 displayed green fluorescence, indicating that the intron was shared (Fig. 4B).
Fig. 4.
Identification of incorrect splicing in ARF18 from zy72360 and comparison of inhibitory activity in ARF18 alleles. (A) Comparison of the ARF18 partial CDS and genomic sequences originating from zy72360 and R1. (B) The sixth intron from R1 and the corresponding sequence from the zy72360 gene were inserted into the GFP gene, which was driven by the CaMV 35S promoter and transformed into tobacco leaves. A is the intron from zy72360, and B is the intron from R1. (Scale bar, 100 μm.) (C) Effector genes that consisted of the CaMV 35S promoter encoding full-length ARF18 proteins were cotransfected into Arabidopsis suspension cell protoplasts with a DR5(7×)-GUS reporter gene. GUS activities were measured from protoplasts treated with 10 μM NAA. The data are representative of three independent experiments. All data are expressed as mean ± SD. **P = 0.01.
Previous reports have shown that Q-rich MRs in ARFs function as activation domains (29). This finding suggests that ARF18 might function as a repressor because it does not possess a Q-rich MR. To confirm our inference, we detected the transcriptional activity of ARF18 by using a protoplast assay system. In this system, ARF18 genes from zy72360 and R1 were fused with the CaMV 35S promoter as effector plasmids (Fig. 4C), and the β-glucuronidase (GUS) gene under the control of auxin-responsive direct repeat DR5(7×) was used as reporter. Luciferase (LUC) driven by the CaMV 35S promoter was used as internal reference. After the plasmids were introduced into the protoplasts, GUS activity was measured to determine ARF18 transcriptional activity. Fig. 4C shows that under the N-acetyl-aspartate (NAA) treatment the effector gene encoding the full-length ARF18 repressed the transcription of GUS, whereas the effector gene from zy72360 did not differ significantly from that of the control without an effector.
ARF18 from zy72360 Lost Its Binding Activity Because of Failure in Forming a Homodimer.
Because the 55 amino acids were shared between the DBD and the MR, it was essential to determine which domain had lost its activity. To identify the inhibition activity of the ARF18 proteins, the allelic genes were constructed into PGBKT7 as bait vectors and were cotransformed with PGADT7 into yeast. PGBKT7-VP16 was used as positive control. The empty vectors PGBKT7 and PGADT7 were used as negative control. The results showed that the ARF18 proteins from the two parents had nearly the same inhibition activity, suggesting that the partial deletion in the MR region did not affect the inhibition activity of ARF18 from zy72360 (Fig. 5A).
Fig. 5.
Comparison of inhibitory and binding activity of two ARF18 proteins. (A) Detection of ARF18 transcriptional activity in a yeast one-hybrid system. ARF18 genes from zy72360 and R1 were constructed as bait vector. +, positive control; −, empty control vector. (B) Detection of interaction in ARF18 proteins by yeast two-hybridization. (C) EMSA indicating that ARF18 from R1 binds to the biotin-labeled DR5 probe. The free probe is visible at the bottom of the gel, and complexes with the ARF18 protein are shifted upward.
The structures of the ARF18 proteins were predicted by using SWISS-MODEL. The results showed that ARF18 from R1 could form homodimers, whereas the protein from zy72360 could form only monomers (Fig. S4C). Yeast two-hybridization showed that R1-ARF18 interacted with itself, but zy72360-ARF18 did not self-interact (Fig. 5B). The revised R1-ARF18 (R1-ARF18r) in which the 55 amino acids were deleted also failed to interact with itself. To detect further differences in binding activity between the two ARF18 proteins, gel mobility shift assays using a labeled DR5(7×) probe were performed. The results showed that under the same conditions R1-ARF18 bound the DR5(7×) probe as expected, but zy72360-ARF18 failed to bind the sequence (Fig. 5C). Based on these results, we concluded that the full-length ARF18 protein from R1 acts as a repressor in regulating transcription, and ARF18 with the exon deletion from zy72360 lost that activity because of its failure to form dimers.
ARF18 Is Expressed Differentially in Various Tissues and Is Expressed Predominantly in the Silique Wall.
Quantitative RT-PCR (qRT-PCR) analysis of various organs of the NIL (SW) and R1 lines indicated that ARF18 was expressed differentially in the root, leaf, stem, bud, silique wall, and ovule. At 25 d after flowering (daf), the expression levels of ARF18 were significantly higher in the silique walls than in the other tissues; the stems and seeds showed very low transcript levels of ARF18 (Fig. 6A). No differences in ARF18 expression level in the two lines were observed, suggesting that the sequence change in the coding region accounted for the functional variation in the two alleles. Further expression analysis was performed using GUS fusion studies in which the ARF18 promoter (promoter length, 1.6 kb) was ligated into pCXGUS-P and transformed in Arabidopsis. The results of ARF18 promoter-GUS expression analysis are shown in Fig. 6B. The seedlings, older leaves, and silique walls of transgenic plants showed high levels of GUS expression. Moderate expression levels were detected in the root, stem, and floral organs, including the stamen and pistil. In the developing seeds, moderate levels of ARF18 expression were observed in the embryo but not in the seed coat. GUS expression driven by the ARF18 promoter basically is consistent with the results of RT-qPCR analysis.
Fig. 6.
ARF18 expression patterns by RT-qPCR and GUS assays. (A) Analysis of ARF18 expression in selected tissues in R1 using RT-qPCR. ENTH was used as the reference. Data are expressed as the means of three biological replicates; error bars indicate SDs. (B) GUS expression patterns(blue staining) at various stages in the ARF18 promoter-GUS transgenic line. GUS was strongly expressed in the cotyledon (cn), seedling (sl), older leaves (lf), and silique wall (sw). Moderate expression levels were detected in the bud (bd) and embryo (sd). No expression was observed in the seed coat (sd). (Scale bars, 5 mm for cotyledon, seedling, and older leaves; 0.8 mm for silique wall and bud; 0.1 mm for embryo.)
ARF18 Might Affect SW by Regulating the Development of the Silique Wall.
To investigate further the mechanism by which ARF18 influences SW, reciprocal cross experiments between NIL (SW) (ARF18−) and R1 (ARF18+) were performed. Pollinating NIL (SW) plants with R1 pollen led to the development of ARF18−/+ embryos within an ARF18−/− seed coat. Similarly, pollinating R1 plants with NIL (SW) pollen led to the development of ARF18+/− embryos within an ARF18+/+ seed coat. No obvious differences existed in the weight between the resulting seeds and the corresponding self-pollinated seeds; this result was suggestive of maternal control of SW in NIL (SW) plants (Fig. S4D).
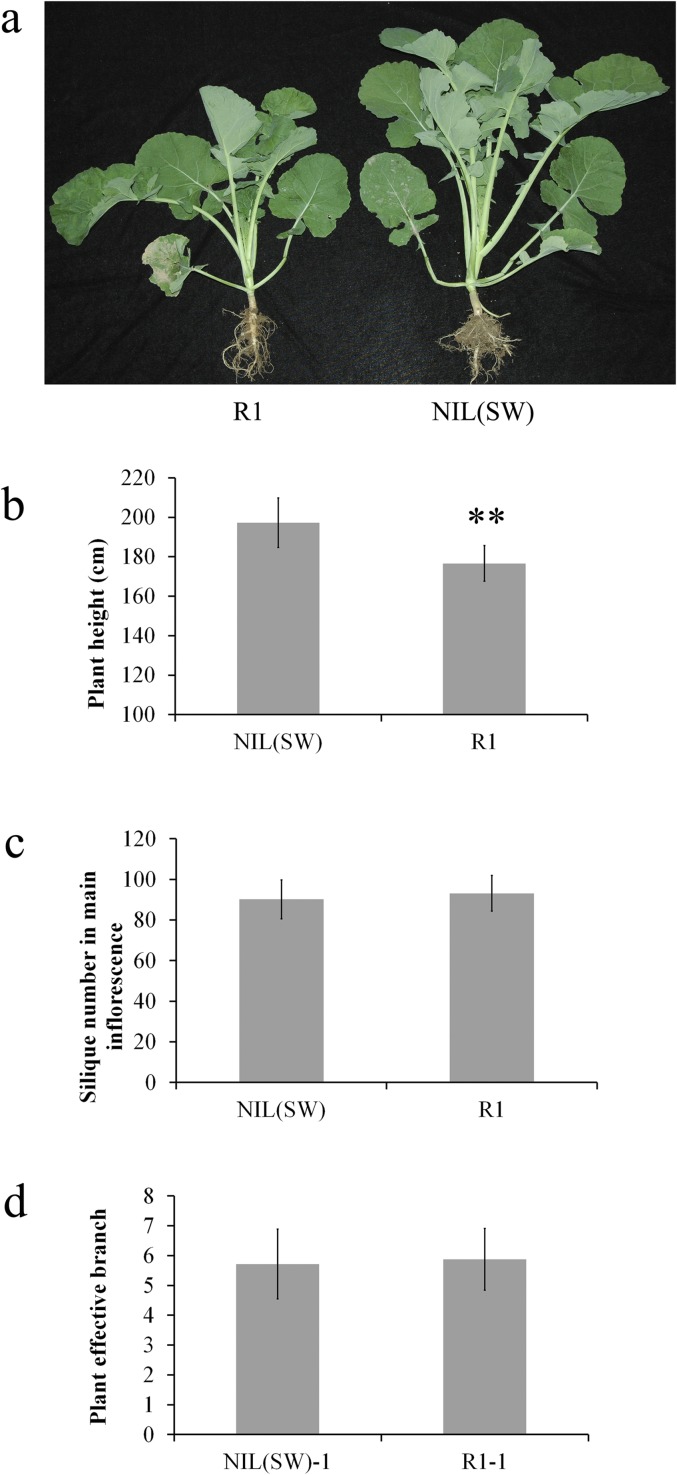
Phenotype comparison between the NIL (SW) and R1 lines showed differences in plant height and leaf size of the seedlings, but no differences in the other characters such as the number of branches or the number of siliques per branch were observed (Fig. S5). SL increased by >30% in the NIL (SW) line (80.8 ± 9.6 mm) as compared with the R1 line (61.2 ± 7.1 mm), but the number of seeds in siliques did not differ (Fig. 7A). Microscopic observation showed that the longer siliques in the NIL (SW) line might result from the increased length of the cells (Fig. 7B). The silique wall is an important source organ that provides photosynthates to the developing seeds. Therefore, by determining the surface area of the silique wall, SL may affect seed filling as a maternal organ by regulating the accumulation of photosynthates.
Fig. S5.
Comparison of leaf size in seedlings (A), plant height (B), silique number (C), and effective branch number in the main inflorescence (D) in NIL (SW) and R1 plants. Data are shown as mean ± SD. **P < 0.01.
Fig. 7.
Analysis of silique phenotype and DEGs in the silique wall and seed of the NIL (SW) and R1 lines. (A) Comparisons of seed number per silique and silique length. (Scale bar, 0.5 cm.) **P = 0.01. (B) Comparison of cell length. (Scale bar, 50 μm.) **P = 0.01. (C) Differentially expressed genes analyses in the silique wall and seed between the NIL (SW) and R1 lines. (D) KEGG enrichment analysis of the regulated genes in the silique wall and seed.
To understand better the mechanism underlying the regulation of SW in ARF18, we compared the differentially expressed genes (DEGs) in the silique wall and seed in the NIL (SW) and R1 lines. In the silique wall in NIL (SW), 2,178 genes were up-regulated, and 1,774 genes were down-regulated (Fig. 7C). An overview of DEGs in the seed showed that expression levels of fewer genes were affected in the seed (702 up-regulated and 625 down-regulated) than in the silique wall (Fig. 7C). DEGs between the silique wall and seeds were detected also: 158 genes were up-regulated, and 69 genes were down-regulated in the silique wall and seed (Fig. 7C).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for DEGs showed the top 10 pathways affected in the silique wall and seed, respectively (Fig. 7D). Comparative analysis showed that except for the common pathways including cell wall transport and hormone metabolism that occur in both the silique wall and the seed, some special pathways such as nitrogen metabolism, carbohydrate metabolism, and development that might influence silique development were affected in the silique wall. In the seed, pathways that could affect seed filling (e.g., glycolysis, lipid metabolism, and amino acid metabolism, among others) were affected. Evidently, genes in the plant hormone metabolism were enriched in both the silique wall and seed (Table S3). Among these genes, in addition to the other ARF genes, three different kinds of early auxin-response genes such as Aux/IAA, small auxin up-regulated RNA (SAUR), and GH3, were differentially expressed in the NIL (SW) and R1 lines. However, significantly fewer auxin-response genes were detected in the seed than in the silique wall (Table 1).
Table S3.
Regulated genes participating in plant hormone signal transduction pathways in silique wall
| Gene ID | l-SD-RPKM | S-SD-RPKM | Log2 ratio, S/L | P value | FDR | KEGG |
| Fna_40824 | 0.001 | 2.46688702 | 11.26847593 | 1.03E-09 | 1.20E-08 | K14493|1|5e-20|98.2|ppp:PHYPADRAFT_118478|gibberellin receptor GID1 |
| Fna_63742 | 3.871002181 | 15.08487235 | 1.962323463 | 1.44E-29 | 5.09E-28 | K14493|1|3e-64|246|ppp:PHYPADRAFT_118478|gibberellin receptor GID1 |
| Fna_45250 | 6.321888566 | 0.821962085 | −2.943211853 | 1.16E-15 | 2.16E-14 | K14517|1|1e-22|107|ath:AT5G47220|ethylene-responsive transcription factor 2 |
| Fna_76636 | 2.099818808 | 0.451887168 | −2.216230348 | 0.00011182 | 0.00065537 | K14517|1|9e-23|107|ath:AT5G47220|ethylene-responsive transcription factor 2 |
| Fna_22508 | 9.029323824 | 4.334690184 | −1.058689071 | 1.44E-05 | 9.99E-05 | K14516|1|1e-93|342|ath:AT3G23240|ethylene-responsive transcription factor 1 |
| Fna_31543 | 0.579436535 | 4.497065288 | 2.956261271 | 4.86E-18 | 1.03E-16 | K14432|1|1e-146|520|ath:AT2G36270|ABA responsive element binding factor |
| Fna_02912 | 3.061549709 | 0.417634534 | −2.873949191 | 5.25E-11 | 6.89E-10 | K14432|1|1e-123|442|ath:AT1G49720|ABA responsive element binding factor |
| Fna_20531 | 1.813025212 | 0.039287785 | −5.528174354 | 2.31E-13 | 3.68E-12 | K14508|1|8e-37|155|rcu:RCOM_1047190|regulatory protein NPR1 |
| Fna_59274 | 3.290857888 | 7.530550836 | 1.194291671 | 3.50E-08 | 3.48E-07 | K14503|1|1e-120|433|ath:AT1G75080|brassinosteroid resistant 1/2 |
| Fna_84506 | 1.167648585 | 0.242905355 | −2.265139948 | 8.97E-07 | 7.45E-06 | K13422|1|5e-19|97.4|vvi:100250607|transcription factor MYC2 |
| Fna_36672 | 3.836745524 | 8.599852705 | 1.16442887 | 5.87E-09 | 6.35E-08 | K13464|1|5e-35|149|ath:AT1G70700|jasmonate ZIM domain-containing protein |
| Fna_34305 | 1.440828791 | 3.496907133 | 1.279180568 | 5.44E-07 | 4.66E-06 | K14489|1|3e-19|97.8|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_35986 | 44.9036726 | 15.37592652 | −1.546160099 | 1.08E-82 | 1.18E-80 | K14489|1|2e-18|94.4|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_48649 | 26.23328535 | 9.3601424 | −1.486796113 | 6.65E-46 | 3.86E-44 | K14489|1|6e-19|96.3|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_00762 | 56.66331914 | 20.57556168 | −1.461483298 | 1.11E-93 | 1.41E-91 | K14489|1|5e-14|80.1|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_83989 | 18.00258003 | 7.407826368 | −1.281081492 | 3.06E-26 | 9.53E-25 | K14489|1|7e-19|96.3|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_17044 | 149.8113679 | 65.5956865 | −1.191474248 | 6.16E-183 | 1.67E-180 | K14489|1|5e-19|96.7|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_07366 | 5.85456058 | 13.50517601 | 1.205879645 | 9.57E-16 | 1.79E-14 | K13415|1|1e-34|147|pop:POPTR_650846|protein brassinosteroid insensitive 1 |
| Fna_52145 | 57.36070837 | 7.721825098 | −2.893049058 | 9.23E-172 | 2.23E-169 | K13415|1|1e-29|131|osa:4324691|protein brassinosteroid insensitive 1 |
| Fna_46526 | 0.109042315 | 4.139833389 | 5.246612706 | 1.85E-20 | 4.46E-19 | K14493|1|7e-65|248|ppp:PHYPADRAFT_118478|gibberellin receptor GID1 |
| Fna_83824 | 3.156332954 | 8.497310741 | 1.428756924 | 2.37E-10 | 2.93E-09 | K14493|1|3e-28|126|ath:AT5G27320|gibberellin receptor GID1 |
| Fna_85565 | 0.956217223 | 0.10469547 | −3.191139367 | 0.00017253 | 0.00097593 | K14493|1|0.0|662|ath:AT3G63010|gibberellin receptor GID1 |
| Fna_35769 | 10.60287819 | 3.81091512 | −1.476246562 | 2.23E-14 | 3.83E-13 | K14493|1|5e-33|142|ppp:PHYPADRAFT_118478|gibberellin receptor GID1 |
| Fna_71813 | 12.42330374 | 5.256434649 | −1.240892402 | 1.03E-13 | 1.68E-12 | K14493|1|1e-175|615|ath:AT3G05120|gibberellin receptor GID1 |
| Fna_69782 | 0.235790511 | 1.018759488 | 2.111235931 | 1.17E-07 | 1.09E-06 | K14510|1|2e-48|196|rcu:RCOM_0382890|serine/threonine-protein kinase CTR1 |
| Fna_12083 | 1.16169959 | 4.182711717 | 1.848201525 | 9.77E-06 | 6.97E-05 | K14496|1|1e-110|398|ath:AT5G05440|abscisic acid receptor PYR/PYL family |
| Fna_30568 | 5.173452054 | 10.85052222 | 1.068565315 | 1.14E-06 | 9.31E-06 | K14496|1|2e-83|308|ath:AT4G17870|abscisic acid receptor PYR/PYL family |
| Fna_17636 | 126.8173364 | 37.51983443 | −1.757026615 | 3.41E-117 | 5.67E-115 | K14496|1|1e-93|342|ath:AT4G01026|abscisic acid receptor PYR/PYL family |
| Fna_55594 | 154.6838127 | 61.64715636 | −1.327215977 | 1.15E-91 | 1.42E-89 | K14496|1|1e-95|348|ath:AT1G01360|abscisic acid receptor PYR/PYL family |
| Fna_47822 | 85.92984077 | 36.93051766 | −1.218345737 | 4.04E-47 | 2.42E-45 | K14496|1|6e-95|346|ath:AT4G01026|abscisic acid receptor PYR/PYL family |
| Fna_62133 | 0.125688569 | 0.686356926 | 2.44910557 | 3.39E-07 | 2.98E-06 | K13415|1|0.0|1018|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_83054 | 0.272200727 | 0.990951183 | 1.864143069 | 9.18E-05 | 0.00054903 | K13416|1|2e-47|191|ath:AT4G33430|brassinosteroid insensitive 1-associated receptor kinase 1 |
| Fna_17261 | 0.623283291 | 2.170417635 | 1.800012732 | 1.16E-08 | 1.22E-07 | K13415|1|5e-45|183|pop:POPTR_562744|protein brassinosteroid insensitive 1 |
| Fna_26567 | 4.160333744 | 14.48601764 | 1.799889865 | 6.84E-97 | 9.25E-95 | K13415|1|7e-87|323|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_25640 | 1.591792969 | 4.714534169 | 1.56646252 | 1.89E-14 | 3.26E-13 | K13415|1|9e-48|192|rcu:RCOM_0478150|protein brassinosteroid insensitive 1 |
| Fna_35624 | 3.252089367 | 9.372120247 | 1.527008561 | 6.24E-24 | 1.75E-22 | K13415|1|5e-66|253|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_60130 | 3.057361542 | 8.706194597 | 1.509755102 | 6.87E-26 | 2.11E-24 | K13415|1|6e-54|213|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_10023 | 7.535954502 | 21.25318048 | 1.495816593 | 5.06E-97 | 6.89E-95 | K13415|1|0.0|2088|ath:AT4G39400|protein brassinosteroid insensitive 1 |
| Fna_19853 | 0.494368756 | 1.371243821 | 1.471825646 | 0.00010221 | 0.00060532 | K13415|1|1e-49|199|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_03950 | 0.807453836 | 2.239656269 | 1.471825646 | 1.79E-10 | 2.24E-09 | K13415|1|0.0|974|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_04307 | 3.613507126 | 9.562970526 | 1.40405909 | 7.96E-28 | 2.66E-26 | K13415|1|2e-55|218|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_06936 | 0.83395033 | 2.110749649 | 1.33972211 | 7.63E-06 | 5.53E-05 | K13415|1|5e-54|213|rcu:RCOM_0478150|protein brassinosteroid insensitive 1 |
| Fna_43573 | 3.246424364 | 8.097333212 | 1.318595249 | 1.70E-15 | 3.13E-14 | K14510|1|1e-57|225|ath:AT5G03730|serine/threonine-protein kinase CTR1 |
| Fna_13420 | 0.789906531 | 1.922938822 | 1.283559009 | 1.01E-07 | 9.48E-07 | K13415|1|0.0|974|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_10027 | 0.813718961 | 1.97490476 | 1.279180568 | 1.39E-06 | 1.12E-05 | K13415|1|2e-61|238|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_67028 | 1.269094894 | 2.9844516 | 1.233665909 | 5.22E-07 | 4.48E-06 | K13415|1|9e-54|212|rcu:RCOM_0478150|protein brassinosteroid insensitive 1 |
| Fna_39336 | 4.415680975 | 10.1169245 | 1.196062935 | 3.60E-18 | 7.73E-17 | K13415|1|5e-67|256|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_66187 | 9.507385638 | 19.3862217 | 1.02791107 | 3.32E-27 | 1.08E-25 | K13415|1|8e-68|259|osa:4324691|protein brassinosteroid insensitive 1 |
| Fna_64146 | 2.487780223 | 4.982846369 | 1.002111054 | 1.99E-10 | 2.48E-09 | K13415|1|1e-103|376|sbi:SORBI_03g032990|protein brassinosteroid insensitive 1 |
| Fna_81727 | 0.536823704 | 0.001 | −9.068304567 | 9.70E-08 | 9.11E-07 | K13416|1|3e-58|228|ath:AT4G33430|brassinosteroid insensitive 1-associated receptor kinase 1 |
| Fna_42548 | 1.272160341 | 0.105858753 | −3.587068043 | 4.85E-06 | 3.62E-05 | K14498|1|0.0|648|ath:AT5G63650|serine/threonine-protein kinase SRK2 |
| Fna_70718 | 1.700860949 | 0.203016787 | −3.066594269 | 1.20E-11 | 1.67E-10 | K13415|1|5e-55|216|ath:AT4G39400|protein brassinosteroid insensitive 1 |
| Fna_33680 | 1.052564937 | 0.186352818 | −2.497800705 | 1.03E-07 | 9.60E-07 | K13416|1|1e-60|236|ath:AT4G33430|brassinosteroid insensitive 1-associated receptor kinase 1 |
| Fna_70657 | 5.434593497 | 0.965683065 | −2.492550444 | 6.13E-34 | 2.53E-32 | K14510|1|0.0|1391|ath:AT5G03730|serine/threonine-protein kinase CTR1 |
| Fna_41717 | 1.503391917 | 0.289583216 | −2.37617126 | 0.0001042 | 0.00061552 | K13415|1|2e-42|173|osa:4324691|protein brassinosteroid insensitive 1 |
| Fna_12057 | 2.067816745 | 0.51498853 | −2.005496131 | 9.31E-09 | 9.87E-08 | K13415|1|7e-65|249|osa:4324691|protein brassinosteroid insensitive 1 |
| Fna_26733 | 10.91669347 | 4.5886529 | −1.250393363 | 8.39E-17 | 1.67E-15 | K14500|1|0.0|749|vvi:100249852|BR-signaling kinase |
| Fna_76943 | 6.435455426 | 3.118712973 | −1.045091467 | 2.23E-08 | 2.27E-07 | K13415|1|5e-62|239|vvi:100252647|protein brassinosteroid insensitive 1 |
| Fna_03532 | 1.71122453 | 0.843122814 | −1.021214365 | 5.75E-05 | 0.00035706 | K13416|1|4e-62|241|ath:AT4G33430|brassinosteroid insensitive 1-associated receptor kinase 1 |
| Fna_43500 | 0.001 | 48.69502654 | 15.57148681 | 1.09E-114 | 1.75E-112 | K14509|1|2e-08|58.5|vvi:100249603|ethylene receptor |
| Fna_25093 | 0.724745891 | 1.972822221 | 1.444713801 | 7.02E-10 | 8.32E-09 | K14489|1|0.0|1884|ath:AT2G01830|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_22498 | 24.51181539 | 11.90298681 | −1.042153703 | 3.94E-42 | 2.08E-40 | K14509|1|0.0|1290|ath:AT3G23150|ethylene receptor |
| Fna_74751 | 3.945654227 | 16.92925755 | 2.101182266 | 3.76E-28 | 1.27E-26 | K13464|1|6e-92|337|ath:AT1G19180|jasmonate ZIM domain-containing protein |
| Fna_35386 | 9.366160947 | 33.66785914 | 1.845842252 | 2.40E-24 | 6.88E-23 | K13464|1|8e-07|52.8|pop:POPTR_712551|jasmonate ZIM domain-containing protein |
| Fna_15237 | 10.80007167 | 34.69639139 | 1.683744736 | 2.24E-22 | 5.90E-21 | K13464|1|5e-08|56.6|pop:POPTR_712551|jasmonate ZIM domain-containing protein |
| Fna_02073 | 11.04740156 | 29.23640989 | 1.40405909 | 1.13E-14 | 1.97E-13 | K13464|1|6e-08|56.2|osa:4347164|jasmonate ZIM domain-containing protein |
| Fna_41698 | 16.54807723 | 43.69312049 | 1.400742548 | 6.95E-51 | 4.46E-49 | K13464|1|1e-101|370|ath:AT1G19180|jasmonate ZIM domain-containing protein |
| Fna_77106 | 12.92706931 | 29.33685602 | 1.182319029 | 5.65E-22 | 1.46E-20 | K13464|1|6e-95|347|ath:AT1G19180|jasmonate ZIM domain-containing protein |
| Fna_04732 | 12.36668137 | 26.20671322 | 1.083478025 | 6.56E-23 | 1.77E-21 | K13464|1|1e-140|499|ath:AT3G17860|jasmonate ZIM domain-containing protein |
| Fna_02775 | 1.285551503 | 3.96970325 | 1.626643755 | 3.49E-09 | 3.86E-08 | K14494|1|4e-30|133|ath:AT2G01570|DELLA protein |
| Fna_43716 | 2.23216033 | 4.923766384 | 1.141321657 | 2.86E-07 | 2.54E-06 | K14494|1|7e-31|135|ath:AT2G01570|DELLA protein |
| Fna_40930 | 2.447209206 | 5.381826696 | 1.136958496 | 1.96E-08 | 2.00E-07 | K14494|1|6e-30|133|ath:AT2G01570|DELLA protein |
| Fna_77939 | 5.046165226 | 10.72018694 | 1.087070714 | 2.76E-16 | 5.34E-15 | K14494|1|2e-29|131|ath:AT2G01570|DELLA protein |
| Fna_85795 | 5.126482533 | 1.677995894 | −1.611230094 | 1.94E-12 | 2.88E-11 | K14494|1|3e-21|104|ath:AT3G03450|DELLA protein |
| Fna_03453 | 0.001 | 10.79759284 | 13.3984221 | 8.60E-22 | 2.21E-20 | K14489|1|5e-16|83.6|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_18999 | 0.001 | 2.362137471 | 11.20587721 | 2.22E-05 | 0.0001489 | K14489|1|7e-09|59.7|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_67681 | 0.001 | 1.190910975 | 10.21784986 | 9.24E-05 | 0.00055279 | K14489|1|3e-12|72.0|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_76841 | 0.101772827 | 4.12849138 | 5.342190366 | 1.76E-11 | 2.41E-10 | K14489|1|1e-12|73.2|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_79459 | 0.465740057 | 10.49616453 | 4.494193459 | 8.20E-33 | 3.25E-31 | K14489|1|6e-12|71.6|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_77145 | 0.452323677 | 3.528625111 | 2.963678742 | 2.01E-06 | 1.59E-05 | K14489|1|7e-12|70.1|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_60675 | 2.7243803 | 6.3515252 | 1.221174965 | 8.61E-05 | 0.0005177 | K14489|1|4e-14|78.6|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_05162 | 40.07842607 | 17.17595547 | −1.222435497 | 4.70E-25 | 1.39E-23 | K14489|1|6e-12|71.2|vvi:100263145|arabidopsis histidine kinase 2/3/4 (cytokinin receptor) |
| Fna_33192 | 6.589607522 | 2.878748832 | −1.19475062 | 2.79E-06 | 2.16E-05 | K14499|1|2e-06|53.5|sbi:SORBI_02g026700|BRI1 kinase inhibitor 1 |
Table 1.
Auxin-response genes including ARF, Aux/IAA, SAUR, and GH3 were regulated in NIL (SW) and R1
| Gene ID | NIL (SW) RPKM | R1 RPKM | Log2 ratio, NIL (SW)/R1 | P value | Tissue | KEGG |
| Fna_59492 | 0.15657358 | 1.661168129 | 3.407285394 | 2.50E-11 | Silique wall | Auxin-response factor |
| Fna_70218 | 0.094105012 | 0.685181657 | 2.864143069 | 0.000106615 | Silique wall | Auxin-response factor |
| Fna_20825 | 3.337202011 | 12.77690534 | 1.936827523 | 1.91E-19 | Silique wall | Auxin-response factor |
| Fna_31741 | 1.331146322 | 4.087832828 | 1.618667034 | 1.53E-16 | Silique wall | Auxin-response factor |
| Fna_56595 | 3.535266632 | 8.669274747 | 1.294092278 | 5.43E-24 | Silique wall | Auxin-response factor |
| Fna_37605 | 3.460516441 | 1.642247956 | −1.075315389 | 4.85E-08 | Silique wall | Auxin-response factor |
| Fna_74610 | 1.043493546 | 10.00968212 | 3.261902577 | 4.85E-17 | Silique wall | Auxin-responsive protein IAA |
| Fna_71197 | 4.669576781 | 11.58219301 | 1.310544739 | 5.09E-05 | Silique wall | Auxin-responsive protein IAA |
| Fna_15898 | 5.899374056 | 12.99909607 | 1.139777512 | 4.36E-09 | Silique wall | Auxin-responsive protein IAA |
| Fna_87374 | 1.750631568 | 0.001 | −10.77365978 | 6.56E-09 | Silique wall | Auxin-responsive protein IAA |
| Fna_37713 | 2.682714694 | 0.079726258 | −5.07249487 | 1.09E-09 | Silique wall | Auxin-responsive protein IAA |
| Fna_34616 | 76.66316166 | 29.54259831 | −1.375736779 | 1.09E-61 | Silique wall | Auxin-responsive protein IAA |
| Fna_10794 | 10.18816752 | 4.279637301 | −1.251334148 | 7.17E-07 | Silique wall | Auxin-responsive protein IAA |
| Fna_78680 | 21.84578229 | 9.782028116 | −1.159149252 | 7.42E-12 | Silique wall | Auxin-responsive protein IAA |
| Fna_29766 | 14.05158922 | 6.495878046 | −1.113136855 | 2.58E-07 | Silique wall | Auxin-responsive protein IAA |
| Fna_73932 | 10.4476168 | 5.061371644 | −1.045573571 | 4.25E-10 | Silique wall | Auxin-responsive protein IAA |
| Fna_70892 | 1.099146535 | 5.335281168 | 2.279180568 | 7.32E-05 | Silique wall | SAUR family protein |
| Fna_00244 | 6.846535654 | 15.01269593 | 1.132737 | 5.00E-05 | Silique wall | SAUR family protein |
| Fna_10378 | 6.628624935 | 1.62966765 | −2.024131849 | 3.92E-07 | Silique wall | SAUR family protein |
| Fna_61491 | 12.17256579 | 3.13397625 | −1.957567146 | 1.40E-11 | Silique wall | SAUR family protein |
| Fna_23302 | 37.89295131 | 16.18333818 | −1.227420282 | 7.11E-17 | Silique wall | SAUR family protein |
| Fna_50955 | 15.95535292 | 7.375964749 | −1.113136855 | 6.86E-05 | Silique wall | SAUR family protein |
| Fna_74090 | 0.001 | 0.866117073 | 9.758418237 | 2.09E-09 | Silique wall | Auxin responsive GH3 gene family |
| Fna_86647 | 0.08921644 | 4.083123343 | 5.516219766 | 7.25E-37 | Silique wall | Auxin responsive GH3 gene family |
| Fna_02366 | 0.193511714 | 1.207684369 | 2.641750648 | 8.76E-07 | Silique wall | Auxin responsive GH3 gene family |
| Fna_54159 | 9.730243207 | 52.38524289 | 2.428612686 | 1.74E-243 | Silique wall | Auxin responsive GH3 gene family |
| Fna_61300 | 0.328727892 | 1.740711637 | 2.40471145 | 5.05E-09 | Silique wall | Auxin responsive GH3 gene family |
| Fna_72281 | 0.627571431 | 2.14480541 | 1.772995181 | 9.26E-08 | Silique wall | Auxin responsive GH3 gene family |
| Fna_74219 | 6.106369637 | 19.58386937 | 1.681279012 | 1.87E-20 | Silique wall | Auxin responsive GH3 gene family |
| Fna_30176 | 2.085362661 | 0.166852676 | −3.643651571 | 3.91E-14 | Silique wall | Auxin responsive GH3 gene family |
| Fna_72776 | 2.073861386 | 0.359520294 | −2.528174354 | 6.07E-08 | Silique wall | Auxin responsive GH3 gene family |
| Fna_59492 | 8.306 | 3.469 | −1.260 | 1.41E-17 | Seed | Auxin-response factor |
| Fna_17490 | 0.001 | 1.996 | 10.963 | 2.44E-09 | Seed | Auxin-response factor |
| Fna_77259 | 1.739 | 0.584 | −1.574 | 5.80E-06 | Seed | Auxin responsive GH3 gene family |
| Fna_02366 | 2.068 | 0.715 | −1.533 | 1.35E-06 | Seed | Auxin responsive GH3 gene family |
| Fna_55303 | 0.747 | 0.058 | −3.689 | 9.63E-07 | Seed | Auxin responsive GH3 gene family |
| Fna_72776 | 1.558 | 0.458 | −1.767 | 6.92E-05 | Seed | Auxin responsive GH3 gene family |
Discussion
Variation in the Region Upstream of the 3′ Splice Site Induces Nonfunctionalization of the Sixth Intron in zy72360.
In the present study, comparison of ARF18 cDNAs and genome sequences from zy72360 and R1 indicated skipping of the seventh exon in ARF18 from zy72360. By comparing the sixth intron of the ARF18 alleles, extensive variations, including six SNPs and an 11-bp insertion, were detected in the region upstream of the 3′ splicing site in the nonfunctional sixth intron. In plants, although individual introns exhibit extensive variation around highly conserved dinucleotides (GU and AG) at the 5′ and 3′ splice sites (29), branchpoint sequences (YUNAN consensus) that usually are located 18–40 nt upstream of the 3′ splice site of introns play an important role in splicing efficiency (30). Introns are removed via a two-step cleavage–ligation reaction in which the first step involves cleavage at the 5′ splice site with formation of an intron lariat at the branchpoint (31). Because dinucleotides (AG) at the 3′ splice site in the sixth intron show no change, we deduced that the skipping of the seventh exon might have been induced by the variation in the upstream sequence of the 3′ splicing site in the sixth intron. Comparative analysis showed that the possible branchpoints differed between the ARF18 genes from zy72360 and R1, suggesting that the disruption of the branchpoint in the sixth intron induced the loss of splicing function at the 3′ site in zy72360. In fact, exon skipping caused by an insertion in the sequence upstream of the 3′ splice site of the intron has been reported previously (32, 33).
ARF18 Acts as a Repressor of Auxin-Responsive Genes by Forming Homodimers.
By affecting cell division, cell growth, or cell differentiation, auxins are involved in controlling virtually all aspects of plant growth and development (34, 35). Auxin-response factors as transcriptional regulators mediate auxin responses by binding to the auxin-response elements in the promoter region of early auxin-response genes and mediate auxin gene-induction responses (36, 37). In Arabidopsis, the ARF proteins are encoded by a large family of genes that includes 23 members, and each member is thought to play a central role in various auxin-mediated developmental processes (38). The MR determines the ARF protein functions as an activator or repressor of auxin-responsive genes (39). Phylogenetic analysis of Arabidopsis ARF genes has revealed that ARF18 belongs to the class that also includes ARF1, ARF2, ARF9, and ARF11 (39). Of these, ARF1 and ARF2 function as transcription repressors (40). The present study has determined that ARF18 also functions as a repressor. Although the MR is partially deleted in zy72360-ARF18, its inhibitory activity does not decrease significantly compared with that observed in R1-ARF18.
Transcription factor dimerization is a common element of the transcriptional control mechanism. Most of the ARF genes are no exception, and homodimerization also is required for its biological function (29). Dimer interface contacts include hydrophobic interactions between several highly conserved residues. Based on the reports on ARF1 and ARF5, ARF dimerization is induced through the DBD (41). The present study performed yeast hybridization assays to determine that R1-ARF18 interacts with itself, suggesting that ARF18 functions as a homodimer. On the other hand, in zy72360-ARF18 some highly conserved residues were located within the 55 deleted amino acids, resulting in a failure in dimerization and subsequently loss of binding activity.
Functional Differentiation of BnaA.ARF18.a and BnaC.ARF18.a in B. napus.
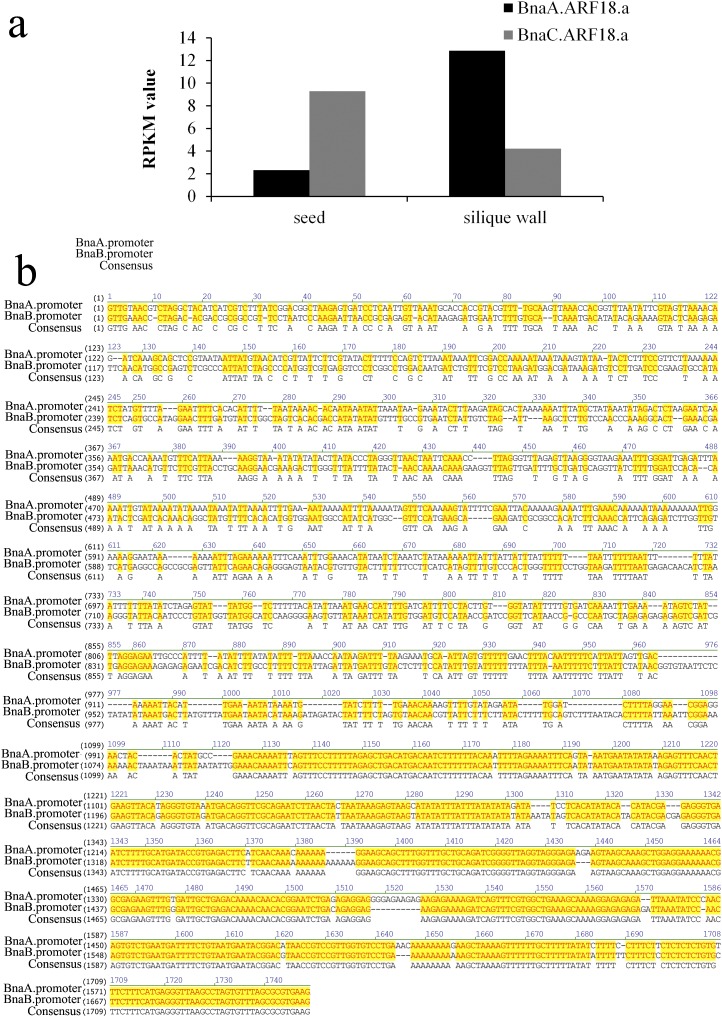
As a tetraploid species, the ARF18 gene of B. napus occurs as two homologs, namely BnaA.ARF18.a and BnaC.ARF18.a (BnaC08g31690D), which are located on chromosomes A9 and C8, respectively. It is assumed that when a gene is duplicated, the two duplicates are almost identical in sequence and have full redundancy (functional overlap) (42). In B. napus, based on transcriptome sequencing data, we observed a higher level of BnaA.ARF18.a expression than of BnaC.ARF18.a in the silique wall. The opposite trend was observed in the seed (Fig. S6A). Promoter sequence comparison also indicated major differences between BnaA.ARF18.a and BnaC.ARF18.a (Fig. S6B). In A. thaliana, more than half of the duplicates were found to have divergence in expression levels (43). Divergence in the expression states may lead to new expression states (neofunctionalization), partitioning of ancestral functions (subfunctionalization), or loss of expression state that leads to pseudogenization (44). Therefore, we believe that subfunctionalization might have been generated between BnaA.ARF18.a and BnaC.ARF18.a. As expected, based on the transcriptome data, the expression level of BnaC.ARF18.a in C8 did differ in the NIL (SW) and R1 lines. In general, BnaA.ARF18.a plays indispensable roles in the development of the silique wall that could not be replaced by BnaC.ARF18.a in rapeseed.
Fig. S6.
Comparative analysis of BnaA.ARF18.a and BnaC.ARF18.a. (A) Expression levels of BnaA.ARF18.a and BnaC.ARF18.a in silique wall and seed in NIL (SW) and R1 lines. (B) Comparison of the promoter sequences of BnaA.ARF18.a and BnaC.ARF18.a from the R1 line.
Silique Wall Genes Exert Maternal Control of SW in B. napus.
The present study showed that the ARF18 gene regulated SW by affecting the development of the silique wall. In fact, several genes have been reported to affect SW by maternal organs. In Arabidopsis, the most commonly reported genes, e.g., TTG2, AP2, ARF2, and DA1, influence seed size by affecting cell expansion and/or cell proliferation in the integuments or seed coat (1). Therefore the integument or seed coat plays a key role in the maternal control of seed size. In rice, the loss of GW2 function increases cell number, resulting in a larger spikelet hull, and increases the grain milk-filling rate, which in turn enhances grain weight (13).
In rapeseed, the silique wall not only is an important sink organ that assimilates the carbohydrates synthesized in the vegetative parts of the plant such as leaves and stem but also is an important source organ that provides photosynthates to the developing seeds (45). Because the functional leaf area declines rapidly during seed filling, silique wall photosynthesis is the main source of nutrition for seed growth (46). Moreover, signals originating from the silique wall coordinate seed filling and regulate reallocation of reserves (45). SL has been significantly correlated with SW variation (47). By determining the surface area of the silique wall, SL may affect SW by regulating the accumulation of photosynthates.
The present study has determined that ARF18 is up-regulated in the silique wall and down-regulated in the seed, suggesting that this gene might regulate SW indirectly by affecting silique development. Further studies showed that the loss of ARF18 activity induced the development of a longer silique by accelerating cell expansion in the silique wall. This change has resulted in more photosynthates in the longer silique which then enter the seed, generating larger seeds.
Methods
Plant Materials.
Rapeseed line zy72360 was crossed with line R1. The resultant F1 plants were self-pollinated to produce F2 seeds, which were backcrossed with R1 plants to produce BC4F1 seeds. We selected several plants in which the region around the SW locus was heterozygous and almost all other regions were homozygous for R1. These plants were used to develop segregating populations for fine mapping and high-resolution mapping of SW by repetitive backcrossing and marker-assisted selection. From the BC4F2 generation, we developed a nearly isogenic line with a small zy72360 chromosomal region containing the SW locus in the R1 genetic background (4,000 lines). The association population consisted of a panel of 380 inbred rapeseed lines. The association population was arranged in a randomized complete block design with three replicates.
Trait Measurement.
Seeds from the main inflorescence were used to represent SW in each plant. The average SW of 10 plants from the center of each plot was used to represent SW in the association population.
Fine Mapping and High-Resolution Mapping.
A BC4F2 population was used for fine mapping of SW, which was based on rough mapping the F2 population. To perform SW QTL analysis, six molecular markers were developed in the target region containing the SW locus to detect recombinants in the 4,000 BC4F2 plants. To determine further the location of the recombination nearest to the SW locus, we developed 11 markers on the basis of the parental sequence and determined the genotypes of the recombinants using these markers. The BC4F2 progeny derived from recombinant plants were used to screen for recombination products. The molecular marker primers are listed in Table S1.
Gene Cloning and Transgenic Analysis.
Total RNA was extracted from various plant tissues in NILs and transgenic lines. Seven candidate genes were named according to Chalhoub et al. (22). The upstream regulatory and coding regions of seven genes were cloned with the cDNAs originating from the R1 and zy72360 lines and were sequenced. Genes with existing SNPs and indels between the parents were ligated into the TOPO vector (Invitrogen) and transferred into the plant binary vector pEarleyGate-100 (Invitrogen). All constructs were introduced into Agrobacterium tumefaciens strain EHA105 and were transferred into Col-0 Arabidopsis by floral dip. The empty pEarleyGate-100 vector also was transformed into Arabidopsis as a control. The genome sequence including the ARF18 gene and its upstream promoter sequence originating from the R1 line was cloned and ligated into the Pbi121 vector (Invitrogen). The construct was transferred into A. tumefaciens strain EHA105 and was transformed into the rapeseed line zy72360 by the hypocotyl transgenic system. The primers used in this experiment are listed in Table S2.
Protein Subcellular Localization and Intron Splicing Identification.
To investigate the cellular localization of ARF18, a 35S GFP-ARF18 fusion construct (ARF18 from R1) was transfected into the tobacco leaves based on the method of Sparkes et al. (48). The sixth intron in the ARF18 genome sequence from R1 and the corresponding sequence from zy72360 were cloned and inserted into the GFP gene by fusion PCR. The two reconstructive GFP genes were ligated into the TOPO vector and then were transferred into the plant binary vector, pEarleyGate-100. The splicing function of introns was detected by using a tobacco transient expression system. Primers used in this experiment are listed in Table S4.
Table S4.
Primers used for vector construction in research
| Primer name | Primer sequence (5′→3′) |
| At/BnARF18RT-QpcrF | GTGAACAAGTATATGGAAGCTAT |
| At/BnARF18RT-QpcrR | CTGTTGTTGGCTCATCCCAT |
| BnaA.ARF18aRT-QpcrF | TCTCTTGGTACAAGATTCAGGA |
| BnaA.ARF18aRT-QpcrR | GATTGTTGAGAAGGTGTTGAAG |
| BnaA.ARF18a promoter F1 | GATCCTCAATTGTTAAATGCACCACCG |
| BnaA.ARF18a promoter F2 | GTCTATTTAGGAGAATTGCCC |
| BnaA.ARF18a promoter R | TCACGCGCTAAACACTAGGC |
| GFPF | ATGGTGAGCAAGGGCGAG |
| GFPR | TTACTTGTACAGCTCGTCCAT |
| GFP1it1R | AGTTTCAGGAAAAGTAGTGGTACGTCAGCTTGCCGTAGGTGGC |
| GFP2it1F | AATGAAATTCATTTGTGTTTGTAGCCTGAAGTTCATCTGCACCAC |
| Intron1F | GCCACCTACGGCAAGCTGACGTACCACTACTTTTCCTGAAACT |
| Intron1R | GTGGTGCAGATGAACTTCAGGCTACAAACACAAATGAATTTCATT |
| GFP1it2R | AAGTTTGAGAATAGCTAATAGTACGTCAGCTTGCCGTAGGTGGC |
| GFP2it2F | TTAAATGCCTTTTGTGAGTCTTAGCCTGAAGTTCATCTGCACCAC |
| Intron2F | GCCACCTACGGCAAGCTGACGTACTATTAGCTATTCTCAAACTT |
| Intron2r | GTGGTGCAGATGAACTTCAGGCTAAGACTCACAAAAGGCATTTAA |
Detection of Inhibitory Activity and Dimers by Using a Yeast Hybrid System.
The inhibitory activity of ARF18 was assessed using a yeast hybrid system. The ARF18 CDS from zy72360 and R1 and VP16 were constructed into PGBKT7 as bait vectors and were cotransformed with PGADT7 into yeast strain MaV203 (Invitrogen). VP16 was used as positive control, and the empty vectors PGBKT7 and PGADT7 were used as negative control. Yeast growth was analyzed at 30 °C on the SD agar plates lacking Leu, Trp, and His and containing various concentrations of 3-amino-1,2,4-triazole as indicated. For dimer detection, the ARF18 CDs from zy72360 and R1 were fused to the BD (PGBKT7) and AD (PGADT7), respectively. pGBKT7-53 and pGADT7-T were used as positive controls. pGBKT7-lam and pGADT7-T were used as a negative controls. All the vectors were transformed into yeast stain AH109 (Clontech).
Detection of Binding Activity by Using Electrophoretic Mobility Shift Assays.
zy72360-ARF18 and R1-ARF18 CDS were subcloned into the expression vector pGEX6P-1 (Amersham Pharmacia Biotech). The GST fusion protein was extracted from bacteria using glutathione-tagged Sepharose (GE Healthcare). The eluted GST fusion protein was assayed by SDS/PAGE. To test ARF18 binding activity, a 5′ biotin-labeled DR5(7×) probe used in the electrophoretic mobility shift assay (EMSA) was synthesized (Genecreate). EMSA was performed with a LightShift Chemiluminescent EMSA kit (Pierce). Briefly, protein extracts in 20 μL of a 1× binding buffer with 2.5% (vol/vol) glycerol, 5 mM MgCl2, 50 ng/L poly(dI-dC), 0.05% Nonidet P-40, 1 mM DTT, and 20 fmol probe were incubated at room temperature for 15 min. For STAT3 supershift analysis, an anti-STAT3 polyclonal antibody (Santa Cruz; 1 g per reaction) was incubated with the nuclear proteins on ice for 20 min before the addition of the labeled oligonucleotides. Reaction products were separated by electrophoresis [5% acrylamide (29:1 acryl/bis)] in 0.5 Tris/borate/EDTA. After electrophoresis, the protein–DNA complexes were transferred onto nylon membranes and detected by using chemiluminescence.
Expression Pattern Analysis.
Total RNA was extracted from rapeseed tissues, including seed, root, stem, leaf, bud, and the silique wall, and from the Arabidopsis silique at 3 daf using the RNeasy Plant Mini kit (Qiagen). The reverse transcription reaction was performed using the First Strand cDNA Synthesis Kit for RT-PCR (Takara). Epsin N-terminal homology (ENTH) and Arabidopsis β-actin1 (At2g37620) were used as references (49, 50). Data were expressed as the mean of three biological replicates ± SD. The ARF18 promoter was ligated into pCXGUS-P. The construct was introduced into A. tumefaciens strain EHA105 and transferred into Col-0 Arabidopsis. GUS activity was visualized by staining all tissues from homozygous transgenic lines overnight in an X-Gluc solution (Biosharp), and the tissues then were cleared in 75% (vol/vol) ethanol. The tissues were imaged directly or captured under a microscope. The primers used are listed in Table S4.
Cell Length Observation.
Twenty daf rapeseed siliques were sampled and fixed in 50% ethanol, 5% glacial acetic acid, and 5% formaldehyde for 16 h, dehydrated in an ethanol series, and photographed with an Olympus compound microscope.
Protoplast Transformation and Transient Expression Assays.
Protoplast from leaves was isolated according to the tape-Arabidopsis sandwich method described by Wu et al. (51). Plasmid DNA was prepared using a QIAfilter Plasmid Midi Kit (Qiagen). The coding sequence of ARF18 under the control of a 35S promoter was amplified from the pEarleyGate-100 vector and subcloned into a PUC19 vector. The DR5GUS reporter construct comprised seven copies of DR5 cloned upstream of a −46 CaMV 35S promoter with a TMV 5′ leader (40). The efficiency of transfection was standardized using a CaMV 35S promoter–LUC construct, which shows no response to auxin (52). Three plasmids expressing a regulatory effector, a specific reporter, and a transfection control reporter were used at a ratio of 5:4:1. Ten micrograms of the plasmid were introduced into Arabidopsis protoplasts by PEG/calcium-mediated transformation. After the protoplasts were cultured and harvested, LUC and GUS assays were performed (53). All transfections were performed in triplicate, and at least three independent transfection assays were performed with protoplasts.
Transcriptome Analysis.
Total RNAs from silique wall (16 daf) and seed (25 daf) were sent to Beijing Genomics Institute, and the transcriptomes were sequenced on an Illumina HiSeqTM 2000 platform. Gene-expression levels were calculated using the reads per kilobase per million (RPKM) method (54). To screen for DEGs, a P value corresponding to a differential gene-expression test based on the method introduced by Audic and Claverie was used (55). False-positive (type I errors) and false-negative (type II) errors were corrected using the false-discovery rate (FDR) method (55). Finally, we chose FDR ≤0.001 and the absolute value of log2 ratio ≥1 as the thresholds to judge the significance of the differences in gene expression in the silique wall and seed between the NIL (SW) and R1 lines. The DEGs were submitted to the website of Bio-Analytic Resource for Plant Biology program (bar.utoronto.ca/ntools/cgi-bin/ntools_classification_superviewer.cgi) for MapMan analysis.
Acknowledgments
This study was supported by Grants 2015CB150200 and 2011CB109300 from the National Key Basic Research Program of China, Grant 2013AA102602 from the National High Technology Research and Development Program of China, Grant 31201241 from the National Natural Science Foundation of China, and Grant 2014020101010066 from the Applied Basic Research Project in Wuhan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI database [accession nos. BnaA09g55520D cDNA from zy72360 (KT000600); BnaA09g55520D cDNA from R1 (KT000602); BnaA09g55570D promoter from zy72360 (KT000601); BnaA09g55570D promoter from R1 (KT000605); BnaA09g55560D from ZY72360 (KT000603); BnaA09g55560D from R1 (KT000604); BnaA09g55580D from zy72360 (KT000607); BnaA09g55580D from R1 (KT000608); and BnaA09g55580D promoter from R1 (KT000606)]. The transcriptome data for silique wall and seed has been deposited in ocri-genomics.org/RNA-seq/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502160112/-/DCSupplemental.
References
- 1.Fang W, Wang Z, Cui R, Li J, Li Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012;70(6):929–939. doi: 10.1111/j.1365-313X.2012.04907.x. [DOI] [PubMed] [Google Scholar]
- 2.Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. Control of seed mass by APETALA2. Proc Natl Acad Sci USA. 2005;102(8):3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133(2):251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 4.Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17(1):52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci USA. 2009;106(47):20115–20120. doi: 10.1073/pnas.0907024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia D, et al. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 2003;131(4):1661–1670. doi: 10.1104/pp.102.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(48):17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang A, et al. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 2010;63(4):670–679. doi: 10.1111/j.1365-313X.2010.04271.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, et al. SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell. 2009;21(1):106–117. doi: 10.1105/tpc.108.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang X, Li W, Zhou Y, Ni M. A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 2013;9(3):e1003347. doi: 10.1371/journal.pgen.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao JS. Genes and QTLs for rice grain quality improvement. In: Yan W, editor. Agricultural and Biological Sciences: Rice-Germplasm, Genetics and Improvement. InTech; Vienna: 2014. Chap 9. [Google Scholar]
- 13.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39(5):623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 14.Wang E, et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40(11):1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- 15.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40(8):1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 16.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18(12):1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 17.Mao H, et al. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA. 2010;107(45):19579–19584. doi: 10.1073/pnas.1014419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci USA. 2012;109(52):21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masterson J. Stomatal size in fossil plants: Evidence for polyploidy in majority of angiosperms. Science. 1994;264(5157):421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 22.Chalhoub B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 23.Rana1 D, et al. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004;40(5):725–733. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- 24.Quijada PA, Udall JA, Lambert B, Osborn TC. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor Appl Genet. 2006;113(3):549–561. doi: 10.1007/s00122-006-0323-1. [DOI] [PubMed] [Google Scholar]
- 25.Udall JA, Quijada PA, Lambert B, Osborn TC. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor Appl Genet. 2006;113(4):597–609. doi: 10.1007/s00122-006-0324-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang P, et al. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.) Theor Appl Genet. 2012;125(2):285–296. doi: 10.1007/s00122-012-1833-7. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Shi J, Wang X, Liu G, Wang H. A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.) BMC Plant Biol. 2014;14:114. doi: 10.1186/1471-2229-14-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Østergaard L, King GJ. Standardized gene nomenclature for the Brassica genus. Plant Methods. 2008;4:10. doi: 10.1186/1746-4811-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson CG, Clark G, Davidson D, Smith P, Brown JWS. Mutation of putative branchpoint consensus sequences in plant introns reduces splicing efficiency. Plant J. 1996;9(3):369–380. doi: 10.1046/j.1365-313x.1996.09030369.x. [DOI] [PubMed] [Google Scholar]
- 30.Brown JWS, Smith P, Simpson CG. Arabidopsis consensus intron sequences. Plant Mol Biol. 1996;32(3):531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- 31.Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 1996;10(5):771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- 32.Kosaki A, Nelson J, Webster NJG. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J Biol Chem. 1998;273(17):10331–10337. doi: 10.1074/jbc.273.17.10331. [DOI] [PubMed] [Google Scholar]
- 33.Ganguly A, Dunbar T, Chen P, Godmilow L, Ganguly T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum Genet. 2003;113(4):348–352. doi: 10.1007/s00439-003-0986-5. [DOI] [PubMed] [Google Scholar]
- 34.Benková E, et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 35.Scarpella E, Marcos D, Friml J, Berleth T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006;20(8):1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guilfoyle TJ, Hagen G. Auxin response factors: Recent advances in auxin biology. J Plant Growth Regul. 2001;20(3):281–291. [Google Scholar]
- 37.Korasick DA, et al. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA. 2014;111(14):5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004;135(3):1738–1752. doi: 10.1104/pp.104.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9(11):1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boer DR, et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell. 2014;156(3):577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 42.Rutter MT, Cross KV, Van Woert PA. Birth, death and subfunctionalization in the Arabidopsis genome. Trends Plant Sci. 2012;17(4):204–212. doi: 10.1016/j.tplants.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moghe GD, Shiu SH. The causes and molecular consequences of polyploidy in flowering plants. Ann N Y Acad Sci. 2014;1320:16–34. doi: 10.1111/nyas.12466. [DOI] [PubMed] [Google Scholar]
- 45.Bennett EJ, Roberts JA, Wagstaff C. The role of the pod in seed development: Strategies for manipulating yield. New Phytol. 2011;190(4):838–853. doi: 10.1111/j.1469-8137.2011.03714.x. [DOI] [PubMed] [Google Scholar]
- 46.King SP, Lunn JE, Furbank RT. Carbohydrate Content and Enzyme Metabolism in Developing Canola Siliques. Plant Physiol. 1997;114(1):153–160. doi: 10.1104/pp.114.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chay P, Thurling N. Identification of genes con trolling pod length in spring rapeseed, Brassica napus L. and their utilization for yield improvement. Plant Breed. 1989;103(1):54–62. [Google Scholar]
- 48.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1(4):2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 49.Yang H, et al. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene. 2014;538(1):113–122. doi: 10.1016/j.gene.2013.12.057. [DOI] [PubMed] [Google Scholar]
- 50.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu FH, et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi: 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo S, et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun. 2013;4:1566. doi: 10.1038/ncomms2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 54.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 55.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]