Significance
Computational tools applied to any human genome sequence identify hundreds of genetic variants predicted to disrupt the function of individual proteins as the result of a single codon change. These tools have been trained on disease mutations and common polymorphisms but have yet to be tested against an unbiased spectrum of random mutations arising de novo. Here we perform such a test comparing the predicted and actual effects of de novo mutations in 23 genes with essential functions for normal immunity and all possible mutations in the TP53 tumor suppressor gene. These results highlight an important gap in our ability to relate genotype to phenotype in clinical genome sequencing: the inability to differentiate immediately clinically relevant mutations from nearly neutral mutations.
Keywords: de novo mutation, immunodeficiency, evolution, nearly neutral, cancer
Abstract
Each person’s genome sequence has thousands of missense variants. Practical interpretation of their functional significance must rely on computational inferences in the absence of exhaustive experimental measurements. Here we analyzed the efficacy of these inferences in 33 de novo missense mutations revealed by sequencing in first-generation progeny of N-ethyl-N-nitrosourea–treated mice, involving 23 essential immune system genes. PolyPhen2, SIFT, MutationAssessor, Panther, CADD, and Condel were used to predict each mutation’s functional importance, whereas the actual effect was measured by breeding and testing homozygotes for the expected in vivo loss-of-function phenotype. Only 20% of mutations predicted to be deleterious by PolyPhen2 (and 15% by CADD) showed a discernible phenotype in individual homozygotes. Half of all possible missense mutations in the same 23 immune genes were predicted to be deleterious, and most of these appear to become subject to purifying selection because few persist between separate mouse substrains, rodents, or primates. Because defects in immune genes could be phenotypically masked in vivo by compensation and environment, we compared inferences by the same tools with the in vitro phenotype of all 2,314 possible missense variants in TP53; 42% of mutations predicted by PolyPhen2 to be deleterious (and 45% by CADD) had little measurable consequence for TP53-promoted transcription. We conclude that for de novo or low-frequency missense mutations found by genome sequencing, half those inferred as deleterious correspond to nearly neutral mutations that have little impact on the clinical phenotype of individual cases but will nevertheless become subject to purifying selection.
The genome sequence of any particular person contains extensive protein-altering genetic variation and de novo point mutations (1), of which only a minority are unambiguously deleterious and introduce premature stop codons or disrupt normal mRNA splicing. When considering a person with a suspected genetic illness, the first clinical question is whether any of the mutations identified in their genome sequence involve essential genes whose disruption is known to cause a phenotype resembling that of the patient in question. The most numerous class of protein-altering mutations is missense mutations, where a single codon is altered to encode a different amino acid. On average, 2% of people carry a missense mutation in any given gene (2). Hence, by chance, missense mutations will often be found in genes that are seemingly relevant to a person’s disease phenotype, and the next key clinical question is whether or not these substitutions alter the function of the corresponding protein. Short of mutational studies of all possible amino acid substitutions coupled with comprehensive functional assays, the sheer number and diversity of missense mutations present in each person’s genome means that their functional importance must presently be addressed primarily by computational inference.
Many tools now exist that use diverse information to make inferences of the functional importance of single amino acid substitutions (3, 4). The majority of better-performing tools use a protein multiple sequence alignment and judge the importance of residues depending on their conservation across available homologous sequences. Notably, these tools are sensitive to the sequence choice in the input alignment (5). Some examples of commonly used tools that use a sequence conservation approach, with diverse methods to calculate conservation, are SIFT (6), MutationAssessor (7), MAPP (8), AlignGVGD (9), PANTHER (10), and GERP (11). Protein structural information is also included in other tools to judge whether an amino acid substitution may importantly alter protein stability or catalysis, but few commonly used tools rely just on these data alone (3).
The widely used PolyPhen2 and CADD tools (12, 13) integrate a number of different information sources, including sequence- and structure-based features (and in the case of CADD, the results of other tools), and use a machine learning approach to categorize variants as benign or deleterious. It is worth noting that inference tools that integrate diverse information, such as PolyPhen2 and SNAP (14), do not necessarily have efficacy better than simpler tools (3). Interestingly, there is further utility in integrating the predictions of individual methods (15), even those that are very similar or that already integrate wide ranges of information, potentially due to minimization of outlier effects.
Functional inferences of the severity of protein disruption only weakly predict disease incidence, disease severity, or clinical outcome. For example, functional inferences of missense variants in the cystic fibrosis gene, CFTR, are not well correlated with disease incidence or severity (16). Similarly, the functional inference of mutation severity in the tumor suppressor gene, TP53, does not correlate significantly with patient clinical outcome (17). An emerging consensus has formed that functional inference scores lack the sensitivity and specificity for their clinical use (18–20). The human genome of any particular individual likely contains many apparently unambiguous disease-associated variants (1), and these very often occur in people without symptoms (21). Variable penetrance is most frequently cited to explain the presence of deleterious variants in asymptomatic individuals, although a prevalent viewpoint is that functional inference tools overcall pathogenic variants (22). Most validation of the various predictive algorithms have used test sets of selected mutations that are known or likely to be disease causing (such as recurrent mutations inferred to be driver mutations in cancer) and likely to be benign (such as germ-line polymorphisms present at high frequency in the human population). What remains to be tested, in essential genes that produce a clear mutant phenotype, is how often do predicted deleterious variants produce the expected mutant phenotype? This question sidesteps the thicket of variable penetrance issues and allows direct appraisal of whether functional inferences accurately predict disruption of the gene/protein in question.
Here we addressed this question by breeding to homozygosity and phenotyping mice with 1 of 33 potentially disruptive de novo point mutations residing in 23 essential immune system genes. Should a mutation disrupt the function of the gene in which it occurs, we expected to observe a previously characterized immune system phenotype specific to that gene. We found that only 4 of 20 missense mutations predicted to be “probably damaging” or “possibly damaging” by PolyPhen2 produced the expected mutant phenotype. This apparent overcalling of deleterious variants by computational tools was not simply due to masking of immune defects in vivo, because when the algorithms were compared with systematic in vitro phenotyping of all possible missense mutations in TP53, only half of the mutations predicted to be deleterious caused a clear reduction of transcription-enhancing activity. Interestingly, the large set of apparent false-positive TP53 inferences has a mean distribution of measured transcriptional activities that is 14.2% lower than the set of TP53 mutations predicted to be benign, consistent with population genetics models that predict a large class of nearly neutral mutations. We present evidence that the apparent overcalling of deleterious mutations by functional inference tools is not a failure of the inference tools per se, but elucidates a critical gap between these inferences and our understanding of how mutations yield phenotypes measured in individuals as opposed to small differences in fecundity compounded over hundreds of generations in large populations of competing individuals.
Results
Identification of Induced De Novo Mutations in Essential Immune Genes.
To identify large numbers of de novo point mutations that have yet to be subject to phenotypic selection, we previously developed a system to exome sequence and accurately identify single nucleotide, protein-altering variants in the progeny of C57BL/6 laboratory mice exposed to the spermatogonial point-mutagen N-ethyl-N-nitrosourea (ENU) (23). As part of a larger project aimed at producing an ENU-induced mouse mutant for each gene in the mouse genome, we developed a resource of mice with identified mutations and corresponding exome sequence data (databases.apf.edu.au/mutations and mutagenetix.org/incidental/incidental_list.cfm). First generation (G1) offspring from male mice treated with ENU have a mean of 45 nonsynonymous de novo mutations (23), although the functional effect of each mutation is largely unknown. Mutations induced by exposure to ENU are likely to have diverse functional consequences, from benign to severely deleterious. This diversity replicates that of spontaneous nonsynonymous de novo mutations that arise in humans at an average rate of 0.75 mutations per child (24). It is also likely to replicate the functional spectrum of inherited nonsynonymous variants in recessive genes that occur at frequencies below 1% in the human population.
To gain an understanding of the functional effects of these phenotypically unselected point mutations, we propagated 33 mutations within 23 genes and bred mouse pedigrees to bring these to homozygosity. Mutations were propagated if they produced a nonsynonymous alteration in genes that, when rendered null, were already known to cause a fully penetrant, well-characterized phenotype in the mouse immune system that was readily detectable by flow cytometry of peripheral blood lymphocytes (details of these genes and the expected phenotypes are given in Table S1). Loss-of-function mutations in 11 of the 23 studied genes (BTK, DCLRE1C, DOCK8, IL2RA, IL7R, JAK3, LIG4, PRKDC, PTPRC, RAG1, and RAG2) are also already known to cause human immune deficiency with Mendelian inheritance.
Table S1.
Expected cellular phenotypes of null mutants
| Gene symbol | Expected phenotype | Gene name |
| BCL2 | Low B and T cells (56, 57) | B cell leukemia/lymphoma 2 |
| BTK | Reduced mature B cells (58–60) | Bruton agammaglobulinemia tyrosine kinase |
| CD22 | Low IgM expression on mature B cells (61–63) | CD22 antigen |
| CD74 | Low CD4 T cells (64, 65) | CD74 antigen |
| DCLRE1C | Absence of B and T cells (66) | DNA cross-link repair 1C |
| DOCK8 | Absence of MZB cells in spleen in ENU mutant mice (67) | dedicator of cytokinesis 8 |
| ETS1 | Reduced T cells (68, 69) | E26 avian leukemia oncogene 1 |
| HNRNPL | Reduced T cells in conditional knockout (70) | heterogeneous nuclear ribonucleoprotein L |
| IL2RA | Activated (increased CD44hi) T cells and enlargement of spleen and LN (71) | interleukin 2 receptor, alpha chain |
| IL7R | Low lymphocytes (72) | interleukin 7 receptor |
| ITCH | Activated (increased CD44hi) T cells (73) | itchy, E3 ubiquitin protein ligase |
| ITPKB | Absence of T cells (74) | inositol 1,4,5-trisphosphate 3-kinase B |
| JAK3 | Low lymphocytes (75, 76) | Janus kinase 3 |
| LIG4 | Low B and T cells in ENU mutant (48) | ligase IV, DNA, ATP-dependent |
| PRKDC | Absence of B and T cells (77–79) | protein kinase, DNA activated, catalytic polypeptide |
| PTPN6 | Early death, immunodeficiency, autoimmunity in spontaneous mutants (80, 81); reduced B cells, down-regulation of IgM and IgD on mature B cells in ENU mutants | protein tyrosine phosphatase, nonreceptor type 6 |
| PTPRC | Low B and T cells (82) | protein tyrosine phosphatase, receptor type, C |
| RAG1 | Absence of B and T cells (83) | recombination activating gene 1 |
| RAG2 | Absence of B and T cells (84) | recombination activating gene 2 |
| RASGRP1 | Low mature T cells (85) | RAS guanyl releasing protein 1 |
| SATB1 | Small mice, early death, low and abnormal T cells (86) | special AT-rich sequence binding protein 1 |
| TBX21 | Low NK and NK T cells (87) and Th1 response (88) | T-box 21 |
| TNFAIP3 | Increased granulocytes and activated (CD44hi) T cells in ENU mutant | tumor necrosis factor, alpha-induced protein 3 |
Mouse-Specific Calculation of Mutation Functional Impact.
The majority of tools that infer the functional consequences of missense sequence variants are understandably built to analyze human sequence data. By default, the inferences made with these tools for model organisms, such as the mouse, using human-specific data sources are inherently of lower and mixed accuracy. We substituted the internal data sources within PolyPhen2 to produce mouse-specific values (see Materials and Methods for details). Table S2 shows PolyPhen2 values for the 33 de novo mutations in immune genes, calculated using both human- and mouse-specific implementations, and human inferences for the mouse mutations made with CADD. Although the mouse- and human-specific PolyPhen2 values are generally well correlated, several values cannot be calculated in humans due to lack of conservation between mouse and human amino acids at the position in question (which is also a limitation when calculating CADD scores for mouse mutant orthologs from human sequence information). Of the values that can be calculated, several PolyPhen2 values change between deleterious and benign categories (Il7r, Lig4, Rasgrp1, and Tnfaip3) depending on the species of database used. Human-specific PolyPhen2 and CADD scores are also highly similar.
Table S2.
Comparison of scores calculated with mouse- and human-specific PolyPhen2 and with human-specific CADD
| Gene symbol | Mutant phenotype mouse | Mouse (mm9 assembly) | Human (GRCh37 assembly) | |||||||||||||||||||
| Chr | Coordinate | Base substitution | UniProt identifier | Amino acid substitution | Polyphen2 | Chr | Coordinate | Strand | Ref nt | Ref codon | Var nt | Ref AA | Var AA | Polyphen2* | CADD† | |||||||
| Category | Score | Category | Score | C score | Phred-like score | |||||||||||||||||
| Bcl2 | 1 | 108608935 | T/C | Q4VBF6 | T175A | Poss. Dam. | 0.924 | 18 | 60985368 | −1 | A | ACT | G | T | A | Poss. Dam. | 0.898 | 1.254779 | 14.68 | |||
| Btk | Yes | X | 131093803 | A/G | P35991 | Y152H | Prob. Dam. | 0.998 | X | 100617615 | −1 | T | TAT | C | Y | H | Prob. Dam. | 0.985 | 5.052784 | 27 | ||
| Cd22 | 7 | 31661166 | T/C | Q3UP36 | M157V | Benign | 0 | 19 | 35826965 | 1 | C | CTC | L** | ‡ | ‡ | |||||||
| Cd74 | 18 | 60970980 | A/T | P04441 | I203F | Poss. Dam. | 0.434 | 5 | 149782843 | −1 | A | ATC | T | I | F | Benign | 0.006 | 1.847686 | 17.91 | |||
| Dock8 | 19 | 25125999 | A/G | Q8C147 | K26E | Poss. Dam. | 0.894 | 9 | 271649 | 1 | A | AAA | F** | ‡ | ‡ | |||||||
| Dock8 | 19 | 25248733 | A/T | Q8C147 | N1567Y | Poss. Dam. | 0.904 | 9 | 432235 | 1 | A | AAT | T | N | Y | Poss. Dam. | 0.578 | 4.777098 | 26 | |||
| Dock8 | 19 | 25257466 | A/G | Q8C147 | T1748A | Benign | 0.001 | 9 | 441301 | 1 | A | ACA | G | T | A | Benign | 0.05 | 1.291052 | 14.89 | |||
| Ets1 | Yes | 9 | 32560393 | C/T | P27577 | P334L | Prob. Dam. | 1 | 11 | 128333513 | −1 | C | CCA | T | P | L | Prob. Dam. | 0.983 | 2.002118 | 19.09 | ||
| Hnrnpl | 7 | 29598392 | C/A | Q8R081 | A118D | Prob. Dam. | 1 | 19 | 39337980 | −1 | C | GCC | T | A | V | Benign | 0.079 | 2.390845 | 22 | |||
| Il2ra | 2 | 11604726 | T/C | P01590 | V230A | Poss. Dam. | 0.763 | 10 | 6061417 | −1 | A | ATA | I** | ‡ | ‡ | |||||||
| Il7r | Yes | 15 | 9454864 | T/G | P16872 | T56P | Prob. Dam. | 0.985 | 5 | 35861037 | 1 | A | ACC | C | T | P | Prob. Dam. | 0.917 | 0.914559 | 12.47 | ||
| Itch | 2 | 155034818 | A/T | Q8C863 | H563L | Prob. Dam. | 0.972 | 20 | 33067458 | 1 | A | CAT | T | H | L | Poss. Dam. | 0.692 | 2.276928 | 21.5 | |||
| Itpkb | 1 | 182263124 | T/C | B2RXC2 | L228P | Benign | 0 | 1 | 226924492 | −1 | A | CCC | P** | ‡ | ‡ | |||||||
| Jak3 | 8 | 76731835 | A/G | Q62137 | I663V | Poss. Dam. | 0.705 | Absent in GRCh37§ | ||||||||||||||
| Lig4 | 8 | 9972775 | T/C | Q8BTF7 | Y335C | Prob. Dam. | 0.985 | 13 | 108862613 | −1 | A | ATG | M** | Prob. Dam. | 0.923 | 4.648035 | 25.7 | |||||
| Lig4 | 8 | 9973305 | A/T | Q8BTF7 | N158K | Benign | 0.013 | 13 | 108863143 | −1 | T | AAT | A | N | L | Benign | 0.016 | 1.494994 | 15.95 | |||
| Prkdc | 16 | 15664332 | A/G | P97313 | D382G | Prob. Dam. | 0.997 | 8 | 48849046 | −1 | A | GAC | G | D | G | Benign | 0.059 | 3.592267 | 23.6 | |||
| Prkdc | 16 | 15697319 | T/C | P97313 | I1010T | Poss. Dam. | 0.935 | 8 | 48817433 | −1 | T | ATA | C | I | T | Poss. Dam. | 0.894 | 4.722739 | 25.9 | |||
| Prkdc | 16 | 15809737 | G/T | P97313 | V3389L | Benign | 0.097 | 8 | 48733488 | −1 | A | TAC | T | Y | F | Benign | 0.142 | 2.284577 | 21.5 | |||
| Prkdc | 16 | 15790559 | A/T | P97313 | Y2044F | Benign | 0.346 | 8 | 48711903 | −1 | G | GTG | C | V | L | Benign | 0.06 | 1.065498 | 13.5 | |||
| Prkdc | 16 | 15816851 | T/C | P97313 | V3589A | Benign | 0.007 | Absent in GRCh37§ | ||||||||||||||
| Ptpn6 | 6 | 124682374 | A/T | P29351 | D90E | Benign | 0.015 | 12 | 7061284 | 1 | C | GAC | A | D | E | Poss. Dam. | 0.511 | 0.916457 | 12.48 | |||
| Ptprc | 1 | 139975069 | T/C | P06800 | K921R | Benign | 0.322 | 1 | 198713286 | 1 | A | AAA | G | K | R | Benign | 0.274 | 2.360073 | 21.9 | |||
| Rag1 | Yes | 2 | 101482545 | T/C | P15919 | E803G | Poss. Dam. | 0.879 | 11 | 36597271 | 1 | A | GAG | G | E | G | Prob. Dam. | 0.988 | 5.433662 | 29.2 | ||
| Rag2 | 2 | 101470774 | A/G | P21784 | D424G | Benign | 0.151 | 11 | 36614448 | −1 | G | CCT | P** | ‡ | ‡ | |||||||
| Rasgrp1 | 2 | 117110866 | T/C | Q9Z1S3 | K659R | Poss. Dam. | 0.712 | 15 | 38786866 | −1 | A | AAG | G | K | R | Benign | 0.389 | 2.452775 | 22.2 | |||
| Satb1 | 17 | 51944614 | A/G | Q60611 | V99A | Prob. Dam. | 0.999 | 3 | 18458486 | −1 | T | GTG | C | V | A | Prob. Dam. | 0.997 | 2.179183 | 20.7 | |||
| Tbx21 | 11 | 96960237 | T/G | Q9JKD8 | E481A | Prob. Dam. | 0.971 | 17 | 45822581 | 1 | A | GAA | C | E | A | Poss. Dam. | 0.551 | 1.464169 | 15.79 | |||
| Tnfaip3 | 10 | 18724943 | A/G | Q60769 | F394S | Prob. Dam. | 1 | 6 | 138199763 | 1 | T | TTC | C | F | S | Prob. Dam. | 0.999 | 1.65794 | 16.78 | |||
| Tnfaip3 | 10 | 18731461 | T/C | Q60769 | K41E | Benign | 0.049 | 6 | 138192485 | 1 | A | AAA | G | K | E | Benign | 0.01 | 2.538327 | 22.3 | |||
Chr, chromosome; Poss. Dam., possibly damaging; Prob. Dam., probably damaging; Ref AA, reference amino acid; Ref nt, reference nucleotide; Var AA, variant amino acid; Var nt, variant nucleotide.
Human PolyPhen2 values calculated using orthologous human protein from UniProt and were only calculated where the reference amino acid was the same between species. Human calculations performed using public PolyPhen2 web server at genetics.bwh.harvard.edu/pph2/bgi.shtml.
CADD scores obtained from public web server at cadd.gs.washington.edu.
Absent values are a consequence of the mouse and human orthologs having a different amino acid at the variant site.
Nucleotide position unable to mapped from mm9 to hg19 with LiftOver (genome.ucsc.edu/cgi-bin/hgLiftOver).
We also calculated functional impact inferences using the sequence homology-based methods SIFT (6) GERP (11), MutationAssessor (7), and PANTHER (10), using mouse input sequences (Table 1 and Table S3) and generated weighted average scores from these diverse measures using Condel (15). Table 1 shows that the functional inference scores for each of the de novo mutations are generally in agreement. There are small differences between functional inference tools regarding the deleterious/benign cutoff used by each different tool, but these are not a predominant feature of the data (Table S3). Occasionally, a single tool produces a discordant inference to the other tools, such as the benign call by PolyPhen2 of the Ptpn6 variant at chr6:124682374. Potentially one tool is making a better call than each of the other tools, although as Condel validation shows, removal of these outlier functional inferences improves the accuracy of the inferences overall (15).
Table 1.
Predicted and observed effect of unselected de novo ENU mutations in 23 essential immune genes
| Gene symbol | Num hom tested | Mut phen obs? | UniProt identifier | AA change | Polyphen category | Polyphen score | SIFT cat. | GERP score* | Mut Assessor cat. | PANTH subPSEC† | Condel cat. | CADD Phred-like score‡ |
| Dclre1c | 3 | Yes | Q8K4J0 | L95Stop | ||||||||
| Dock8 | 1 | Yes | Q8C147 | R1630Stop | ||||||||
| Dock8 | 3 | Yes | Q8C147 | F1885Stop | ||||||||
| Ets1 | 4 | Yes | P27577 | P334L | Prob. Dam. | 1 | Del. | 3.97 | Medium | −2.3993 | Del. | 19.09 |
| Hnrnpl | 5 | No | Q8R081 | A118D | Prob. Dam. | 1 | Del. | 3.74 | Medium | −4.06474 | Del. | 22 |
| Tnfaip3 | 3 | No | Q60769 | F394S | Prob. Dam. | 1 | Del. | 3.27 | Medium | § | Del. | 16.78 |
| Satb1 | 2 | No | Q60611 | V99A | Prob. Dam. | 0.999 | Toler. | Low | −4.11365 | Neutral | 20.7 | |
| Btk | 3 | Yes | P35991 | Y152H | Prob. Dam. | 0.998 | Del. | 3.91 | Low | −2.39271 | Del. | 27 |
| Prkdc | 8 | No | P97313 | D382G | Prob. Dam. | 0.997 | Del. | 3.91 | Medium | −3.61416 | Del. | 23.6 |
| Il7r | 3 | Yes | P16872 | T56P | Prob. Dam. | 0.985 | Del. | 2.92 | Low | § | Del. | 12.47 |
| Lig4 | 4 | No | Q8BTF7 | Y335C | Prob. Dam. | 0.985 | Del. | 2.32 | § | 25.7 | ||
| Itch | 1 | No | Q8C863 | H563L | Prob. Dam. | 0.972 | Del. | 3.61 | Medium | −3.98029 | Del. | 21.5 |
| Tbx21 | 7 | No | Q9JKD8 | E481A | Prob. Dam. | 0.971 | Toler. | 3.22 | Low | −0.97263 | Neutral | 15.79 |
| Prkdc | 4 | No | P97313 | I1010T | Poss. Dam. | 0.935 | Del. | 3.17 | Medium | −1.55087 | Del. | 25.9 |
| Bcl2 | 3 | No | Q4VBF6 | T175A | Poss. Dam. | 0.924 | Toler. | 3.04 | Neutral | −2.00435 | Neutral | 14.68 |
| Dock8 | 5 | No | Q8C147 | N1567Y | Poss. Dam. | 0.904 | Del. | 3.66 | Medium | −4.15438 | Del. | 26 |
| Dock8 | 5 | No | Q8C147 | K26E | Poss. Dam. | 0.894 | Del. | 3.92 | Low | § | Del. | ¶ |
| Rag1 | 1 | Yes | P15919 | E803G | Poss. Dam. | 0.879 | Del. | 3.83 | High | −2.75501 | Del. | 29.2 |
| Il2ra | 2 | No | P01590 | V230A | Poss. Dam. | 0.763 | Del. | 1.89 | Low | −2.25488 | Del. | ¶ |
| Rasgrp1 | 4 | No | Q9Z1S3 | K659R | Poss. Dam. | 0.712 | Toler. | 3.49 | Low | −1.80793 | Neutral | 22.2 |
| Jak3 | 2 | No | Q62137 | I663V | Poss. Dam. | 0.705 | Del. | Low | −3.23261 | Del. | || | |
| Cd74 | 1 | No | P04441 | I203F | Poss. Dam. | 0.434 | Toler. | −2.02 | Medium | −2.04615 | Neutral | 17.91 |
| Prkdc | 5 | No | P97313 | V3389L | Benign | 0.346 | Toler. | 3.73 | Medium | § | Neutral | 21.5 |
| Ptprc | 3 | No | P06800 | K921R | Benign | 0.322 | Toler. | 2.64 | Low | −0.99775 | Neutral | 21.9 |
| Rag2 | 2 | No | P21784 | D424G | Benign | 0.151 | Toler. | 3.51 | Medium | −2.32166 | Neutral | ¶ |
| Prkdc | 2 | No | P97313 | Y2044F | Benign | 0.097 | Toler. | 3.35 | Medium | −2.36465 | Neutral | 13.5 |
| Tnfaip3 | 1 | No | Q60769 | K41E | Benign | 0.049 | Toler. | Neutral | −0.78466 | Neutral | 22.3 | |
| Ptpn6 | 4 | No | P29351 | D90E | Benign | 0.015 | Del. | −5.96 | Medium | § | Neutral | 12.48 |
| Lig4 | 1 | No | Q8BTF7 | N158K | Benign | 0.013 | Toler. | −0.667 | § | 15.95 | ||
| Prkdc | 3 | No | P97313 | V3589A | Benign | 0.007 | Toler. | −1.75 | Low | −1.69134 | Neutral | || |
| Dock8 | 4 | No | Q8C147 | T1748A | Benign | 0.001 | Toler. | 2.51 | Neutral | −1.77255 | Neutral | 14.89 |
| Cd22 | 1 | No | Q3UP36 | M157V | Benign | 0 | Toler. | 2.65 | Neutral | −0.93903 | Neutral | ¶ |
| Itpkb | 2 | No | B2RXC2 | L228P | Benign | 0 | Toler. | −1.04 | Neutral | § | Neutral | ¶ |
AA, amino acid; cat., category; Del., deleterious; Mut Assessor cat., MutationAssessor category; Mut phen obs?, mutant phenotype observed?; Num hom, number of homozygotes; Poss. Dam., possibly damaging; Prob. Dam., probably damaging; Toler., tolerated.
Larger GERP scores denote variants more likely to be deleterious.
Smaller subPSEC scores denote increasingly deleterious variants.
Phred-like scores calculated using coordinates of mouse mutation lifted over to the orthologous human protein.
Missing values were not contained in the alignment created by the HMM.
Mouse and human orthologs have a different amino acid at the variant site.
Absent in GRCh37.
Table S3.
Predicted and observed effect of unselected de novo ENU mutations in 23 essential immune genes
| Gene symbol | Number homozygotes tested | Mutant phenotype observed? | Coordinate (mm9)* | Base substitution | UniProt identifier | Amino acid position | Gene symbol | Number homozygotes tested | GERP | MutationAssessor | PANTHER* | Condel | |||||
| Category | Score | Category | Score | Category | Score | subPSEC | Pdel. | Category | Score | ||||||||
| Dclre1c | 3 | Yes | Chr2:3351056 | T/A | Q8K4J0 | L95Stop | NA | NA | NA | NA | NA | NA | NA | ||||
| Dock8 | 1 | Yes | Chr19:25249949 | A/T | Q8C147 | R1630Stop | NA | NA | NA | NA | NA | NA | NA | ||||
| Dock8 | 3 | Yes | Chr19:25262899 | G/T | Q8C147 | F1885Stop | NA | NA | NA | NA | NA | NA | NA | ||||
| Ets1 | 4 | Yes | Chr9:32560393 | C/T | P27577 | P334L | Prob. Dam. | 1 | Del. | 0 | 3.97 | Medium | 2.685 | −2.3993 | 0.35418 | Del. | 0.638 |
| Hnrnpl | 5 | No | Chr7:29598392 | C/A | Q8R081 | A118D | Prob. Dam. | 1 | Del. | 0 | 3.74 | Medium | 2.19 | −4.06474 | 0.7436 | Del. | 0.612 |
| Tnfaip3 | 3 | No | Chr10:18724943 | A/G | Q60769 | F394S | Prob. Dam. | 1 | Del. | 0 | 3.27 | Medium | 1.955 | −3.25635 | 0.56374 | Del. | 0.6 |
| Satb1 | 2 | No | Chr17:51944614 | A/G | Q60611 | V99A | Prob. Dam. | 0.999 | Toler. | 0.6 | Low | 1.285 | −4.11365 | 0.75281 | Neutral | 0.446 | |
| Btk | 3 | Yes | ChrX:131093803 | A/G | P35991 | Y152H | Prob. Dam. | 0.998 | Del. | 0 | 3.91 | Low | 1.32 | −2.39271 | 0.35268 | Del. | 0.567 |
| Prkdc | 8 | No | Chr16:15664332 | A/G | P97313 | D382G | Prob. Dam. | 0.997 | Del. | 0.04 | 3.91 | Medium | 2.35 | −3.61416 | 0.64889 | Del. | 0.612 |
| Il7r | 3 | Yes | Chr15:9454864 | T/G | P16872 | T56P | Prob. Dam. | 0.985 | Del. | 0.01 | 2.92 | Low | 1.445 | Del. | 0.569 | ||
| Lig4 | 4 | No | Chr8:9972775 | T/C | Q8BTF7 | Y335C | Prob. Dam. | 0.985 | Del. | 0 | 2.32 | ||||||
| Itch | 1 | No | Chr2:155034818 | A/T | Q8C863 | H563L | Prob. Dam. | 0.972 | Del. | 0 | 3.61 | Medium | 3.105 | −3.98029 | 0.72716 | Del. | 0.654 |
| Tbx21 | 7 | No | Chr11:96960237 | T/G | Q9JKD8 | E481A | Prob. Dam. | 0.971 | Toler. | 0.53 | 3.22 | Low | 1.04 | −0.97263 | 0.11636 | Neutral | 0.442 |
| Prkdc | 4 | No | Chr16:15697319 | T/C | P97313 | I1010T | Poss. Dam. | 0.935 | Del. | 0 | 3.17 | Medium | 2.6 | −1.55087 | 0.19013 | Del. | 0.62 |
| Bcl2 | 3 | No | Chr1:108608935 | T/C | Q4VBF6 | T175A | Poss. Dam. | 0.924 | Toler. | 0.33 | 3.04 | Neutral | 0.395 | −2.00435 | 0.2698 | Neutral | 0.439 |
| Dock8 | 5 | No | Chr19:25248733 | A/T | Q8C147 | N1567Y | Poss. Dam. | 0.904 | Del. | 0 | 3.66 | Medium | 2.67 | −4.15438 | 0.76031 | Del. | 0.618 |
| Dock8 | 5 | No | Chr19:25125999 | A/G | Q8C147 | K26E | Poss. Dam. | 0.894 | Del. | 0.01 | 3.92 | Low | 1.585 | Del. | 0.558 | ||
| Rag1 | 1 | Yes | Chr2:101482545 | T/C | P15919 | E803G | Poss. Dam. | 0.879 | Del. | 0 | 3.83 | High | 3.71 | −2.75501 | 0.43906 | Del. | 0.666 |
| Il2ra | 2 | No | Chr2:11604726 | T/C | P01590 | V230A | Poss. Dam. | 0.763 | Del. | 0.01 | 1.89 | Low | 1.04 | −2.25488 | 0.32189 | Del. | 0.504 |
| Rasgrp1 | 4 | No | Chr2:117110866 | T/C | Q9Z1S3 | K659R | Poss. Dam. | 0.712 | Toler. | 0.24 | 3.49 | Low | 0.805 | −1.80793 | 0.23289 | Neutral | 0.436 |
| Jak3 | 2 | No | Chr8:76731835 | A/G | Q62137 | 663V | Poss. Dam. | 0.705 | Del. | 0.04 | Low | 0.96 | −3.23261 | 0.55789 | Del. | 0.482 | |
| Cd74 | 1 | No | Chr18:60970980 | A/T | P04441 | I203F | Poss. Dam. | 0.434 | Toler. | 0.13 | −2.02 | Medium | 1.965 | −2.04615 | 0.27811 | Neutral | 0.462 |
| Prkdc | 5 | No | Chr16:15809737 | G/T | P97313 | V3389L | Benign | 0.346 | Toler. | 0.31 | 3.73 | Medium | 2.28 | Neutral | 0.424 | ||
| Ptprc | 3 | No | Chr1:139975069 | T/C | P06800 | K921R | Benign | 0.322 | Toler. | 0.18 | 2.64 | Low | 0.805 | −0.99775 | 0.11897 | Neutral | 0.37 |
| Rag2 | 2 | No | Chr2:101470774 | A/G | P21784 | D424G | Benign | 0.151 | Toler. | 0.06 | 3.51 | Medium | 1.995 | −2.32166 | 0.33663 | Neutral | 0.421 |
| Prkdc | 2 | No | Chr16:15790559 | A/T | P97313 | Y2044F | Benign | 0.097 | Toler. | 0.16 | 3.35 | Medium | 2.175 | −2.36465 | 0.3463 | Neutral | 0.399 |
| Tnfaip3 | 1 | No | Chr10:18731461 | T/C | Q60769 | K41E | Benign | 0.049 | Toler. | 0.18 | Neutral | 0.205 | −0.78466 | 0.09838 | Neutral | 0.284 | |
| Ptpn6 | 4 | No | Chr6:124682374 | A/T | P29351 | D90E | Benign | 0.015 | Del. | 0.02 | −5.96 | Medium | 2.45 | Neutral | 0.425 | ||
| Lig4 | 1 | No | Chr8:9973305 | A/T | Q8BTF7 | N158K | Benign | 0.013 | Toler. | 0.21 | −0.667 | ||||||
| Prkdc | 3 | No | Chr16:15816851 | T/C | P97313 | V3589A | Benign | 0.007 | Toler. | 0.49 | −1.75 | Low | 0.895 | −1.69134 | 0.21271 | Neutral | 0.249 |
| Dock8 | 4 | No | Chr19:25257466 | A/G | Q8C147 | T1748A | Benign | 0.001 | Toler. | 1 | 2.51 | Neutral | −0.555 | −1.77255 | 0.22663 | Neutral | 0.072 |
| Cd22 | 1 | No | Chr7:31661166 | T/C | Q3UP36 | M157V | Benign | 0 | Toler. | 0.18 | 2.65 | Neutral | 0.345 | −0.93903 | 0.11295 | Neutral | 0.282 |
| Itpkb | 2 | No | Chr1:182263124 | T/C | B2RXC2 | L228P | Benign | 0 | Toler. | 0.16 | −1.04 | Neutral | −0.55 | Neutral | 0.24 | ||
Del., deleterious; NA, not applicable; Pdel., Pdeleterious; Prob. Dam., probably damaging; Poss. Dam., possibly damaging; Toler., tolerated.
Missing values were not contained in the alignment created by the HMM.
In Vivo Phenotypic Consequences of Nonsense and Missense Mutants.
For each mutation, heterozygous G1 animals were bred with unrelated mice and heterozygous mutant offspring identified by allele-specific genotyping. These mutant offspring were intercrossed to yield third-generation (G3) offspring, of which 25% were expected to be homozygous for the mutation. Peripheral blood, or the spleen when necessary for particular mutants (e.g., Dock8), was collected from homozygous mice, and leukocyte subsets were analyzed by flow cytometry. A panel of antibodies was used for flow cytometry capable of detecting abnormalities in lymphocyte subsets that characterize mice with well-defined, homozygous loss-of-function mutations in each of the 23 genes (Table S1). Particular attention was also paid to the detection of subtle hypo- and hypermorphic alterations within the lymphocyte subsets as detectable by flow cytometry of blood samples, such as the compensating shift toward the CD44hi subset of T cells that occurs when thymic T-cell output or peripheral T-cell survival is subtly decreased. We nevertheless recognize that these in vivo phenotyping tests may fail to detect some hypomorphic alleles due to compensating processes in the immune system and lack of exposure to pathogenic microbes in the environment where these mice were raised.
Three of the 33 de novo mutations taken to homozygosity introduced premature stop codons, two in Dock8 and one in Dclre1c (also called Artemis), and all three resulted in the expected immune system in vivo blood cell phenotype (Table 1). Introduction of a premature stop codon is rarely ambiguous because these eliminate a portion of the protein and often cause the transcript to be degraded by nonsense-mediated decay unless the premature stop codon occurs within 55 nucleotides 5′ to the terminal intron (25). All three of the nonsense mutations studied here bear this out.
By contrast with the nonsense mutations, only 4 of 30 missense variants (13%, in Btk, Ets1, Il7r, and Rag1) screened for an expected gene-specific loss-of-function blood cell phenotype had a detectable change in the expected lymphocyte subpopulations relative to WT mice analyzed in parallel (Table 1). This result is consistent with previous evidence comparing the frequency of overt null mutations to missense mutations in sets of unphenotyped incidental ENU mutations to sets of ENU mutations known to cause a mouse phenotype, where it was estimated that only 21% of missense mutations cause a measurable individual phenotype (26). Of the four missense mutations here that produced a phenotype, three (Btk, Ets1, and Il7r) were inferred by PolyPhen2 to be probably damaging and one (Rag1) as possibly damaging. One missense variant (Il7r), which produced the expected mutant phenotype in mice, was predicted to be a benign change with human-specific PolyPhen2, whereas it was predicted probably damaging by the mouse-specific calculation (Table S2). Conversely, none of the 12 missense mutations predicted to be benign had a measurable blood cell immune phenotype by flow cytometry. Hence, from this set of tested mutations in essential immune genes, none of the inference tools appear to generate a high rate of false-negative calls. In contrast, 10 of the 30 de novo missense mutations were called as probably damaging by Polyphen2, yet only 3 of these (30%) resulted in the expected phenotype. A further nine missense mutations were inferred as possibly damaging, yet only one of these (11%) produced the expected mutant phenotype. Fifteen mutations received a Polyphen2 score of 0.85 or greater, yet only 4 of these (27%) were sufficient to cause a discernable immune cell abnormality in individual homozygous mice. PolyPhen2 probably damaging predictions appeared the most reliable indicator of a mutant phenotype, although SIFT predictions (20%) and Condel aggregated predictions (23%) were of similar utility. Table 1 and Table S2 include Phred-like scores calculated with the CADD method (13) for each orthologous human mutation corresponding to each mouse mutation. The CADD scores correctly prioritized only two of the four mutations with a measurable in vivo phenotype (Btk and Rag1), assigning these the highest two scores, whereas the other two received relatively low scores. By contrast, Polyphen2 correctly predicted all four as probably damaging. CADD assigned a score of greater than 20 to 13 missense mutations, yet only 2 of these (15%) had a measurable phenotype. Thus, use of CADD did not improve the prediction of in vivo immune cell phenotypes.
Functional Inferences for All Potential Mutations in 23 Immune Genes.
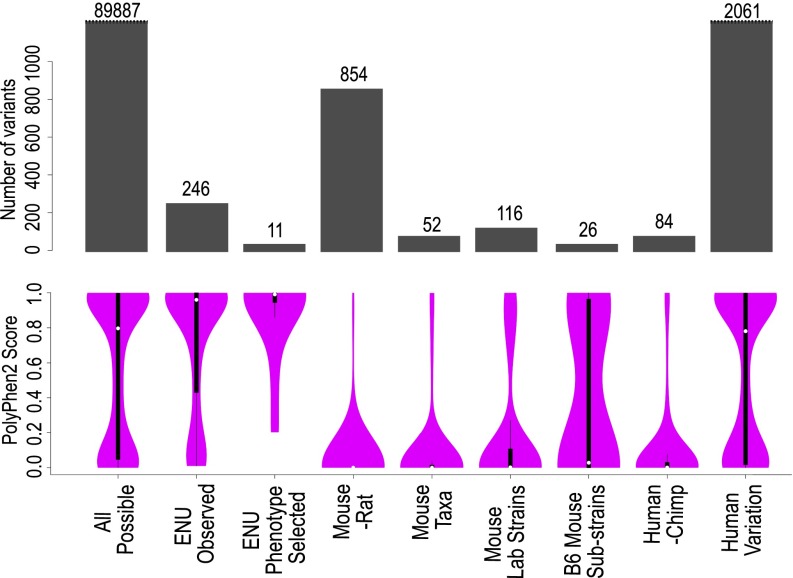
The disparity between the predicted and observed effects of the missense mutations above led us to ask what the range of functional inferences was like for all possible missense mutations in the same set of essential immune genes. We determined the exhaustive set of all 89,887 possible single missense mutations in the 23 essential immune system genes studied above and calculated their PolyPhen2 scores (labeled All possible in Fig. 1). These potential mutations produce a characteristic “hourglass” shape previously observed (23) where the range of Polyphen2 scores is concentrated toward being either deleterious or benign and less likely to be of intermediate effect. Half of all potential missense variants in these genes receive a PolyPhen2 score greater than 0.85.
Fig. 1.
Spectrum of functional consequences predicted by PolyPhen2 for different sources of missense variants in 23 essential immune system genes. PolyPhen2 scores were calculated for the following sets of missense variants: All possible, the complete set of 89,887 possible amino acid substitutions caused by single missense changes in the 23 mouse immune system genes listed in Table 1; ENU observed, 388 mostly unphenotyped, de novo mutations found in the 23 genes by exome sequencing of 2,081 G1 mice; ENU phenotype selected, mutations in the 23 genes discovered by flow cytometric screening of thousands of G3 offspring of ENU-exposed mice; Mouse-rat, missense variants between the C57BL/6J mouse and Brown Norway rat genome sequences; Mouse taxa, missense variants between the four wild-derived, inbred strains representing Mus spretus, Mus musculus musculus, Mus musculus domesticus, and Mus musculus castaneus; Mouse Lab strains, missense variants between the genomes of 14 inbred laboratory mouse strains; B6 mouse substrains, missense variants between the genomes of inbred strains C57BL/6J and C57BL/6N; Human-Chimp, missense variants in the orthologous 23 immune genes between the reference human and chimpanzee genome; Human variation, missense variants in the same immune genes detected by population-scale human exome sequencing (36). The barplots in gray indicate the number of variants present in each set. Purple violin plots are kernel density plots representing the distribution of PolyPhen2 scores for each variant set. The white dot indicates the median PolyPhen2 value for each set. The black bars are box-and-whisker plots: thick black bar extends from the first to the third quartile; thin black lines extend to the lowest and highest data points within 1.5 times the interquartile range.
As opposed to all possible mutations, we also analyzed all missense mutations generated by ENU in the same set of 23 essential immune genes but independent of any phenotypic testing. These ENU-induced mutations were identified by exome sequence analysis of 2,081 G1 offspring of ENU-treated C57BL/6 mice and this allowed us to consider the impact of the characteristic T/A → A/T and A/T → C/G substitutions produced by ENU. Of a total of 136,970 ENU-induced coding variants across the mouse genome, 388 nonsynonymous variants were identified in the 23 essential immune genes (ENU observed in Fig. 1). Only 33 of these were bred to homozygosity and tested for in vivo immune phenotypes as described above, and the remaining 355 represent de novo mutations of unknown phenotype. Comparison of kernel density plots in Fig. 1 between these variant sets indicates that the ENU Observed set tended toward higher PolyPhen2 scores than the All Possible set. Restricting the All Possible set to just T/A → A/T and A/T → C/G substitutions did not replicate this effect; hence, this appears not to be due to the characteristic pattern of ENU-induced mutations. The disparity in sample sizes between these variant sets is large (89,887 All Possible vs. 246 ENU Observed). Iterative random sampling (with replacement) of the All Possible variant set to produce a subset of the same size as the ENU Observed set indicated that the median PolyPhen2 value of the ENU Observed set is not significantly higher than the All Possible set at P = 0.02.
For comparison, we also computed Polyphen2 scores for an independent set of ENU-induced mutations in the same 23 genes, but representing alleles discovered by phenotype-driven blood cell screening followed by meiotic mapping and DNA sequencing (ENU phenotype selected in Fig. 1 and Table S4). These alleles were identified using the same flow cytometric panel by screening peripheral blood samples from thousands of G3 progeny from hundreds of G1 mice, where the G1 parents had not been exome sequenced to detect their ENU-induced mutations. When G3 animals were identified with discernible abnormalities in their lymphocyte subsets, and these were proven heritable by identifying the same immune defect in first- or second-degree relatives from later generations of the same pedigree, exon and exome sequencing followed by allele-specific genotyping was used to identify the causative mutation (23, 26). The phenotypically selected set comprised 19 mutations in 12 of the 23 genes studied here: Bcl2, Btk, Cd22, Il7r, Jak3, Lig4, Prkdc, Ptpn6, Ptprc, Rasgrp1, Satb1, and Tnfaip3 (Table S4). Eleven (58%) were missense mutations, 4 (21%) were stop-gain or nonsense mutations (compared with 9% of unphenotyped mutations in the same genes), and 3 (16%) were mutations that disrupted exon splice sites. Compared with the distribution of all possible missense mutations, the PolyPhen2 scores for phenotypically selected missense mutations were strongly biased toward higher values: 82% received scores greater than 0.85.
Table S4.
Predicted effect of phenotypically selected ENU mutations in 23 essential immune genes
| Gene symbol | Coordinate (mm10) | Base substitution | UniProt identifier | Amino acid position | Polyphen2 | SIFT | GERP | PANTHER | Condel | PANTHER | |||||
| Category | Score | Category | Score | Category | Score | subPSEC | Pdel. | Category | Score | ||||||
| Btk | ChrX:134558685 | C to T | P35991 | Q196Stop | NA | NA | NA | NA | 3.91 | NA | NA | NA | NA | NA | NA |
| Btk | ChrX:134570797 | G to A | P35991 | Intronic | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Cd22 | Chr7:30870310 | T to A | P35329 | C511Stop | NA | NA | NA | NA | −0.765 | NA | NA | NA | NA | NA | NA |
| Prkdc | Chr16:15714375 | T to C | P17313 | Intronic | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Prkdc | Chr16:15777109 | T to A | P17313 | Y3442Stop | NA | NA | NA | NA | 2.21 | NA | NA | NA | NA | NA | NA |
| Ptprc | Chr1:139980367 | T to A | P06800 | Intronic | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Ptprc | Chr1:140007727 | T to C | P06800 | Intronic | NA | NA | NA | NA | 0 | NA | NA | NA | NA | NA | NA |
| Rag1 | Chr2:101845708 | C to T | P15919 | R328Stop | NA | NA | NA | NA | −0.906 | NA | NA | NA | NA | NA | NA |
| Jak3 | Chr8:71680943 | T to C | Q62137 | L398P | Prob. Dam. | 1 | Not Tol. | 0.00 | −3.3 | Medium | 2.545 | −5.78447 | 0.94183 | Deleterious | 0.635 |
| Jak3 | Chr8:71685302 | G to T | Q62137 | G825V | Prob. Dam. | 1 | Not Tol. | 0.00 | 3.08 | High | 4.145 | −6.22242 | 0.96167 | Deleterious | 0.717 |
| Ptpn6 | Chr6:124722095 | C to T | P29351 | T462M | Prob. Dam. | 1 | Not Tol. | 0.03 | −4.72 | High | 3.61 | −4.82869 | 0.86161 | Deleterious | 0.689 |
| Ptprc | Chr1:138099437 | T to C | P06800 | F526S | Prob. Dam. | 1 | Not Tol. | 0.01 | 0.292 | Medium | 2.095 | * | * | Deleterious | 0.607 |
| Tnfaip3 | Chr10:19005539 | T to A | Q60769 | I325N | Prob. Dam. | 1 | Not Tol. | 0.00 | −0.294 | Low | 1.04 | −3.71158 | 0.67075 | Deleterious | 0.556 |
| Rasgrp1 | Chr2:117287984 | A to G | Q9Z1S3 | R519G | Prob. Dam. | 0.991 | Not Tol. | 0.01 | 1.23 | Low | 1.935 | * | * | Deleterious | 0.597 |
| Satb1 | Chr17:51782585 | T to A | Q60611 | F393L | Prob. Dam. | 0.984 | Not Tol. | 0.00 | 3.63 | Low | 1.875 | −2.081 | 0.28516 | Deleterious | 0.594 |
| Lig4 | Chr7:13311208 | A to G | Q8BTF7 | Y288C | Prob. Dam. | 0.975 | Not Tol. | 0.04 | 3.83 | Low | 1.805 | −5.10454 | 0.89134 | Deleterious | 0.592 |
| Ptpn6 | Chr6:124732942 | C to T | P29351 | V2M | Poss. Dam. | 0.913 | Not Tol. | 0.00 | 2.91 | Neutral | 0.68 | * | * | Deleterious | 0.517 |
| Ptprc | Chr1:138101016 | T to C | P06800 | C463R | Benign | 0.56 | Tolerated | 0.11 | 1.59 | Low | 1.095 | * | * | Deleterious | 0.469 |
| Bcl2 | Chr1:106711797 | T to C | P10417 | S24P | Benign | 0.203 | Not Tol. | 0.00 | −0.17 | Low | 1.355 | −2.05643 | 0.28018 | Neutral | 0.411 |
NA, not applicable; Not Tol., not tolerated; Pdel., Pdeleterious; Poss. Dam., possibly damaging; Prob. Dam., probably damaging.
Position not aligned in the HMM.
Functional Inference of Rodent Interspecies and Interstrain Variants.
Given the distribution of scores for all possible missense mutations, we next asked what range of PolyPhen2 scores would be calculated for inherited nonsynonymous variants between mouse and rat in the same 23 essential immune genes. Alignments of these mouse genes with orthologous rat genes (27) were obtained from Ensembl Compara (28), and PolyPhen2 scores were calculated for all missense variants between the two rodent species, which have been separate for 12–24 My (29, 30). Individual neutrally evolving nucleotide positions within the mouse and rat genomes have been mutated on average between 0.15 and 0.2 times since species divergence (27), so it would be expected that between 9,427 and 12,569 missense variants are likely to have arisen in the 62.8 Kb of coding sequence within the 23 essential immune genes during this time. Because the rat sequence comes from highly inbred individuals of the Brown Norway strain (27), the set of mouse-rat missense immune gene variants is expected to have been subject to many intensified generations of purifying selection to remove deleterious variants. Fig. 1 shows that functional inference scores calculated with PolyPhen2 for mouse-rat variants were almost exclusively benign. In the small number of cases where a deleterious prediction was made, investigation of the orthologous mouse and rat sequences demonstrated that the deleterious variants originated from areas of low homology in the pairwise protein sequence alignment. In the investigated subset of these low-homology protein regions, it was apparent that the UniProt reference sequences chosen for the Compara mouse/rat alignment were not the identical splice form, and the deleterious substitutions lay within short regions due to alignment of nonorthologous exons. Even without excluding these seemingly nonhomologous nucleotides and assuming that all variants are nonrecent and have been subject to extensive purifying selection, the deleterious overcall rate of PolyPhen2 is still just 4.68% (deleterious variants/total variants = 40/854 = 0.0468). Similarly, among other functional inference tools, the rates of deleterious overcalling were determined: MutationAssessor, 3.30% (25/758 = 0.0330); SIFT, 2.05% (13/634 = 0.0205; excluding low confidence predictions); PANTHER, 4.44% (15/338 = 0.0444; Pdel > 0.5). Overall, these tools appear remarkably accurate by this measure, and these values are likely an overestimate as some of the variants will be of recent origin or spurious due to local misalignment.
As the likely evolutionary age of a mutation decreases, the time for purifying selection to occur on deleterious variants is diminished and the number of deleterious variants as a proportion of the total variants is expected to be greater. Much genomic data exist for mouse strains, especially the recently diverged laboratory strains of Mus musculus (31, 32). Missense variants between mouse strains in the 23 immune genes above were collected from the Sanger Institute mouse genomes resource (31). The divergences between the four sequenced wild-derived mouse strains, representing four divergent mouse taxa (Mus spretus, Mus musculus musculus, Mus musculus domesticus, and Mus musculus castaneus), are much less than the mouse-rat divergence and are estimated to be not greater than 1.6 My (for musculus/spretus) (33). Each of the four sequenced, wild-derived mouse strains has nevertheless undergone many generations of laboratory-based inbreeding before genome sequencing, potentially exerting strong purifying selection against deleterious missense mutations that might have arisen since strain/subspecies divergence. Fig. 1 shows the range of PolyPhen2 scores for 52 missense variants in the 23 essential immune genes between the mouse subspecies (Mouse Taxa): only 5.7% (3/52) of those observed between mouse taxa received a PolyPhen2 score greater than 0.85 compared with 50% of all possible missense variants in the same immune genes. This result implies that most amino acid substitutions in the 23 immune genes that receive a PolyPhen2 score of greater than 0.85 are indeed sufficiently deleterious to be removed over many generations by purifying selection, either during the divergence of wild-mouse strains or during fixation to homozygosity by laboratory inbreeding.
We extended this analysis to missense variations of very recent origin that exist between inbred strains of laboratory mice. Between the 14 Mus musculus laboratory strains for which a full genome sequence has been obtained (31) 116 missense variants were identified within the set of 23 essential immune genes. The divergences between laboratory Mus strains are complicated, but a large number of the variants identified between strains will be of much more recent origin than those that exist between Mus taxa—of the order of a hundred years. The range of PolyPhen2 scores for these variants (Fig. 1, Mouse Lab Strains) includes 12.9% (15/116) with a PolyPhen2 score of 0.85 or greater, representing an apparently increased fraction of possibly damaging or probably damaging variants than the interspecies Mus variants, but still much lower than the random set of all possible variants. The range also includes a great many benign variants that may predate the strain divergences or are evidence for the effect of purifying selection even over a few hundred generations, especially during the inbreeding conducted over the last century to produce these strains. The inbred C57BL/6J and C57BL/6N substrains have only been genetically separate for ∼220 generations, since 1951 (32), and their genomes have only 32 missense variants in the 23 essential immune genes studied. Although the number of variants is small and the median score is benign, these variants will be mostly of very recent origin, and a higher proportion is computationally inferred to be deleterious (Fig. 1, B6 Mouse Substrains).
Functional Inference of Human Missense Variants in 23 Immune Genes.
Because missense mutations in many of the immune genes studied above are already known to cause devastating human immune deficiency or autoimmune disorders, we performed a parallel set of analyses of human missense variants in the same genes. Within these 23 genes, PolyPhen2 scores were calculated for the 84 missense variants identified between the human reference genome and the chimpanzee genome sequence (34) [Pan troglodytes; human-chimp divergence 5–7 Mya (35)]. The range of scores was similar to those for missense variants between mouse taxa: the median score being benign (0.001), and only 5.6% (5/84) receiving a score greater than 0.85 (Fig. 1, Human-Chimp). However, a very different distribution was observed when we calculated PolyPhen2 scores for the set of all missense variants within these same genes detected in 6,503 human genomes by exome sequencing (36) (data release ESP6500SI-V2). The distribution of scores for all observed human variants was similar to the distribution observed for all potential missense variants in these immune genes, with approximately half (987/2,061 = 47.9%) receiving a score of 0.85 or greater (Fig. 1, Human Variation). Most of these human population variants are likely to be of relatively recent origin, because those with a minor allele frequency of 5% or greater account for 1.7% (35/2,026) of the total variants. Of these more prevalent and presumably older missense mutations, only 20% (7/35) have a PolyPhen2 score >0.85.
Systematic Comparison of Prediction and in Vitro Phenotype for Mutations in TP53.
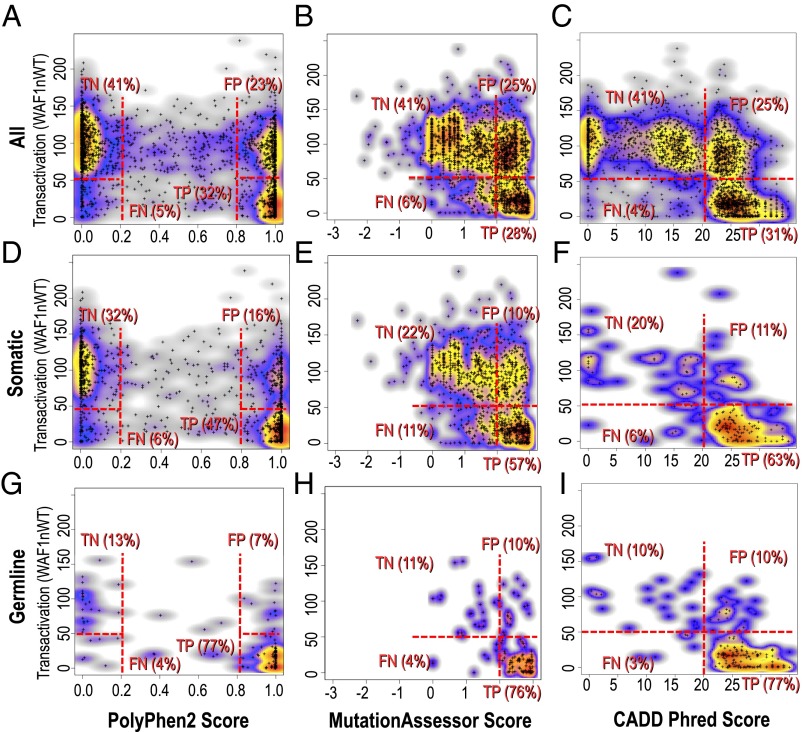
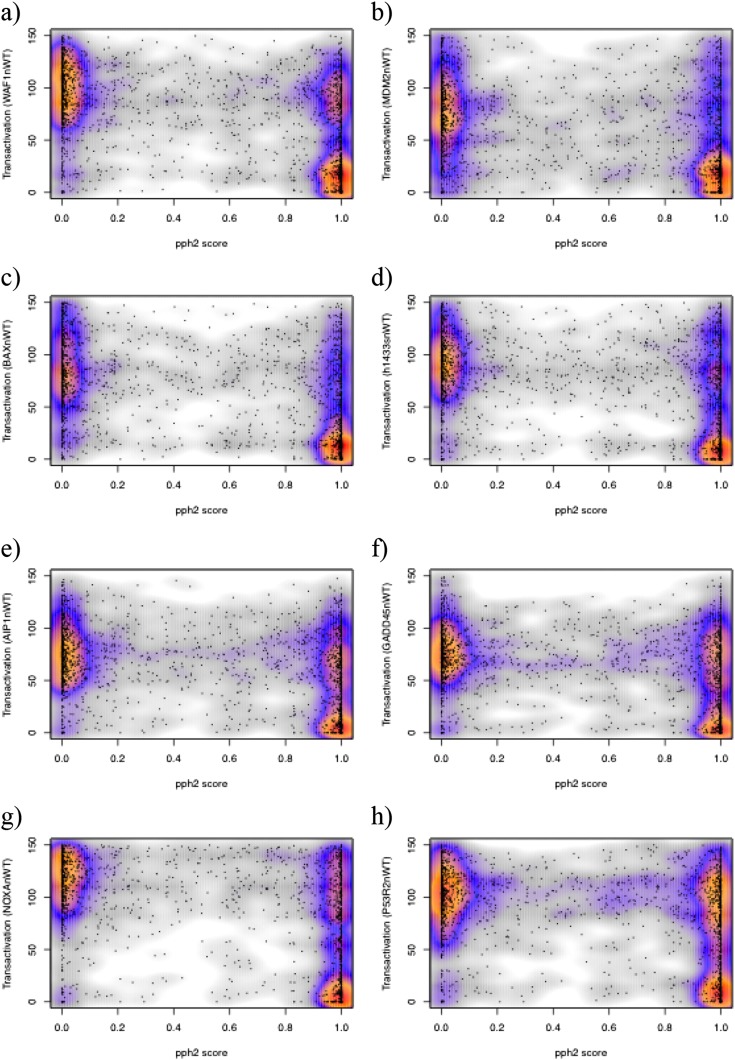
As noted above, a limitation of the in vivo immune cell phenotyping tests is the potential for clinically significant partial loss-of-function mutations to be masked by compensating processes, such as lymphocyte homeostatic expansion to counter diminished lymphocyte differentiation, and by shielding of the mice in the laboratory from normal exposure to a wide range of pathogens and dietary fluctuations. We therefore sought an independent approach to compare computational inferences with experimentally measured effect in a random set of unselected missense mutations, using experimental testing in vitro by quantitative assays with the least possibility for compensation or environmental masking. A comprehensive mutation dataset meeting these criteria exists for the human tumor suppressor gene and protein, TP53 (also called p53) (37). Loss-of-function mutations in TP53 are the most frequent somatic mutation in human cancer and cause highly penetrant immune system cancers when experimentally bred to homozygosity in the germ line of mice. WT TP53 suppresses tumor development by blocking cell division and inducing apoptosis, primarily by binding to defined DNA sequence motifs in numerous target genes and enhancing their transcription into mRNA (38). The transcription-enhancing activity of WT TP53, and each possible single amino acid substitution arising from single nucleotide mutations, has been systematically quantified by expressing each mutant in yeast in vitro and measuring transcription-enhancing activity against different TP53-binding enhancer sequences from the human TP53 target genes p21WAF1, MDM2, BAX, 14–3-3σ, p53AIP1, GADD45, NOXA, and p53R2. Each TP53 target sequence has been inserted into an enhancerless reporter gene encoding a fluorescent protein (37). When WT or mutant TP53 was expressed, transcription and mean accumulation of the fluorescent protein in a population of yeast cells was measured with a spectrophotometer (37).
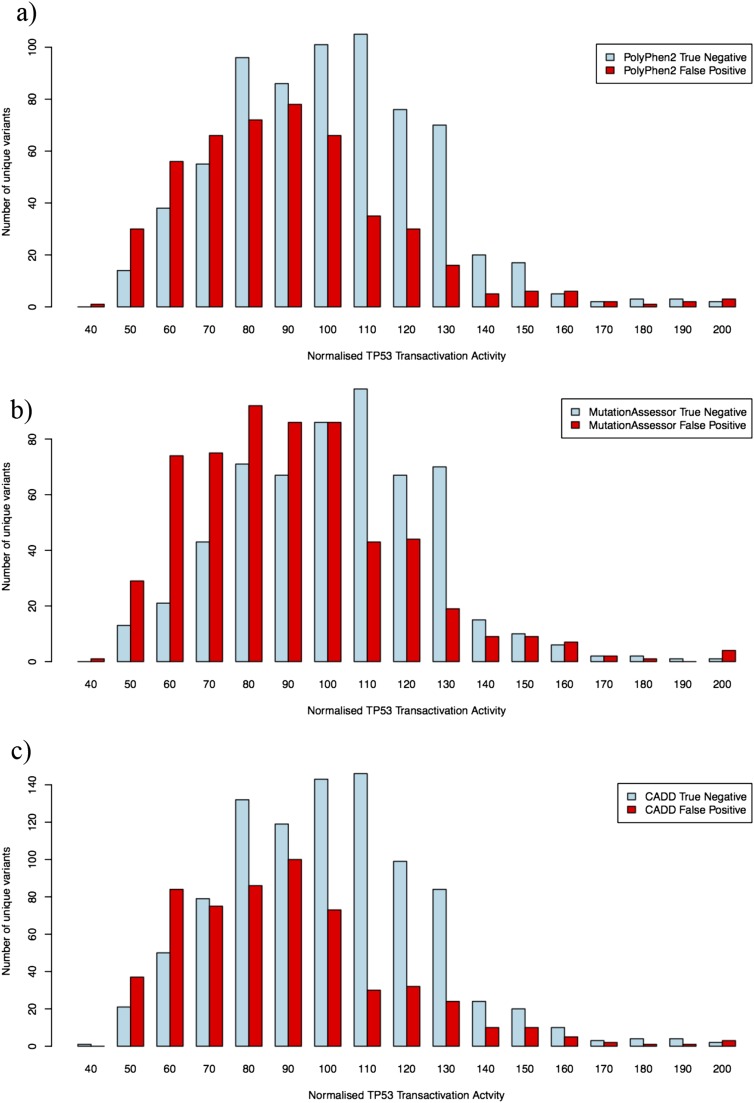
We calculated the PolyPhen2 score for all 2,314 potential TP53 missense mutations arising from single nucleotide substitutions and plotted the score for each mutation against its measured p21WAF1 transcriptional enhancer activity, the latter normalized to the activity of WT TP53 (Fig. 2A). The transactivation (TA) assay results are similar among the differing TP53 target binding sequences (Table S5), as are the plots of PolyPhen2 score against TA activity (Fig. S1). Fig. 2A shows that the set of all possible mutations in the resulting plots cluster into three clear density regions. In a first cluster, 640 TP53 mutations (31.6%) had TA activity diminished to less than 50% of WT and received PolyPhen2 scores of 0.8 or greater. The mutations in this first cluster appear to represent true-positive (TP) predictions. A second cluster includes 831 mutations (41.0%) and had good transcriptional activity, greater than 50% of WT, and received a PolyPhen2 score of 0.2 or less. This cluster appears to represent true-negative (TN) predictions. However, the third clear cluster of 462 mutations (22.8%) also had WT or near WT TA activity yet received a PolyPhen2 score of 0.8 or higher. This cluster represents apparent false-positive (FP) predictions of the effect of missense mutations, and its range of PolyPhen2 scores was comparable to the TP predictions (Fig. 2A). Of the 1,102 mutations predicted to be deleterious with score of 0.8 or greater, 42% had good TA activity measured in yeast with a reporter carrying the TP53-binding sequence from p21WAF1. When these 1,102 predicted deleterious mutations were tested for activity against TP53-binding sequences from other target genes, the fraction that were FP predictions ranged from 34% for MDM2 sequences to 61% for P53R2 sequences (Table S5 and Fig. S1). By contrast, false-negative (FN) predictions, where mutants have less than 50% of WT TA activity yet are predicted not to be damaging (PolyPhen2 ≤ 0.2), accounted for only 93 of the 2,026 mutations (4.6%).
Fig. 2.
Comparison of computationally predicted damage with experimentally measured activity of TP53. (A–C) Analysis of all 2,314 amino acid substitutions in human TP53 that can arise from a single nucleotide substitution. The transcription-enhancing activity of each mutant, acting on the p21WAF1 target sequence in yeast (37), is shown on the y axis normalized to the activity of WT TP53 and multiplied by 100. Predicted damage for each possible mutation is plotted on the x axis, calculated by (A) PolyPhen2; (B) MutationAssessor; and (C) CADD. Regions marked with dashed lines denote true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). (D–F) The same analyses performed on 1,191 somatically acquired TP53 missense mutations from human cancers in the TP53 database. (G–I) The same analyses of 172 germ-line TP53 missense mutations found by clinical sequencing, primarily performed when the clinical phenotype was consistent with a germ-line TP53 mutation.
Table S5.
Apparent accuracy of PolyPhen2 predictions for different sets of TP53 mutations
| Variant set | True positive (pph2 ≥ 0.8 and TA ≤ 50) | True negative (pph2 ≤ 0.2 and TA > 50) | False Positive (pph2 ≥ 0.8 and TA > 50) | False negative (pph2 ≤ 0.2 and TA ≤ 50) | % false-positive predictions among mutations with pph2 ≥ 0.8 |
| All possible variants, target gene TP53 binding sequence | |||||
| WAF1 | 640 (31.6%) | 831 (41.0%) | 462 (22.8%) | 93 (4.6%) | 42% |
| MDM2 | 728 (35.9%) | 709 (35.0%) | 373 (18.4%) | 217 (10.7%) | 34% |
| BAX | 660 (32.6%) | 778 (38.4%) | 441 (21.8%) | 147 (7.3%) | 40% |
| 14–3-3σ | 619 (30.6%) | 817 (40.3%) | 482 (23.8%) | 107 (5.3%) | 44% |
| p53AIP1 | 621 (30.6%) | 753 (37.1%) | 482 (23.8%) | 173 (8.5%) | 44% |
| GADD45 | 566 (27.9%) | 809 (39.9%) | 536 (26.5%) | 115 (5.7%) | 49% |
| NOXA | 470 (23.2%) | 869 (42.9%) | 631 (31.2%) | 55 (2.7%) | 57% |
| p53R2 | 433 (21.4%) | 864 (42.7%) | 668 (33.0%) | 60 (3.0%) | 61% |
| Clinically observed variants, WAF1 binding sequence | |||||
| Somatic | 560 (47%) | 377 (31.6%) | 188 (15.8%) | 67 (5.6%) | 25% |
| Germ line | 132 (76.7%) | 22 (12.8%) | 12 (7.0%) | 6 (3.5%) | 8% |
Fig. S1.
Analysis of PolyPhen2 predicted damage scores compared with the measured transactivation activity of all possible TP53 missense mutations using binding sequences from eight different TP53 target genes. The target binding sequences used in each transactivation assay are (A) p21WAF1, (B) MDM2, (C) BAX, (D) 14–3-3σ, (E) p53AIP1, (F) GADD45, (G) Noxa, and (H) p53R2.
We performed parallel analyses of all possible 2,314 TP53 missense mutations using MutationAssessor and CADD (Fig. 2 B and C and Tables S6 and S7), yielding a similar clustering of all possible mutations into three main groups of TN, FP, and TN predictions. However, unlike PolyPhen2 that produces a simple bimodal distribution of scores, CADD and MutationAssessor gave a broader spread of scores, and a trimodal distribution in the case of CADD. Both of these measures appeared to spread out scores at the upper end of the deleterious range so that the distribution of scores for FPs tended to be lower than the range of scores for TPs. This was especially pronounced for CADD scores: 45% of mutations with a score of 20 or greater were FP predictions (retaining at least 50% of WT activity), but only 19.5% of mutations with a score of 30 or greater were FP by the same measure.
Table S6.
Apparent accuracy of MutationAssessor predictions for different sets of TP53 mutations
| Variant set | True positive (MA ≥ 2 and TA ≤ 50) | True negative (MA < 2 and TA ≥ 50) | False positive (MA ≥ 2 and TA ≥ 50) | False negative (MA < 2 and TA ≤ 50) | % false-positive predictions among mutations with MA ≥ 2 |
| All possible variants, target gene TP53 binding sequence | |||||
| WAF1 | 651 (28.2%) | 935 (40.5%) | 581 (25.2%) | 142 (6.1%) | 20% |
| MDM2 | 765 (33.1%) | 796 (34.5%) | 466 (20.2%) | 283 (12.3%) | 38% |
| BAX | 678 (29.4%) | 877 (38.0%) | 554 (24.0%) | 200 (8.7%) | 27% |
| 14-3-3σ | 632 (27.4%) | 929 (40.3%) | 599 (26.0%) | 148 (6.4%) | 20% |
| p53AIP1 | 626 (27.1%) | 848 (36.7%) | 605 (26.2%) | 233 (10.1%) | 28% |
| GADD45 | 579 (25.1%) | 923 (40.0%) | 653 (28.3%) | 155 (6.7%) | 19% |
| NOXA | 484 (21.0%) | 999 (43.3%) | 747 (32.4%) | 78 (3.4%) | 9% |
| p53R2 | 449 (19.4%) | 998 (43.2%) | 783 (33.9%) | 79 (3.4%) | 9% |
| Clinically observed variants, WAF1 binding sequence | |||||
| Somatic | 93 (57.1%) | 35 (21.5%) | 17 (10.4%) | 18 (11%) | 51% |
| Germ line | 138 (75.4%) | 20 (10.9%) | 18 (9.8%) | 7 (3.8%) | 28% |
Table S7.
Apparent accuracy of CADD predictions for different sets of TP53 mutations
| Variant set | True positive (Phred ≥ 20 and TA ≤ 50) | True negative (Phred < 20 and TA ≥ 50) | False positive (Phred ≥ 20 and TA ≥ 50) | False negative (Phred < 20 and TA ≤ 50) | % false-positive predictions among mutations with Phred ≥ 20 |
| All possible variants, target gene TP53 binding sequence | |||||
| WAF1 | 708 (30.7%) | 941 (40.8%) | 573 (24.8%) | 85 (3.7%) | 45% |
| MDM2 | 832 (36.0%) | 812 (35.2%) | 449 (19.5%) | 215 (9.3%) | 35% |
| BAX | 731 (31.7%) | 879 (38.1%) | 550 (23.8%) | 147 (6.4%) | 43% |
| 14-3-3σ | 671 (29.1%) | 916 (39.7%) | 610 (26.5%) | 109 (4.7%) | 48% |
| p53AIP1 | 702 (30.4%) | 870 (37.7%) | 582 (25.2%) | 156 (6.8%) | 45% |
| GADD45 | 617 (26.7%) | 909 (39.4%) | 666 (28.9%) | 116 (5.0%) | 52% |
| NOXA | 508 (22.0%) | 971 (42.1%) | 773 (33.5%) | 54 (2.3%) | 60% |
| p53R2 | 470 (20.4%) | 968 (42.0%) | 812 (35.2%) | 57 (2.5%) | 63% |
| Clinically observed variants, WAF1 binding sequence | |||||
| Somatic | 102 (62.6%) | 34 (20.9%) | 18 (11.0%) | 9 (5.5%) | 15% |
| Germ line | 140 (76.5%) | 19 (10.4%) | 19 (10.4%) | 5 (2.7%) | 12% |
Evidence for Nearly Neutral TP53 Mutations.
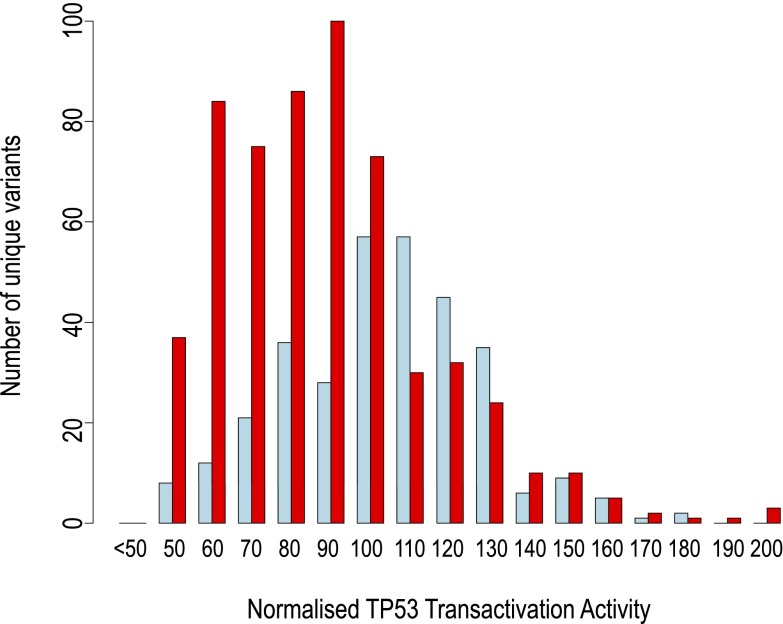
As there are a large number of unique TP53 mutations in both the FP (CADD score > 20; TA activity > 50%) and the group of TN mutations with CADD score < 5 but TA activity > 50%, it was possible to compare the distribution of transcriptional activity values between these groups in aggregate (Fig. 3). This comparison showed that the mean activity of the FP set of mutations was 86% of the mean activity in the TN set, with the difference being statistically significant. A smaller difference, although still statistically significant, was observed in the groups of TN and FP mutants resolved by Polyphen2 and MutationAssessor scores (Fig. S2). Hence a large fraction of mutations predicted to be deleterious by the various algorithms may not be FP functional inferences with no actual effect, but instead appear to be nearly neutral mutations of small effect (39). Fig. 2C also shows that many intermediate points lie between the CADD TN and FP categories (Phred-like score > 5 and < 20). When these intermediate values are included in the TN category, this category remains significantly distinct from the FP category (Fig. S2).
Fig. 3.
Distribution of measured TP53 transactivation activity from true-negative and false-positive TP53 mutations. From all possible TP53 missense mutations, the distribution of measured transcriptional activity values is shown for those inferred as true negative (TN, CADD score < 5 and transactivation activity >50; blue, n = 322) or as false positive (FP, CADD score > 20 and transactivation activity >50; red, n = 573). The mean normalized activity was 108.8 for the TN set and 93.4 for the FP set. The distribution of values for each set is unlikely to be the same (Kolmogorov–Smirnov D = 0.3427 P < 2.2e−16; Wilcox W = 57,261.5, P < 2.2e−16).
Fig. S2.
Distributions of residual TP53 transactivation activities of PolyPhen2-, MutationAssessor-, and CADD-assigned true-negative and false-positive mutations. Measured TP53 transactivation activity for true-negative (TN; blue) and false-positive (FP; red) categories of TP53 missense mutations identified in Fig. 2 A–C using inference scores calculated with (A) PolyPhen2, (B) MutationAssessor, and (C) CADD. For PolyPhen2, the mean normalized activity was 106.1 for the TN set and 94.6 for the FP set (Kolmogorov–Smirnov D = 0.2385, P = 2.454e−14; Wilcox W = 210,343.5, P = 3.541e−16). For MutationAssessor, the mean normalized activity was 104.1 for the TN set and 95.4 for the FP set (Kolmogorov–Smirnov D = 0.1792, P = 2.204e−10; Wilcox W = 327,022.5, P = 3.364e−12). For CADD, when intermediate values (score >5 and <20) are included with the TN category, the mean normalized activity was 105.3 for the TN set and 93.4 for the FP set (Kolmogorov–Smirnov D = 0.2401, P < 2.2e−16; Wilcox W = 190,543.5, P < 2.2e−16).
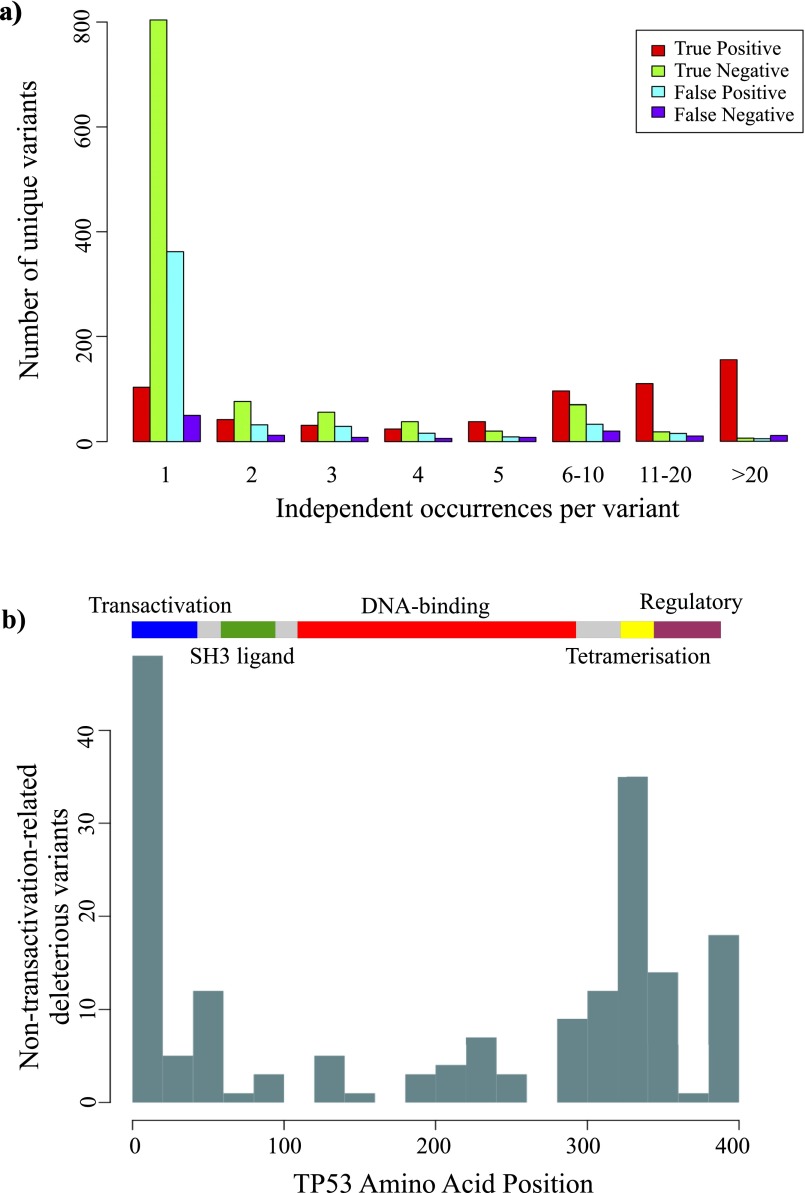
Some apparent FP mutations might have lost some other function critical for TP53 in human cells not measured in the yeast assay. To explore this possibility, we prepared comparable plots for a set of 1,191 distinct amino acid substitutions identified in human cancers and obtained from the curated TP53 mutation database (IARC TP53 Database, R17) (40). Compared with the 42% FP rate found for all mutations with PolyPhen2 score > 0.8, in the cancer-selected somatic mutation set, this was reduced to 25% (Fig. 2C and Table S5). The decrease in FP predictions is likely to be due to the cancer mutation set being enriched for mutations that diminish TP53 transcriptional activity to less than 50% of WT (667/1,192, 53%) compared with the full set of mutations (733/2,026, 36%), consistent with previous analysis using a cutoff of 20% of WT activity (40); 82.8% of the TP subset of TP53 mutations was found in two or more independent cases of cancer (reported in the TP53 mutation database), whereas only 27.7% of the FP nearly neutral subset was found recurrently (Fig. S3A). The small FN subset also had a higher fraction found recurrently in cancer (60.0%), emphasizing the importance of experimental testing to identify this small but important subset of mutations. TN mutations (26.1%) were no more likely to be found recurrently in cancer than FP mutations, suggesting that these subsets represent passenger mutations that confer no neoplastic advantage.
Fig. S3.
Incidence and location of apparent false-positive TP53 somatic mutations. (A) Unique TP53 somatic mutations found in human cancers, separated into categories as shown in Fig. 2D and ranked by the number of independent occurrences in separate cancer cases. Red, true positive; green, true negative; blue, false positive; purple, false negative. (B) The ideogram shows the major functional domains of the TP53 protein. Plotted against this are the number of false-positive mutations in 20 amino acid bins: from the set of all possible TP53 mutations, those that were predicted to be deleterious with Polyphen2 score >0.8 yet actually retained >40% of WT transactivation activity. Unlike most oncogenic TP53 mutations that occur in the DNA-binding domain (red), false-positive mutations appear concentrated in the extremities of the transactivation and tetramerization domains.
The same analysis was performed on a phenotypically selected set of 172 TP53 missense mutations found by sequencing germ-line DNA in patients with types of tumors and age of onset suggesting a loss-of-function TP53 germ-line mutation (IARC TP53 Database, R17) (40). Only 8% of those that were predicted to be deleterious by PolyPhen2 retained >50% activity in the yeast assay, representing a 5.25-fold lower fraction of FP predictions than for all possible mutations. The particularly low rate of FP predictions in the germ-line set is likely to reflect the high proportion of these mutations that lack transcriptional activity, especially in families with highly penetrant cancer meeting the criteria of familial Li-Fraumeni syndrome, where 92% of mutations have less than 20% of WT transcriptional activity (40). In other words, when the clinical phenotype is sufficiently clear and specific for profound loss-of-function mutations in a particular gene, simply finding a rare missense mutation in that gene is already highly predictive, so that computational predictions pose a lower risk of FPs.
The higher concordance between prediction and actual effect among cancer-selected somatic mutations and in phenotypically selected germ-line mutations in TP53 implies that the yeast transcriptional assay measures the relevant TP53 function for tumor suppression. However, it does not rule out the possibility that some FP predictions might disrupt other evolutionarily conserved TP53 functions. Interestingly, FP mutations were concentrated outside the core DNA-binding domain residues 102–292 (41) (Fig. S3B), in contrast to the 95% of oncogenic mutations recorded in TP53 that lie within the DNA-binding domain (42). An example of a functionally important mutation that lies outside the DNA-binding domain and does not have a direct effect on transactivation is the E388A substitution that disrupts conjugation of SUMO1 to K396 in TP53 (43). SUMO1 conjugation at this site has a regulatory effect on TP53 activity (44), but the transactivation assays in yeast (37) indicate this has little effect on the core transactivation mechanism. The PolyPhen2 score for this change (0.893) predicts it is probably damaging, but as the change has little effect of transactivation (p21WAF1 assay value = 105.5), this data point is classified as a FP inference in Fig. 2A.
Discussion
The discordance between the predicted and actual effect of missense mutations revealed here creates the potential for many FP conclusions in clinical whole genome sequencing. Due to the large number of candidate missense variants revealed by whole exome or genome sequencing of any individual, computational prediction of the functional importance of each mutation is widely used to prioritize candidates for further analysis. However, current functional inference tools rely heavily on sequence conservation information. As we have found, these functional predictions do not effectively differentiate between mutations that are immediately clinically relevant, because they ablate or markedly reduce function of an essential protein, and those that are nearly neutral because they only decrease the function of the corresponding protein by 10% (39). FN predictions were also a small but significant subset of mutations in the systematic analysis of TP53. Hence, for interpretation of a clinical genome sequence at present, it is essential to measure experimentally the consequence of any missense mutation thought to be causal.
Two lines of experimental evidence from our work indicate that this lack of differentiation has the potential for overcalling causative mutations. First, 73% of predicted deleterious missense mutations arising from de novo single nucleotide substitutions in 23 essential immune system genes did not sufficiently disrupt protein function to produce an observable defect in the expected subset of blood lymphocytes in individual homozygous mice. Absence of a phenotype in vivo may occur by compensation: for example, a lymphocyte production defect can be compensated by lymphocyte homeostatic expansion. A discernable in vivo phenotype may also require an environmental cofactor such as malnutrition or exposure to a particular infectious agent. For that reason, we turned to the TP53 gene for a complementary analysis of predicted versus actual effects of all possible single nucleotide, missense mutations, taking advantage of a dataset where the experimentally determined phenotype of each mutation has been measured in a simple in vitro assay. Even under these simplified conditions, 42% of the missense TP53 mutations that were predicted to be deleterious caused little or no measurable decrease in the protein’s transcription-enhancing activity.
The plots in Fig. 2, especially those of CADD Phred-like scores, show that mutations that are computationally predicted to be deleterious have a bimodal distribution. Half drastically decreases protein activity to a mean of 17% of WT, and it is this group that is of interest in clinical cases with onset in childhood or adolescence as suspected monogenic or oligogenic disease. The other half, despite most being scored just as likely to be deleterious, actually only slightly decrease protein activity, to a mean of 86% of WT. Discerning the subtle shift in activity caused by these nearly neutral mutations required us to consider the FP mutants as a group, comprising hundreds of independent measurements. Because they have effects on protein activity that are barely measurable in the laboratory, these nearly neutral mutations are unlikely to cause monogenic diseases but may be relevant to diseases that have a more complex basis involving interaction of many weakly damaged genes and environmental factors.
Presumed nearly neutral FP mutations were frequent among unselected sets of missense mutations (de novo ENU mutations, all possible TP53 mutations) but were rare in sets of mutations that had been subject to phenotypic selection. When applied to missense variants in the 23 essential immune system genes between inbred mouse and rat species, or between inbred mouse taxa, Polyphen2 only predicted 4.7% and 5.7%, respectively, to be deleterious. Because 50% of random mutations in the same 23 genes are predicted to be deleterious, the much lower frequencies among mutations that have become fixed in these species/strains implies that most of the FP predictions in the random mutation sets are indeed deleterious over evolutionary scales. This feature was not restricted to mouse mutations, because a similarly low rate of 5.6% was predicted to be deleterious in the set of variants in the 23 essential immune genes between the consensus human and chimpanzee genome sequences compared with 48% of the predominantly rare variants in the same 23 genes present in a population of 6,503 people for whom exome sequences had been obtained.
At first sight, it appears paradoxical that nearly neutral mutations in essential immune genes will be efficiently removed from the species’ gene pool despite having no easily discernible impact on the immune system of individual homozygotes. One possible explanation is that these subtle variants are individually sufficient to cause a serious problem in conjunction with particular environmental stressors such as malnutrition and pathogenic microbes. Another explanation comes from decades of research into the evolution of protein molecules and the nearly neutral theory of molecular evolution (39, 45). Mathematical models predict that slightly disadvantageous nearly neutral alleles will be lost over many generations through random drift in large populations, even when these alleles reduce fecundity by as little as 1% (39). For essential immune genes, that small difference in fecundity could result from one extra bout of influenza per lifetime, which would not be perceived as clinically significant. In addition to genetic drift in large populations, truncating selection has the potential to remove nearly neutral mutations from the gene pool more rapidly (46). This model considers that, as the number of slightly damaging alleles increases in the population, by Poisson distribution a subset of individuals will inherit a larger burden of nearly neutral alleles affecting a critical function (46), for example, in RAG1, RAG2, LIG4, DCLRE1C, and PRKDC and other genes required for VDJ recombination. Although none of these mutations would be of clinical consequence individually, the chance inheritance of three or more subtle defects in the same pathway may be sufficient to cause recurrent infections or Omenn’s syndrome-like autoimmune manifestations. Previously, we demonstrated experimentally how three heterozygous loss-of-function mutations in sequential steps in a biochemical pathway—affecting Lyn kinase, its substrate CD22, and the CD22-binding tyrosine phosphatase SHP1—only precipitate B-cell deficiency in individuals that inherit all three but not any pair or single mutation (47). Some of the 23 immune genes studied here have essential roles outside the immune system, and those other functions may be more sensitive to disruption by apparently FP mutations. For example LIG4, DCLRE1C, and PRKDC are critical for DNA damage repair in all cells, and it is conceivable that small decreases in their activity could subject these variants to purifying selection over evolutionary timescales because of more rapid aging of stem cells (48).
The same question about subtle loss-of-function arises for the apparent FP predictions among TP53 mutations. These apparent FP predictions were frequent in the set of random TP53 mutations (42%) but much lower in sets of TP53 mutations that had been positively selected for TP53 loss-of-function either as a result of being found by selective resequencing in cancer cells (25%) or in the germ line of young cancer patients with suspected Li-Fraumeni syndrome (8%). Most of the TP somatic mutations were found recurrently in different cancers, consistent with these being driver mutations that confer a growth advantage for neoplastic cells. By contrast, most of the apparent FP somatic mutations that retained greater than 50% of transactivation activity were singleton observations, as would be expected if these were random passenger mutations. These singleton FPs might nevertheless represent driver mutations that provide a more subtle growth advantage. A 10% decrease in TP53 transcriptional activity is difficult to distinguish from WT in individual laboratory tests but may nevertheless confer a selective advantage when this effect is compounded over hundreds of cell divisions or when combined with other partial loss-of-function mutations in the TP53 tumor suppression pathway that may have arisen earlier in the evolution of the neoplastic cell clone.
A similar explanation may apply to the 10% of apparent FP predictions for germ-line TP53 mutations. Indeed one of this set came from a clinical case that did not display the age of onset and pattern of sarcomas typical of Li-Fraumeni syndrome. Instead this individual displayed a syndrome of familial adenomatous polyposis (FAP) that was at the severe end of the spectrum and carried an additional germ-line mutation in the APC tumor suppressor gene that is typically inactivated in FAP (40).
In clinical genome sequencing, the risk of FP calling of missense mutations is likely to be highest when the clinical phenotype does not match a distinct Mendelian syndrome but could be explained by defects in hundreds of genes, for example, in sporadic cases of autoimmune disease or in common variable immune deficiency. Here it will be particularly critical to validate computational predictions experimentally by biochemical tests and recreating the mutations in animals (49). By contrast, the FP problem demonstrated here is minimized when the clinical phenotype can be refined sufficiently that only one or two genes could explain it—for example, when a person develops multiple cancers at an early age including uncommon sarcomas typical of Li-Fraumeni syndrome. Our findings underscore the importance of acquiring two additional types of information to interpret missense variation identified by clinical genome sequencing: (i) direct experimental measurement of the consequences of a candidate mutation, using as specific and sensitive an assay as possible; and (ii) much more specific human phenotyping, capable of narrowing the set of candidate genes to a handful related to a particular biochemical pathway or syndrome. Experimental analysis of the connection between nearly neutral mutations and in vivo immune or cancer phenotypes poses a major challenge and will likely require very large numbers of replicate or iterative measurements.
Materials and Methods
Generation of Random Mouse Mutants, Sequencing of Mouse Exomes, and Variant Calling.
Generation of pedigrees from mice treated with ENU, sequencing of exomes of these mice, and computational identification of ENU-induced point mutants were performed as previously described (23). This research was approved by the animal experimentation and ethics committee of the Australian National University (protocol number A2014/61).
Detailed Description of Mouse-Specific PolyPhen2 Changes.
PolyPhen2 (12) scores were calculated from a local mouse-specific installation of PolyPhen version 2.1.0. Local installation required using the mouse specific UniProt (50) and Pfam (51) databases during setup followed by the subsequent mapping of all mouse UniProt protein sequences to mouse assembly (mm9) coordinates. This mapping allows mouse variant genomic coordinates to be converted directly to UniProt amino acid locations giving PolyPhen2 access to mouse-specific protein annotations (often different from the human homolog annotations), thus often resulting in more accurate PolyPhen2 scores. The mapping was accomplished by searching the CCDS (consensus coding sequence; ref. 52) gene sets for exact UniProt protein sequence matches and was successful for 93% of all mouse UniProt entries. The remaining 7% of nonmapping UniProt entries were provided as input to PolyPhen2 as a UniProt protein FASTA-formatted file and relative protein coordinates (which is a less specific input option available with PolyPhen2). To calculate many thousands of PolyPhen2 scores in a timely manner, UniProt-specific PolyPhen2 calculations were cached, hence avoiding duplicate calculations for variants occurring in UniProt entries previously encountered in other samples.
Calculation of Functional Inferences.
Mouse PolyPhen2 scores were calculated as described above. SIFT (6) scores were obtained from the Variant Effect Predictor (53) and from the SIFT Web server (www.siftdna.org). MutationAssessor (mutationassessor.org) (7) and PANTHER (www.pantherdb.org) (10) scores were calculated for mouse-specific UniProt entries using public webservers provided by the method authors. CADD scores (13), which are presently only calculated from human data, were determined by mapping the coordinate of each mouse mutant to the orthologous base in the human genome with liftOver [via the University of California, Santa Cruz (UCSC) genome browser] (54), providing that the orthologous amino acid in the human reference was identical to that in the mouse reference . Scores for these humanized mutations were obtained from the CADD Web server (cadd.gs.washington.edu). GERP (11) scores were derived from the UCSC Genome Browser (54). Condel weighted average scores were calculated using PolyPhen2, SIFT, and MutationAssessor scores using the Perl code provided by the Condel authors (15).
Allele-Specific Genotyping.
Competitive allele-specific genotyping was performed using the KASP system (Kbioscience). A pair of WT and mutant allele-specific oligonucleotide primers were designed to anneal to sequence flanking the variant site. These primers were conjugated with a tail sequence that contained a FRET cassette labeled with allele-specific dyes. With these primers, sample DNA was amplified with a thermal cycler and dye values read with a FluoStar Optima fluorescent microplate reader (BMG Labtechnologies). Homozygous and heterozygous genotypes were distinguished by whether one or two fluorescent signals, respectively, were detected.
Mouse Phenotyping.
Mouse phenotypes were appraised for most mutations by eight-color flow cytometry on peripheral blood. Two hundred microliters of blood was collected by retro-orbital bleeding into tubes containing 20 μL heparin (Sigma: 1,000 U/mL in PBS). Red blood cells were lysed, and samples were stained in 96-well plates alongside WT C57BL/6 mouse controls as previously described (55). Granulocytes were enumerated by forward and side scatter of laser light. The antibodies used to detect B, T, and NK populations were as follows: anti-B220 (RA3-6B2; BD Pharmingen), anti-IgM (R6-60.2; BD Pharmingen), anti-IgD (11-26c.2a; BioLegend), anti-CD3 (17A2; eBioscience), anti-CD4 (RM4-5; BioLegend), anti-CD44 (IM7; BioLegend), anti-KLRG1 (2F1; eBioscience), and anti-NK1.1 (PK136; BD Pharmingen). To detect splenic marginal zone B (MZB) cells in Dock8 mutant mice, spleens were harvested, and single cell suspensions were stained with the following antibodies to distinguish MZB cells (B220+, IgM high, CD21 high, CD23 negative) from follicular B cells (B220+, IgM intermediate, CD21 intermediate, CD23 positive): anti-B220 (RA3-6B2; BD Pharmingen), anti-IgM (II/41; eBioscience), anti-CD21 (7E9; BioLegend), and anti-CD23 (B3B4; Biolegend). Samples were acquired using a LSR II flow cytometer (BD Bioscience) and analyzed using FlowJo software (FlowJo LLC).
Acknowledgments
We thank the National Computational Infrastructure (Australia) for continued access to significant computation resources and technical expertise; the staff of the Australian Phenomics Facility for animal husbandry, DNA preparation, exome sequencing, and genotyping; and the staff of the Microscopy and Cytometry Resource Facility for help with flow cytometry. This work has been funded by National Institutes of Health Grant AI100627, Australian National Health and Medical Research Council Grant 585490, Wellcome Trust Grant 082030/B/07/Z, and the National Collaborative Research Infrastructure Strategy (Australia).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 11426.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511585112/-/DCSupplemental.
References
- 1.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews TD, Sjollema G, Goodnow CC. Understanding the immunological impact of the human mutation explosion. Trends Immunol. 2013;34(3):99–106. doi: 10.1016/j.it.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z. Assessment of computational methods for predicting the effects of missense mutations in human cancers. BMC Genomics. 2013;14(Suppl 3):S7. doi: 10.1186/1471-2164-14-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 5.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum Mutat. 2011;32(6):661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 7.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15(7):978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathe E, et al. Computational approaches for predicting the biological effect of p53 missense mutations: A comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34(5):1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: Evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci USA. 2004;101(43):15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper GM, et al. NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24(20):2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman R, et al. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin Genet. 2010;77(5):464–473. doi: 10.1111/j.1399-0004.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 17.Masica DL, et al. Predicting survival in head and neck squamous cell carcinoma from TP53 mutation. Hum Genet. 2015;134(5):497–507. doi: 10.1007/s00439-014-1470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good BM, Ainscough BJ, McMichael JF, Su AI, Griffith OL. Organizing knowledge to enable personalization of medicine in cancer. Genome Biol. 2014;15(8):438. doi: 10.1186/s13059-014-0438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manolio TA, et al. Implementing genomic medicine in the clinic: The future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm HL. Disease-targeted sequencing: A cornerstone in the clinic. Nat Rev Genet. 2013;14(4):295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassa CA, Tong MY, Jordan DM. Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. Hum Mutat. 2013;34(9):1216–1220. doi: 10.1002/humu.22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley CM, Sunyaev SR, Greenblatt MS, Oetting WS. Clinically relevant variants - identifying, collecting, interpreting, and disseminating: the 2013 annual scientific meeting of the Human Genome Variation Society. Hum Mutat. 2014;35(4):505–510. doi: 10.1002/humu.22516. [DOI] [PubMed] [Google Scholar]
- 23.Andrews TD, et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: An immediate source for thousands of new mouse models. Open Biol. 2012;2(5):120061. doi: 10.1098/rsob.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14(10):1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 26.Bergmann H, et al. B cell survival, surface BCR and BAFFR expression, CD74 metabolism, and CD8- dendritic cells require the intramembrane endopeptidase SPPL2A. J Exp Med. 2013;210(1):31–40. doi: 10.1084/jem.20121076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbs RA, et al. Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428(6982):493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 28.Hubbard T, et al. Ensembl 2005. Nucleic Acids Res. 2005;33(Database issue):D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adkins RM, Gelke EL, Rowe D, Honeycutt RL. Molecular phylogeny and divergence time estimates for major rodent groups: Evidence from multiple genes. Mol Biol Evol. 2001;18(5):777–791. doi: 10.1093/oxfordjournals.molbev.a003860. [DOI] [PubMed] [Google Scholar]
- 30.Springer MS, Murphy WJ, Eizirik E, O’Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100(3):1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon MM, et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013;14(7):R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol Phylogenet Evol. 2004;33(3):626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Filipski A, Swarna V, Walker A, Hedges SB. Placing confidence limits on the molecular age of the human-chimpanzee divergence. Proc Natl Acad Sci USA. 2005;102(52):18842–18847. doi: 10.1073/pnas.0509585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tennessen JA, et al. Broad GO; Seattle GO; NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato S, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100(14):8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 40.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 41.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science. 1994;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 42.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1(1):68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18(22):6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 46.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1(1):40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 47.Cornall RJ, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8(4):497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 48.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447(7145):686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 49.Casanova J-L, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: Lessons from primary immunodeficiencies. J Exp Med. 2014;211(11):2137–2149. doi: 10.1084/jem.20140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruitt KD, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19(7):1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yabas M, et al. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol. 2011;12(5):441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayama K, et al. Disappearance of the lymphoid system in Bcl-2 homozygous mutant chimeric mice. Science. 1993;261(5128):1584–1588. doi: 10.1126/science.8372353. [DOI] [PubMed] [Google Scholar]
- 57.Veis DJ, Sentman CL, Bach EA, Korsmeyer SJ. Expression of the Bcl-2 protein in murine and human thymocytes and in peripheral T lymphocytes. J Immunol. 1993;151(5):2546–2554. [PubMed] [Google Scholar]
- 58.Hendriks RW, et al. Inactivation of Btk by insertion of lacZ reveals defects in B cell development only past the pre-B cell stage. EMBO J. 1996;15(18):4862–4872. [PMC free article] [PubMed] [Google Scholar]
- 59.Kerner JD, et al. Impaired expansion of mouse B cell progenitors lacking Btk. Immunity. 1995;3(3):301–312. doi: 10.1016/1074-7613(95)90115-9. [DOI] [PubMed] [Google Scholar]
- 60.Khan WN, et al. Defective B cell development and function in Btk-deficient mice. Immunity. 1995;3(3):283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 61.O’Keefe TL, Williams GT, Davies SL, Neuberger MS. Hyperresponsive B cells in CD22-deficient mice. Science. 1996;274(5288):798–801. doi: 10.1126/science.274.5288.798. [DOI] [PubMed] [Google Scholar]
- 62.Otipoby KL, et al. CD22 regulates thymus-independent responses and the lifespan of B cells. Nature. 1996;384(6610):634–637. doi: 10.1038/384634a0. [DOI] [PubMed] [Google Scholar]
- 63.Sato S, et al. CD22 is both a positive and negative regulator of B lymphocyte antigen receptor signal transduction: Altered signaling in CD22-deficient mice. Immunity. 1996;5(6):551–562. doi: 10.1016/s1074-7613(00)80270-8. [DOI] [PubMed] [Google Scholar]
- 64.Bikoff EK, et al. Defective major histocompatibility complex class II assembly, transport, peptide acquisition, and CD4+ T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177(6):1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viville S, et al. Mice lacking the MHC class II-associated invariant chain. Cell. 1993;72(4):635–648. doi: 10.1016/0092-8674(93)90081-z. [DOI] [PubMed] [Google Scholar]
- 66.Rooney S, et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol Cell. 2002;10(6):1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 67.Randall KL, et al. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat Immunol. 2009;10(12):1283–1291. doi: 10.1038/ni.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bories JC, et al. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377(6550):635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- 69.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377(6550):639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 70.Gaudreau M-C, Heyd F, Bastien R, Wilhelm B, Möröy T. Alternative splicing controlled by heterogeneous nuclear ribonucleoprotein L regulates development, proliferation, and migration of thymic pre-T cells. J Immunol. 2012;188(11):5377–5388. doi: 10.4049/jimmunol.1103142. [DOI] [PubMed] [Google Scholar]
- 71.Willerford DM, et al. Interleukin-2 receptor α chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3(4):521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 72.Peschon JJ, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180(5):1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fang D, et al. Dysregulation of T lymphocyte function in itchy mice: A role for Itch in TH2 differentiation. Nat Immunol. 2002;3(3):281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 74.Pouillon V, et al. Inositol 1,3,4,5-tetrakisphosphate is essential for T lymphocyte development. Nat Immunol. 2003;4(11):1136–1143. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- 75.Park SY, et al. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3(6):771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 76.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270(5237):794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 77.Gao Y, et al. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9(3):367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 78.Kurimasa A, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19(5):3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taccioli GE, et al. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9(3):355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 80.Shultz LD, et al. Mutations at the murine motheaten locus are within the hematopoietic cell protein-tyrosine phosphatase (Hcph) gene. Cell. 1993;73(7):1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- 81.Tsui HW, Siminovitch KA, de Souza L, Tsui FW. Motheaten and viable motheaten mice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993;4(2):124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- 82.Byth KF, et al. CD45-null transgenic mice reveal a positive regulatory role for CD45 in early thymocyte development, in the selection of CD4+CD8+ thymocytes, and B cell maturation. J Exp Med. 1996;183(4):1707–1718. doi: 10.1084/jem.183.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 84.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 85.Dower NA, et al. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1(4):317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 86.Alvarez JD, et al. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14(5):521–535. [PMC free article] [PubMed] [Google Scholar]
- 87.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 88.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295(5553):338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]