Significance
This study addresses the evolutionary dynamics of antimalarial drug resistance after changes in drug use. We show that chloroquine resistance in Plasmodium falciparum from French Guiana was lost after sustained drug removal, whereas the resistance marker PfCRT K76T remained fixed in the parasite population. This phenotypic reversion was caused by the acquisition of a single additional C350R substitution in PfCRT. This genetic change also impaired susceptibility to piperaquine, suggesting that piperaquine pressure drove the expansion of this allele. These findings have important implications for understanding drug resistance evolution when standard resistance alleles reach fixation and can lose their utility as markers because of adaptive changes at other amino acid positions or loci in the genome.
Keywords: malaria, drug resistance, evolution, Plasmodium falciparum, PfCRT
Abstract
In regions with high malaria endemicity, the withdrawal of chloroquine (CQ) as first-line treatment of Plasmodium falciparum infections has typically led to the restoration of CQ susceptibility through the reexpansion of the wild-type (WT) allele K76 of the chloroquine resistance transporter gene (pfcrt) at the expense of less fit mutant alleles carrying the CQ resistance (CQR) marker K76T. In low-transmission settings, such as South America, drug resistance mutations can attain 100% prevalence, thereby precluding the return of WT parasites after the complete removal of drug pressure. In French Guiana, despite the fixation of the K76T allele, the prevalence of CQR isolates progressively dropped from >90% to <30% during 17 y after CQ withdrawal in 1995. Using a genome-wide association study with CQ-sensitive (CQS) and CQR isolates, we have identified a single mutation in pfcrt encoding a C350R substitution that is associated with the restoration of CQ susceptibility. Genome editing of the CQR reference strain 7G8 to incorporate PfCRT C350R caused a complete loss of CQR. A retrospective molecular survey on 580 isolates collected from 1997 to 2012 identified all C350R mutant parasites as being CQS. This mutation emerged in 2002 and rapidly spread throughout the P. falciparum population. The C350R allele is also associated with a significant decrease in piperaquine susceptibility in vitro, suggesting that piperaquine pressure in addition to potential fitness costs associated with the 7G8-type CQR pfcrt allele may have selected for this mutation. These findings have important implications for understanding the evolutionary dynamics of antimalarial drug resistance.
The emergence of drug-resistant pathogens is a major threat to human health, and Plasmodium falciparum has shown its capacity to develop resistance to every drug that has been deployed against it on a large scale. Although Africa carries by far the heaviest burden of malaria, parasite resistance to chloroquine (CQ), sulfadoxine-pyrimethamine (1), and more recently, artemisinins (2) first emerged in southeast Asia and South America (the latter only for CQ and sulfadoxine-pyrimethamine), where transmission intensity is low. This evolving situation underscores the importance of understanding the evolutionary dynamics of antimalarial drug resistance in distinct transmission contexts. In Central or South America, where clonal propagation can occur (3), the population genetic diversity is reduced, with high rates of inbreeding (4). As a consequence, strong drug pressure can ultimately lead to fixation of drug resistance alleles, even if they generate a fitness cost to parasites in the absence of drug pressure. Thus, whereas in high-transmission African settings, changes in drug pressure are known to alter the relative proportions of mutant and WT alleles of drug resistance determinants, less is known about how drug-selective forces operate in low-transmission contexts where resistance alleles are already fixed. Here, we have examined the evolution of P. falciparum CQ resistance (CQR) in French Guiana after successive changes in therapeutic policy.
French Guiana is a French overseas territory in South America that is included in the Guiana Shield, a pre-Cambrian geological formation including Guyana, Surinam, French Guiana, and parts of Venezuela, Brazil, and Colombia. The region is endemic for both P. falciparum and Plasmodium vivax (5). In the past 15 y, this population of ∼250,000 inhabitants has experienced a greater than 75% decrease in annual cases of malaria: ∼4,500 cases in 2000 compared with fewer than 500 cases in 2014. French Guiana is currently in a control phase and on track to soon enter a preelimination phase (fewer than 5% of febrile patients test positive for Plasmodium), a situation akin to that in 55 other countries around the world (6). Treatment of uncomplicated P. falciparum malaria cases with the former first-line antimalarial CQ was officially abandoned in 1995 because of poor clinical efficacy. Quinine (QN) plus doxycycline became the subsequent treatment of choice until 2007, with sporadic use of halofantrine and mefloquine (MQ). Artemether-lumefantrine, an artemisinin-based combination therapy (ACT), was widely implemented in 2008. Although the artemether–lumefantrine combination has been the official first-line treatment since 2008, illegal gold miners living in the forest are known to self-medicate their fevers with dihydroartemisinin (DHA) -piperaquine (PPQ) -trimethoprim (TMP; DHA + PPQ + TMP; Artecom) tablets.
In South America, the prevalence of the CQR marker K76T in the drug efflux transmembrane protein PfCRT has remained high, despite CQ having been abandoned for the treatment of P. falciparum infections (7–9). This situation contrasts with multiple settings in Africa, where the majority of molecular studies (10–13) have documented an increase in the prevalence of WT pfcrt after CQ withdrawal. An analysis of the diversity of microsatellites flanking pfcrt in Malawi showed that the return to CQ susceptibility was caused by a reexpansion of the WT PfCRT haplotype (11). Such a difference between South America and other endemic regions in response to CQ withdrawal is generally attributed to other amino acid changes in PfCRT and PfMDR1 (both present on the parasite digestive vacuole membrane), which accompany the PfCRT K76T substitution. Specifically, the SVMNT haplotype of South American parasites at PfCRT amino acids 72–76 has been found to be less deleterious than the CVIET haplotype present elsewhere (14, 15). Another explanation for the virtual fixation of CQR alleles in South American parasites is that CQ continues to be used to treat P. vivax infections and therefore, may exert a residual pressure. Lastly, it is thought that the earlier campaign of adding low-dose amodiaquine (AQ) to table salt proved highly effective at creating a selective sweep of AQ and CQR-conferring mutant pfcrt after it had arisen in the late 1950s and eliminating any vestigial WT pfcrt.
We have previously reported that a significant proportion of P. falciparum parasites from French Guiana have a CQ IC50 below the adopted resistance threshold of 100 nM, despite elevated prevalence of K76T, suggesting a reduced predictive value of this marker, at least in this region of South America (16). In this study, we pursued a genotypic and phenotypic survey, including whole-genome sequencing and gene-editing experiments, which led us to identify a molecular marker of drug susceptibility. These findings have important implications for understanding drug resistance evolution in a low-endemicity context where drug resistance alleles can reach fixation, a scenario that will become increasingly dominant as successful control efforts progressively reduce the burden of malaria.
Results
CQ Susceptibility Has Returned to French Guiana After Drug Withdrawal.
We tested the in vitro CQ susceptibility of 1,207 P. falciparum isolates collected between 1994 and 2012 and compared these data with those previously reported for the 1983–1987 period (17). The general trend is that CQ susceptibility followed changes in drug policy (Fig. 1) and can be divided into three main periods. It took 5 y after the official CQ withdrawal in 1995 to observe a decrease in the prevalence of in vitro CQR isolates from ∼90% to ∼40% (Fig. 1B). Of note, such a high prevalence of CQR at ∼90% in the 1990s was already reported in the mid-1980s. From 2000 to 2006, a consistent but small increase in the prevalence of CQR isolates was observed (up to 70% in 2006), potentially because of an overlap between the mechanisms of drug response to CQ and the first-line therapies used at that time (notably QN). This trend toward a resurgence of CQR, however, reversed concomitantly with the progressive introduction of ACT as first-line therapy in 2007. Within 2 y, the prevalence of CQR isolates dropped and thereafter, remained stable at ∼25% (2008–2012).
Fig. 1.
Evolution of P. falciparum in vitro CQ susceptibility in French Guiana from 1983 to 2012. Lines above graphs indicate the successive change in drug recommendations in French Guiana throughout the study period. (A) Box and whisker plots showing the distribution of CQ IC50 values. Whiskers show the 10th and 90th percentiles, boxes represent interquartile range, and the horizontal line is the median. Isolate numbers per year are listed above the x axis. (B) Observed percentage of CQR isolates over time defined by an IC50 > 100 nM. AR, artemether; DX, doxycycline.*1983–1987 data are from ref. 17.
Mutant PfCRT and PfMDR1 Haplotypes Are Fixed in the Parasite Population.
We genotyped a large panel of patient isolates collected between 1997 and 2012 (Table 1). Of 1,054 isolates genotyped for the pfcrt gene, the mutant haplotype SVMNT was present in 97.5%, a far greater proportion than the mean percentage of in vitro CQR parasites (48.6%) observed in the same period. Haplotypes other than SVMNT were very rare (n = 26 of 1,054). For 18 of these 26 patients for whom we had information, 15 had traveled to West Africa (5 CVIET and 10 CVMNK) and 2 had traveled to Haiti (CVMNK) in the previous month, leaving a single patient harboring a CVMNK haplotype that could have been transmitted locally. A similar allelic fixation could be observed for the pfmdr1 gene, because 86.3% of 861 genotyped isolates displayed the South American resistance-associated NFCDY haplotype (amino acids 86, 184, 1,034, 1,042, and 1,246) (Table 1). Altogether, the sum of PfCRT/PfMDR1 haplotypes SVMNT/NFCDY (7G8-type; n = 688) and SVMNT/NFSDY (n = 108) was present in 99.0% of the isolates sampled during this 16-y period. These results confirmed that the standard molecular markers, notably PfCRT K76T and to a lesser degree, PfMDR1, are no longer predictive of CQR in French Guiana.
Table 1.
Prevalence of PfCRT and PfMDR1 haplotypes of isolates collected in French Guiana (1997–2012)
| Haplotype | n | Frequency (%) |
| PfCRT (n = 1,054) | ||
| SVMNT | 1,028 | 97.5 |
| CVMNK | 17 | 1.6 |
| CVIET | 9 | 0.9 |
| PfMDR1 (n = 861) | ||
| NFCDY | 743 | 86.3 |
| NFSDY | 113 | 13.1 |
| NFCDD | 2 | 0.2 |
| NFSND | 2 | 0.2 |
| NYSND | 1 | 0.1 |
Haplotypes denote amino acids at positions 72–76 for PfCRT and positions 86, 184, 1,034, 1,042, and 1,246 for PfMDR1.
Genome-Wide Association Study Identifies PfCRT C350R as the Single Mutation Associated with CQ Susceptibility.
To identify the genomic variants associated with the reversion of CQR in mutant isolates harboring PfCRT K76T, we performed whole-genome sequence analysis of 54 culture-adapted isolates obtained from distinct malaria-infected patients in French Guiana between 2009 and 2013. We then performed a genome-wide association study (GWAS) on a final dataset of 35 isolates [18 CQ-sensitive (CQS) and 17 CQR parasites] containing 34,196 SNPs (Figs. S1 and S2). Strikingly, a single locus on chromosome 7 achieved genome-wide significance for association with CQS: a Tgt/Cgt nonsynonymous substitution in exon 10 of the pfcrt gene that leads to the replacement of a cysteine by an arginine at codon 350 (C350R) (Fig. 2A).
Fig. S1.
Principal component analysis of a pairwise distance matrix between the genomes of parasites collected in French Guiana from 2008 to 2012. This two-component graph shows the relatively homogeneous distribution of the parasite genomes included in the study and the absence of population stratification.
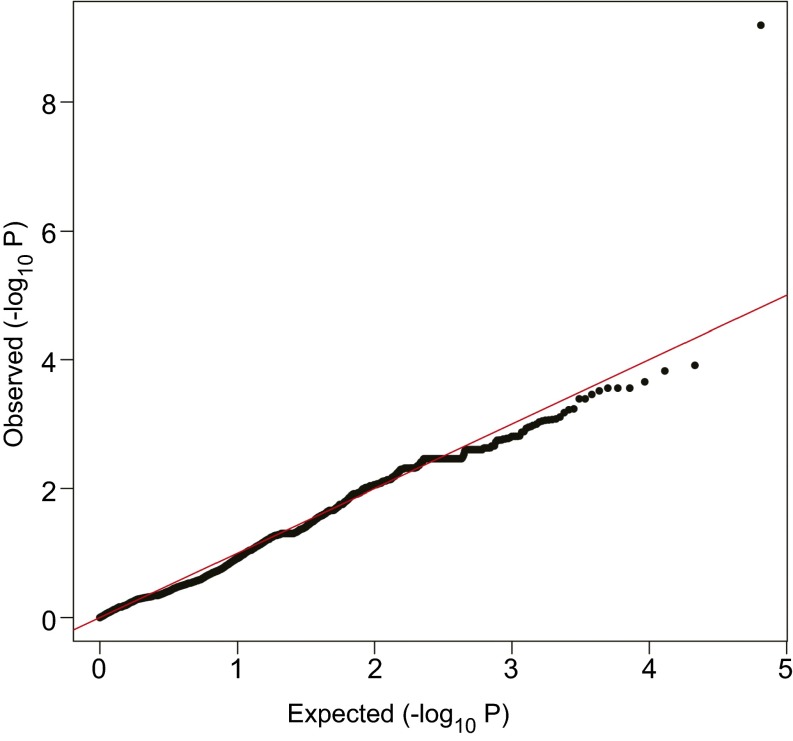
Fig. S2.
Quantile–quantile plots of expected vs. observed P values of individual SNPs associated with the CQ-susceptible phenotype. This plot was generated after correction for inflated P values. The SNP with the most important deviation from the expected P value (upper right corner) corresponds to the PfCRT C350R mutation found in CQ-susceptible parasites.
Fig. 2.
PfCRT C350R is the determinant of in vitro CQR reversion. (A) Signal of SNPs associated with CQ susceptibility. The Manhattan plot shows P values for SNPs with minor allele frequencies >10%. The horizontal red line indicates the P value significance threshold. After adjusting P values for genome-wide inflation (Fig. S2), a single SNP (red dot) on chromosome 7 displayed a highly significant association with CQ susceptibility (P < 1E-11; PfCRT C350R; arrow). (B) pfcrt-edited clones and control reference lines 7G8 and D10 were assayed for their susceptibilities to CQ and its active in vivo metabolite monodesethyl-CQ (mdCQ). IC50 values are represented as means ± SEM. Each line was assayed on at least eight separate occasions in duplicate (Table S1). (C) IC50 values for P. falciparum isolates from French Guiana (means ± SEM). Horizontal dashed lines in B and C denote the resistance threshold of 100 nM. ***P < 0.0001 relative to (B) 7G87G8 or (C) PfCRT C350 isolates (Student t test). (D) Decreasing prevalence of CQR isolates from 1997 to 2012 and its inverse relationship with the increasing prevalence of isolates harboring PfCRT C350R (continuous line). Only isolates carrying the PfCRT SVMNT haplotype at amino acids 72–76 were included in the analysis.
Genome Editing and Molecular Surveys Show That PfCRT C350R Is Responsible for a Loss of CQR.
To assess the impact of PfCRT C350R, we edited the pfcrt gene in the CQR reference strain 7G8. This Brazilian strain carries the canonical South American PfCRT (SVMNT) and PfMDR1 (NFCDY) haplotypes that are shared by the vast majority of isolates from French Guiana. 7G8 parasites that expressed either the recombinant 7G8 allele (7G87G8; transfection control) or the 7G8 variant containing the C350R mutation (7G8C350R) were generated using pfcrt-specific zinc finger nucleases as previously described (18) (Fig. S3). Recombinant clones were obtained by limiting dilution and validated using PCR and sequencing. Drug assay data showed that the PfCRT C350R mutation caused a 24-fold reduction in the mean CQ IC50 values from 380 nM in the reference 7G87G8 clone to 16 nM in the 7G8C350R variant (P < 0.0001, Student t test) (Fig. 2B). A similar reversal of resistance to monodesethyl-CQ, the active in vivo metabolite of CQ, was observed. Parallel studies with parental 7G8 parasites yielded a mean CQ IC50 value of 386 nM, indicating that the expression of the cDNA pfcrt locus (lacking most introns) did not itself cause a significant shift in CQ IC50 values. Of note, the 7G8C350R clone showed a similar mean CQ IC50 value as the reference CQS strain D10 (Papua New Guinea; mean IC50 of 19 nM) that harbors the PfCRT CVMNK WT sequence.
Fig. S3.
Site-specific genome editing strategy to introduce pfcrt modifications into the 7G8 reference line; 7G8 parasites were electroporated with a donor plasmid (pfcrt-hdhfr) encoding a desired compacted (exons 2–13) pfcrt haplotype and pressured with 2.5 nM WR99210. Transformed parasites were subsequently electroporated with a zinc finger nuclease (ZFN) -encoding plasmid (pZFNcrt-bsd) and pressured for 6 d with 2.5 µg/mL BSD + 2.5 nM WR99210 followed by culture in 2.5 nM WR99210. Our pfcrt-specific ZFNs were linked using a viral 2A peptide that permits translation of the two separate ZFNs from a single transcribed mRNA. Binding of these customized DNA binding ZFNs to adjacent sites of intron 1 results in pairing of their split FokI endonucleases, which as an obligate heterodimer form, causes a double-stranded break in intron 1. These events are repaired using the donor template present on the second plasmid that contains 5′ and 3′ regions of homology with DNA resection and donor template strand invasion-dependent DNA repair and resynthesis, thus exchanging the endogenous pfcrt allele with the plasmid-borne variant that contains the entire coding sequence that removes all introns except for intron 1. The presence of ZFN binding site mutations on the plasmid sequences ensures that both the plasmid and the subsequently gene-edited locus are refractory to the ZFNs that can only cleave the original genomic copy. Editing of pfcrt was assessed by PCR analysis of bulk cultures. Cultures showing editing were cloned by limiting dilution and flow cytometrically assessed for growth after 20 d by SYBR Green I and MitoTracker Deep Red (Invitrogen) staining. Presence of the desired pfcrt sequence was validated by PCR and RT-PCR.
In parallel, we examined the association of the C350R mutation with CQ susceptibility in PfCRT K76T mutant isolates in a large set of isolates collected between 1997 and 2012. Exons 2 and 10 were sequenced from 580 isolates. Biallelic samples (C350 + 350R; n = 7) were excluded from the analysis because of a putatively biased IC50 value. All PfCRT C350R mutant isolates (n = 168) were found to be CQS, with a mean CQ IC50 of 23 nM. This value is significantly lower than the mean CQ IC50 value of 160 nM observed with PfCRT C350 parasites (n = 405; P < 0.0001) (Fig. 2C). Altogether, these results provide compelling evidence that the emergence of the PfCRT C350R mutation is primarily responsible for the recent gain of CQ susceptibility in PfCRT K76T isolates from French Guiana.
PfCRT C350R Emerged in 2002 and Then, Rapidly Spread Throughout the Parasite Population in French Guiana.
Only 54% of the CQS isolates with the PfCRT SVMNT sequence harbored the additional C350R mutation in the combined dataset. However, this general picture obscures important temporal disparities. Although it took 5 y after the official withdrawal of CQ to observe a significant decrease of CQR (i.e., in 2000), interestingly, the first PfCRT C350R isolate was not detected until 2 y later in 2002 (Fig. 2D). Thereafter, the prevalence of PfCRT C350R in the total parasite population increased rapidly from 2.7% in 2002 up to 58% in 2012. We note that 100% of the CQR isolates harbored K76T and the C350 residue. Among the SVMNT CQS isolates (defined as CQ IC50 < 100 nM), the prevalence of PfCRT C350R increased from 5% in 2002 to 83% in 2012; the few other CQS C350 isolates nonetheless showed a significantly higher mean CQ IC50 than the CQS C350R isolates (62 vs. 23 nM, respectively; P < 0.001).
Significant Impact of the PfCRT C350R Mutation on Decreased PPQ Susceptibility.
Because PfCRT is known to modulate P. falciparum response to diverse antimalarial drugs, we investigated whether the C350R mutation had an impact on other drug susceptibilities in both the 7G8 genome-edited lines and field isolates collected during the 2008–2012 period (Fig. 3). For all tested compounds, the 7G87G8 modified clone displayed IC50 values similar to the 7G8 parental line, confirming that the pfcrt gene editing did not affect drug responses. Introduction of the PfCRT C350R mutation had no effect on pyronaridine response but significantly increased 7G8 susceptibilities to monodesethyl-AQ (mdAQ), QN, MQ, and lumefantrine (LUM) by 2- to 4-fold (P < 0.001) and induced a moderate but significant 1.6-fold increase in artesunate susceptibility (3.5 vs. 2.2 nM for 7G87G8 and 7G8C350R, respectively; P < 0.05). A significant effect of C350R on PPQ response was also observed, with the 7G8C350R clone showing a 1.4-fold decreased susceptibility (mean PPQ IC50 of 26 vs. 37 nM for 7G87G8 and 7G8C350R, respectively; P < 0.001) (Fig. 3A and Table S1).
Fig. 3.

Impact of PfCRT C350R on antimalarial drug responses (means ± SEM). IC50 values are shown for (A) genome-edited and reference lines (assayed on four to nine separate occasions in duplicate) (Table S1) and (B) French Guianan field isolates from 2008 to 2012. All isolates analyzed in B have the PfCRT SVMNT haplotype at amino acids 72–76. Genome-edited lines or field isolates harboring PfCRT C350R show concordant results in their higher susceptibilities to CQ and mdAQ, contrasting with a decreased susceptibility to PPQ. Asterisks denote the significance level of the difference (A) between 7G8C350R and 7G87G8 or (B) between PfCRT C350R and C350 isolates (Student t test). nd, not determined. *P < 0.05; **P < 0.001; ***P < 0.0001.
Table S1.
Detailed antimalarial drug susceptibilities in genome-edited and reference lines
| Drug and IC values (nM) | D10 | 7G8 | 7G87G8 | 7G8C350R |
| CQ | ||||
| Mean IC50 ± SEM | 19.5 ± 1.5 | 386.2 ± 24.8 | 379.6 ± 24.4 | 16.2 ± 1.8 |
| Mean IC90 ± SEM | 35.1 ± 3.8 | 694.0 ± 67.3 | 676.9 ± 65.7 | 46.3 ± 4.4 |
| Nb of assays | 8 | 8 | 8 | 8 |
| P value vs. 7G87G8 | <0.0001 | |||
| mdCQ | ||||
| Mean IC50 ± SEM | 35.1 ± 3.2 | 1,571.0 ± 228.0 | 1,559.5 ± 146.7 | 53.3 ± 5.7 |
| Mean IC90 ± SEM | 89.0 ± 10.9 | 2,273.2 ± 241.1 | 2,242.1 ± 237.8 | 179.1 ± 16.5 |
| Nb of assays | 8 | 9 | 9 | 9 |
| P value vs. 7G87G8 | <0.0001 | |||
| mdAQ | ||||
| Mean IC50 ± SEM | 24.2 ± 1.2 | 77.9 ± 9.0 | 73.9 ± 5.5 | 21.1 ± 1.5 |
| Mean IC90 ± SEM | 34.8 ± 3.3 | 122.9 ± 9.2 | 121.3 ± 9.1 | 34.6 ± 4.0 |
| Nb of assays | 8 | 8 | 8 | 8 |
| P value vs. 7G87G8 | <0.0001 | |||
| QN | ||||
| Mean IC50 ± SEM | 35.1 ± 2.2 | 275.4 ± 23.4 | 285.1 ± 24.2 | 76.4 ± 5.1 |
| Mean IC90 ± SEM | 151.8 ± 39.1 | 768.0 ± 83.5 | 847.6 ± 92.2 | 260.5 ± 19.6 |
| Nb of assays | 8 | 7 | 7 | 7 |
| P value vs. 7G87G8 | <0.0001 | |||
| LUM | ||||
| Mean IC50 ± SEM | 6.6 ± 1.6 | 2.3 ± 0.4 | 2.4 ± 0.5 | 0.6 ± 0.1 |
| Mean IC90 ± SEM | 19.8 ± 3.4 | 9.4 ± 0.9 | 9.7 ± 0.9 | 2.7 ± 0.3 |
| Nb of assays | 7 | 4 | 4 | 4 |
| P value vs. 7G87G8 | <0.01 | |||
| MQ | ||||
| Mean IC50 ± SEM | nd | 9.0 ± 0.9 | 11.1 ± 0.7 | 6.0 ± 0.9 |
| Mean IC90 ± SEM | nd | 27.4 ± 3.8 | 30.7 ± 3.9 | 20.4 ± 2.0 |
| Nb of assays | nd | 4 | 4 | 4 |
| P value vs. 7G87G8 | <0.01 | |||
| AS | ||||
| Mean IC50 ± SEM | 3.7 ± 0.3 | 3.4 ± 0.4 | 3.5 ± 0.4 | 2.2 ± 0.2 |
| Mean IC90 ± SEM | 8.0 ± 1.1 | 6.3 ± 0.9 | 6.7 ± 0.9 | 5.5 ± 0.4 |
| Nb of assays | 7 | 7 | 7 | 7 |
| P value vs. 7G87G8 | <0.05 | |||
| PPQ | ||||
| Mean IC50 ± SEM | 22.2 ± 2.6 | 25.6 ± 2.1 | 26 ± 2.1 | 36.5 ± 2.4 |
| Mean IC90 ± SEM | 43.1 ± 5.2 | 57.4 ± 3.6 | 60.0 ± 3.7 | 77.9 ± 7.2 |
| Nb of assays | 8 | 9 | 9 | 9 |
| P value vs. 7G87G8 | <0.001 | |||
| PND | ||||
| Mean IC50 ± SEM | 6.1 ± 0.3 | 4.7 ± 0.4 | 4.9 ± 0.4 | 4.5 ± 0.5 |
| Mean IC90 ± SEM | 10.1 ± 1.0 | 7.6 ± 0.5 | 8.7 ± 0.5 | 7.7 ± 0.5 |
| Nb of assays | 8 | 9 | 9 | 9 |
| P value vs. 7G87G8 | ns |
Student t tests were used to compare 7G87G8 with 7G8C350R mean IC50 values. AS, artesunate; mdCQ, monodesethyl-CQ; Nb, number; nd, not determined; ns, not significant; PND, pyronaridine.
French Guianan isolates showed a quite different pattern of drug susceptibilities (Fig. 3B). As expected, C350R mutant isolates were also found to be highly susceptible to mdAQ. Unlike the 7G8 genome-edited lines, no differences in QN, MQ, and LUM susceptibilities were found. However, a moderate but significant association between the presence of the C350R mutation and a decrease in DHA susceptibility was observed (1.0 vs. 1.5 nM for C350 and C350R, respectively; P < 0.001). C350R mutant isolates also displayed a significantly lower susceptibility to PPQ compared with WT C350 isolates (mean IC50 of 61 vs. 33 nM for C350R and C350, respectively; P < 0.001). Overall, both the gene-edited 7G8C350R line and field isolates harboring a PfCRT SVMNT + C350R haplotype showed concordant results in their higher susceptibilities to CQ and mdAQ in contrast with their decreased susceptibilities to PPQ.
Discussion
In French Guiana, the prevalence of CQR has decreased significantly after CQ withdrawal, a pattern also noted in many other endemic areas worldwide (10–13). Here, we show that this phenotypic reversion is caused by the acquisition of a single mutation in the pfcrt gene that completely abolishes the effect of the K76T resistance mutation as opposed to the alternative scenario of the WT allele reemerging.
Repopulation by an allele is driven by multiple parameters, including initial frequency, population size, and the allele’s selective advantage. Our findings clearly show that the WT PfCRT CVMNK haplotype has been purged from the entire population and that the few cases of imported parasites from Africa or Haiti were not sufficient to initiate colonization. The return to CQ susceptibility was not as straightforward as in Malawi, where CQR has completely disappeared, and it has been proposed that CQ treatment could be reintroduced (19). In addition to the fact that an additional mutation had to emerge and then spread in French Guiana, the differential rate of CQR reversal might be related to the clear differences in transmission intensity, population dynamics, and history of drug use between these two settings. In French Guiana, 18 y after CQ withdrawal, 25% of isolates are still CQR in vitro, precluding the reintroduction of CQ as antimalarial treatment.
Two studies have reported the emergence of polymorphisms nearby C350R in PfCRT after in vitro drug selection experiments on CQR strains. Using the 106/176I line, QN single-step selection experiments generated lines with an additional PfCRT Q352K or Q352R mutation that had acquired QN resistance but simultaneously regained CQ susceptibility (20). Separately, in vitro selection for resistance to the IDI-3783 compound that is selectively active against CQR parasites resulted in acquisition of the Q352R mutation in PfCRT (and concomitantly, a loss of CQR), thus providing a conceptual framework for the development of combination therapies that would exploit this evolutionary trap (21). These mutations occur in pfcrt exon 10, which corresponds to amino acid substitutions in transmembrane domain 9. This domain is postulated to function in substrate binding and translocation, similar to transmembrane domain 1, which contains the K76T substitution (20, 22–24). The functional hypothesis for the regained in vitro CQ susceptibility by acquisition of mutations in exon 10 is the reintroduction of a positive charge (K and R) that would inhibit the binding of CQ and block its extrusion from the digestive vacuole. Therefore, an important finding of our study is the demonstration that these particular adaptive mechanisms observed in individual strains during in vitro experiments can also be selected in natural parasite populations where drug resistance alleles are fixed.
Examining the temporal changes in PfCRT haplotype diversity, we note that, between 2001 and 2003, the majority of CQS field isolates did not harbor the C350R mutation. By 2012, however, almost all of the susceptible parasites harbored the PfCRT C350R mutation, implying strong selection for this variant. These results suggest the existence of additional determinants accounting for the loss of CQR in the early 2000s. In another study that examined how the parasite genetic background dictates the degree of mutant pfcrt-mediated CQR, the WT pfcrt allele in CQS strains was replaced by the mutant 7G8-type pfcrt (25). The 3D77G8- and D107G8-transfected lines exhibited only a modest increase in their CQ IC50 values. Nonetheless, these mutant lines were able to withstand high concentrations of CQ and showed delayed recrudescence in culture after high CQ exposure, indicative of a CQ tolerance phenotype. Interestingly, two CQS isolates from French Guiana with the PfCRT SVMNT haplotype were assayed in the same study. The isolate G224 (exhibiting PfCRT C350) also displayed the characteristics of CQ tolerance, whereas the other isolate, H209 (exhibiting PfCRT C350R), had a more typical CQS phenotype. From these previous results and our own observation that the few CQS isolates with WT PfCRT C350 are significantly less susceptible to CQ than the PfCRT C350R isolates, we can hypothesize that these SVMNT PfCRT C350 isolates with a CQ IC50 < 100 nM had a CQ-tolerant phenotype, having already lost robust resistance. Acquisition of full CQ susceptibility would, therefore, be a multistep process that proceeds by an accumulation of adaptive changes, including mutations acquired at other loci, with PfCRT C350R being the final step of a complete CQR reversal.
The successive implementation over time of various first-line drugs after the official cessation of CQ use (Fig. 1) has created a complex selection landscape operating on the pfcrt gene, which is known to impact the efficacy of multiple antimalarials (20, 26). Our drug profiling data showed a significant loss of mdAQ resistance in field isolates harboring the PfCRT C350R mutation, consistent with earlier reports that highlighted the close correlation between CQ and mdAQ responses of PfCRT SVMNT parasites from South America (14). Our observation that mutant C350R isolates were nominally less susceptible to DHA, however, should be interpreted with caution, notably because in vitro artemisinin susceptibilities, as measured using IC50 values, do not correlate with the clinical phenotype of delayed parasite clearance rates in southeast Asia (27, 28).
Our genome-edited 7G8C350R line displayed increased susceptibility to multiple drugs, including mdAQ (as noted above for C350R-carrying field isolates), QN, MQ, artesunate, and LUM. This result reveals a pleiotropic role for this recently emerged pfcrt allele in modulating parasite susceptibility to clinically important antimalarials. This role differs from what is generally observed in Africa with the PfCRT CVIET haplotype or even the PfCRT SVMNT haplotype in Papua New Guinea (29, 30). This effect of C350R on QN and MQ, however, was not replicated in field isolates, underscoring a complex genetic basis of susceptibility in the field. Another primary determinant of susceptibility to these drugs in P. falciparum is the pfmdr1 gene, which in French Guiana harbors the 86N and 184F residues that are associated with increased LUM susceptibility (31, 32). Of note, all isolates remained highly susceptible to LUM, a finding that argues for maintaining the artemether + LUM combination as first-line therapy in the region.
Finally, our data provide evidence that the pfcrt genotype impacts PPQ response. These results suggest that the PPQ pressure exerted by the erratic consumption of Artecom (DHA + PPQ + TMP) by thousands of illegal gold miners in French Guiana since 2002 might be one of the selective forces that drove the expansion of the PfCRT C350R allele during this same period and as a consequence, the return of CQ susceptibility. In fact, miners on the Guiana Shield readily cross borders between Brazil, French Guiana, and Suriname and regularly automedicate with Artecom (33–35). This clue to the putative mechanism of resistance to PPQ is potentially of considerable importance, because PPQ treatment failure is a major and increasing concern in southeast Asia (36–38). Therefore, the association between PfCRT haplotypes and PPQ treatment failure should be further explored in southeast Asia, where polymorphisms in the pfcrt gene, notably in exon 10, were found to accompany mutations in the pfK13 gene associated with artemisinin resistance (39). Given that increased PPQ pressure is likely under mass drug administration campaigns that use DHA–PPQ, it would be also important to monitor parasite changes at the entire pfcrt locus that may result in reduced PPQ susceptibility.
In conclusion, this work illustrates a unique evolutionary path taken by the pfcrt gene as a consequence of an altered drug policy. In the virtual absence of the competing WT, sensitive pfcrt allele, drug susceptibility evolved through the acquisition of an additional amino acid substitution rather than reversion of an existing mutation, such as the critical CQR marker K76T. The extent of this particular mechanism of CQR reversion, described here for the first time to our knowledge, should be further assessed to determine whether it reflects a local evolutionary event or is also happening in other settings or at larger scales. Finally, we provide evidence that, in regions where P. falciparum parasite populations are small and malaria elimination efforts will be targeted, the usefulness of standard molecular markers for drug resistance will require periodic phenotypic validation to contend with ongoing parasite evolution.
Materials and Methods
Origin and Collection of French Guianan Isolates.
P. falciparum isolates were collected between 1994 and 2013 from symptomatic patients diagnosed throughout the network of hospitals, medical laboratories, and health centers in French Guiana. The GWAS samples were collected during 2009–2013. To ensure clonality before whole-genome sequencing, isolates were either assayed with a subset of 15 SNPs from a 24-SNP barcode (40) and confirmed as monoclonal or cloned by limiting dilution.
Drug Assays.
The phenotypes of the isolates were determined ex vivo (i.e., directly after patient blood sampling) or in vitro (after parasite multiplication in culture for several days). The ex vivo method was applied if, at the time of starting phenotypic analysis, the isolate exhibited (i) a P. falciparum infection without concomitant P. vivax infection, (ii) a parasitemia greater than 0.05%, (iii) an interval of less than 4 d between sampling and analysis, and (iv) a quantity of blood sufficient to run the method. Parasite growth was determined by incorporation of tritiated hypoxanthine using a 42-h standard 96-well candle jar method (41). French Guianan culture-adapted clones selected for whole-genome sequencing were assayed in a controlled gas atmosphere with 5% (vol/vol) CO2, 10% (vol/vol) O2, and 85% (vol/vol) N2. For 7G8 and D10 clones, in vitro 72-h drug dose–response assays were performed, and parasite growth was determined using flow cytometry with SYBR Green I and MitoTracker Deep Red (42). Mean IC50 ± SEM values and numbers of repeats are listed in Table S1. Statistical significance of differences between mean IC50 values was assessed using Student t tests.
DNA Extraction and Genotyping.
Genomic DNA was prepared using the QIAmp DNA Blood Kit (Qiagen). Conditions for PCR and targeted sequencing of pfcrt exon 2 (harboring codons 72–76) and polymorphic regions of pfmdr1 were previously described (16). A 121-bp fragment encompassing the complete exon 10 of pfcrt (harboring codon 350) was PCR-amplified and Sanger-sequenced.
Whole-Genome Sequencing and the GWAS.
DNA samples were made into 200-bp Illumina fragment libraries by the Broad Institute Genomics Platform and sequenced with paired-end 101-bp reads on an Illumina HiSeq2000 instrument using V2 chemistry. This design yielded a 70-fold mean coverage, a level sufficient to perform SNP calling. Reads were processed through the Picard sequencing analysis pipeline at the Broad Institute, generating standard sequencing metrics and demultiplexed sample-specific sequencing read BAM (Binary Alignment Map) files. Reads were aligned to the 3D7 reference assembly (PlasmoDB v9.0) using BWA (43), and SNPs were called using the GATK Unified Genotyper (44). Heterozygous SNP calls were discarded along with all calls exhibiting QUAL (quality scores) < Q30. Principal components analysis on called SNPs led to the exclusion of six confounders because of perfect genome identity (n = 4; implying clonal infection in different patients) and population stratification (n = 2; outliers) to find an unstructured sample of unique individual genomes (Fig. S1). We further filtered the dataset to exclude samples exhibiting either a low fraction of genotypable bases because of low sequencing coverage or SNP sites exhibiting a call rate of less than 50% in the remaining samples and/or a minor allele frequency of less than 10%. Samples were excluded from the GWAS if they exhibited calls for fewer than 90% of the SNP loci matching the criteria just described. The GWAS was performed in PLINK v1.07 (pngu.mgh.harvard.edu/purcell/plink/) (45). To avoid false-positive associations, we applied a genomic control correction to adjust P values for genome-wide inflation (Fig. S2).
Generation of pfcrt-Edited 7G8 Clones.
To generate recombinant isogenic lines differing only at their pfcrt locus, we used a previously validated zinc finger nuclease-mediated genome editing strategy (18). A schematic and a description of this approach are shown in Fig. S3.
Ethical Considerations.
Analyzed samples were all obtained by blood collections performed as part of the standard medical care for any patient presenting with fever on hospital admission in French Guiana. Specimen collection complied with Article L.1211–2 and related articles of the French Public Health Code, which states that biobanking and secondary use for scientific purpose of remaining human clinical samples are justified with the informed consent of the patient. Moreover, in compliance with French legislation (article L.1243–3 and related articles of the French Public Health Code), samples received at the Malaria NRC (National Reference Center) had been registered for research purposes in the NRC Biobank declared to both the French Ministry for Research and a French Ethics Committee, which both approved and registered this thematic biobank (declaration number DC-2010–1223; collection no. 2). No institutional review board approval is required according to the French legislation.
Acknowledgments
We thank Denis Blanchet, Bernard Carme, Magalie Demar, Félix Djossou, Claire Grenier, Michel Joubert, and Rachida Boukhari for contributions to the sample collection. This work was supported by European Commission Grant REGPOT-CT-2011-285837-430 STRonGer (to S.P.), Agence Nationale de la Recherche Investissements d’Avenir Grant ANR-10-LABX-25-01 (to S.P. and L.M.), NIH Grants R01AI50234 (to D.A.F.) and R01AI109023 (to D.A.F.), and the Institut de Veille Sanitaire. Sequencing was supported, in part, by National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services Contract HHSN272200900018C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11432.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507142112/-/DCSupplemental.
References
- 1.Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013;29(10):505–515. doi: 10.1016/j.pt.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Ashley EA, et al. Tracking Resistance to Artemisinin Collaboration (TRAC) Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371(5):411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obaldia N, 3rd, et al. Clonal outbreak of Plasmodium falciparum infection in eastern Panama. J Infect Dis. 2015;211(7):1087–1096. doi: 10.1093/infdis/jiu575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39(1):113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 5.Musset L, et al. Malaria on the Guiana Shield: A review of the situation in French Guiana. Mem Inst Oswaldo Cruz. 2014;109(5):525–533. doi: 10.1590/0074-0276140031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO . World Malaria Report. WHO; Geneva: 2014. [Google Scholar]
- 7.Adhin MR, Labadie-Bracho M, Bretas G. Molecular surveillance as monitoring tool for drug-resistant Plasmodium falciparum in Suriname. Am J Trop Med Hyg. 2013;89(2):311–316. doi: 10.4269/ajtmh.12-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffing S, et al. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 2010;54(4):1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vieira PP, et al. pfcrt Polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon Basin. J Infect Dis. 2004;190(2):417–424. doi: 10.1086/422006. [DOI] [PubMed] [Google Scholar]
- 10.Mekonnen SK, et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar J. 2014;13:244. doi: 10.1186/1475-2875-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laufer MK, et al. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis. 2010;202(5):801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mbogo GW, et al. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg. 2014;91(1):54–61. doi: 10.4269/ajtmh.13-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammed A, et al. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J. 2013;12:415. doi: 10.1186/1475-2875-12-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sá JM, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106(45):18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen I, et al. Balancing drug resistance and growth rates via compensatory mutations in the Plasmodium falciparum chloroquine resistance transporter. Mol Microbiol. 2015;97(2):381–395. doi: 10.1111/mmi.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legrand E, et al. Discordant temporal evolution of Pfcrt and Pfmdr1 genotypes and Plasmodium falciparum in vitro drug susceptibility to 4-aminoquinolines after drug policy change in French Guiana. Antimicrob Agents Chemother. 2012;56(3):1382–1389. doi: 10.1128/AAC.05280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedet JP, Germanetto P, Cordoliani G, Bonnevie O, Le Bras J. In vitro activity of various antimalarials (chloroquine, amodiaquine, quinine and mefloquine) against 32 isolates of Plasmodium falciparum in French Guiana. Bull Soc Pathol Exot. 1988;81(1):88–93. [PubMed] [Google Scholar]
- 18.Straimer J, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat Methods. 2012;9(10):993–998. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frosch AE, et al. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J Infect Dis. 2014;210(7):1110–1114. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper RA, et al. Mutations in transmembrane domains 1, 4 and 9 of the Plasmodium falciparum chloroquine resistance transporter alter susceptibility to chloroquine, quinine and quinidine. Mol Microbiol. 2007;63(1):270–282. doi: 10.1111/j.1365-2958.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 21.Lukens AK, et al. Harnessing evolutionary fitness in Plasmodium falciparum for drug discovery and suppressing resistance. Proc Natl Acad Sci USA. 2014;111(2):799–804. doi: 10.1073/pnas.1320886110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin RE, Kirk K. The malaria parasite’s chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol. 2004;21(10):1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 23.Bellanca S, et al. Multiple drugs compete for transport via the Plasmodium falciparum chloroquine resistance transporter at distinct but interdependent sites. J Biol Chem. 2014;289(52):36336–36351. doi: 10.1074/jbc.M114.614206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Paguio M, Roepe PD. The antimalarial drug resistance protein Plasmodium falciparum chloroquine resistance transporter binds chloroquine. Biochemistry. 2004;43(26):8290–8296. doi: 10.1021/bi049137i. [DOI] [PubMed] [Google Scholar]
- 25.Valderramos SG, et al. Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum. PLoS Pathog. 2010;6(5):e1000887. doi: 10.1371/journal.ppat.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson DJ, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15(6):867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witkowski B, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In vitro and ex vivo drug-response studies. Lancet Infect Dis. 2013;13(12):1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361(5):455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mwai L, et al. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob Agents Chemother. 2009;53(12):5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong RP, et al. In vitro sensitivity of Plasmodium falciparum to conventional and novel antimalarial drugs in Papua New Guinea. Trop Med Int Health. 2010;15(3):342–349. doi: 10.1111/j.1365-3156.2009.02463.x. [DOI] [PubMed] [Google Scholar]
- 31.Sisowath C, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199(5):750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad MD, et al. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014;210(3):344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Theije M, Heemskerk M. Moving frontiers in the Amazon: Brazilian small-scale gold miners in Suriname. Eur Rev Latin Am Carib. 2009;87:5–25. [Google Scholar]
- 34.Evans L, 3rd, et al. Quality of anti-malarials collected in the private and informal sectors in Guyana and Suriname. Malar J. 2012;11:203. doi: 10.1186/1475-2875-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nacher M, et al. Made in Europe: Will artemisinin resistance emerge in French Guiana? Malar J. 2013;12:152. doi: 10.1186/1475-2875-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders DL, Vanachayangkul P, Lon C. US Army Military Malaria Research Program National Center for Parasitology, Entomology, and Malaria Control (CNM) Royal Cambodian Armed Forces Dihydroartemisinin-piperaquine failure in Cambodia. N Engl J Med. 2014;371(5):484–485. doi: 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- 37.Lim P, et al. Decreasing pfmdr1 copy number suggests that Plasmodium falciparum in Western Cambodia is regaining in vitro susceptibility to mefloquine. Antimicrob Agents Chemother. 2015;59(5):2934–2937. doi: 10.1128/AAC.05163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leang R, et al. Evidence of falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: Dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. May 26, 2015 doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47(3):226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniels R, et al. A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar J. 2008;7:223. doi: 10.1186/1475-2875-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob Agents Chemother. 1979;16(6):710–718. doi: 10.1128/aac.16.6.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekland EH, Schneider J, Fidock DA. Identifying apicoplast-targeting antimalarials using high-throughput compatible approaches. FASEB J. 2011;25(10):3583–3593. doi: 10.1096/fj.11-187401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]