Progress in microbial ecology over the last few decades has resulted in a new generation of global change models that contemplate microbial feedbacks and offer predictions of an unparalleled accuracy (1). The microbial components of these models are getting increasingly more realistic. In PNAS, Crowther et al. (2) examine how microbes might respond to long-term warming and N fertilization under field conditions to disentangle the relative role of top-down vs. bottom-up controls. The specific study provides clues to the complexity of brown food webs and in many respects is iconic of how microbial manipulation studies should be conducted in the future.
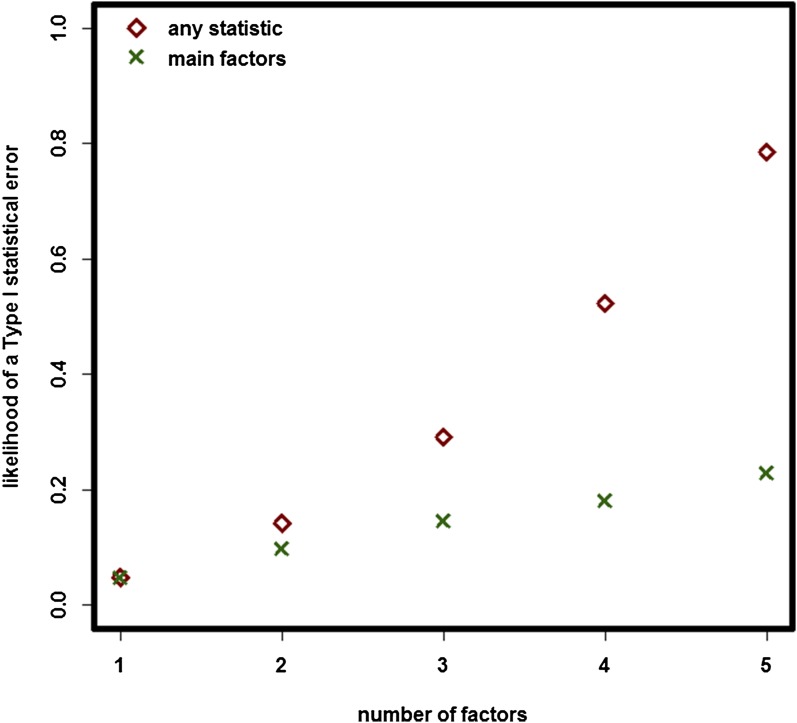
I would like here to raise awareness on a limitation of the abovementioned study: the fact that, due to the experimental design, a four-way factorial design, the experiment was prone to type I statistical errors. The statistical issues arising from complex experimental designs may have been extensively studied in the statistical literature (3), but unfortunately, ignorance of these concerns is common across biologists (4). Data originating from complex studies are commonly interpreted liberally, resulting in what has been termed P hacking: a pronounced mismatch between actual and reported P values (4). Simulations (Fig. 1) have shown that the specific experimental settings resulted in a likelihood of getting a significant result (when considering interaction terms) that was over 50%, whereas the type I likelihood for the four main factors was 18%. A common way to control for type I error likelihood is through a Bonferroni correction (3). The authors implemented a model selection procedure instead based on corrected Akaike information criterion values (2). Unfortunately, this procedure might have only been effective in the cases when the type I errors alluded to high-order interaction terms.
Fig. 1.
Estimates of the likelihood of conducting a type I statistical error (y axis) for experimental designs differing in the number of factors (x axis) assuming that there are six replicates per treatment. The red rhombuses refer to statistical error rates for both main factors and interaction terms, whereas the green symbols are for only main factors.
Despite this shortcoming, the article by Crowther et al. (2) remains a particularly influential study on microbial responses to global change. Many of the effects observed in the study have been strong (some P values are below 0.001), and it is only the subset of the reported P values that were close to the 0.05 threshold that warrant a more cautious interpretation. The merit of reporting incidences of P hacking in prestigious journals mostly lies on raising awareness across biologists that this represents a widespread problem in biology, and it is not limited to articles published in fee-charging open-access journals. It also highlights a possible shortcoming of the existing peer review system in light of the debate over the use of statistical editors in prestigious journals (5). Propagating discussion on current scientific practices may represent a great tool to further improve quality in science.
Footnotes
The author declares no conflict of interest.
References
- 1.Hararuk O, Smith MJ, Luo Y. Microbial models with data-driven parameters predict stronger soil carbon responses to climate change. Glob Change Biol. 2015;21(6):2439–2453. doi: 10.1111/gcb.12827. [DOI] [PubMed] [Google Scholar]
- 2.Crowther TW, et al. Biotic interactions mediate soil microbial feedbacks to climate change. Proc Natl Acad Sci USA. 2015;112(22):7033–7038. doi: 10.1073/pnas.1502956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RA, Levine TR, Lachlan KA, Fediuk TA. The high cost of complexity in experimental design and data analysis type I and type II error rates in multiway ANOVA. Hum Commun Res. 2002;28(4):515–530. [Google Scholar]
- 4.Nuzzo R. Scientific method: Statistical errors. Nature. 2014;506(7487):150–152. doi: 10.1038/506150a. [DOI] [PubMed] [Google Scholar]
- 5.von Wehrden H, Schultner J, Abson DJ. A call for statistical editors in ecology. Trends Ecol Evol. 2015;30(6):293–294. doi: 10.1016/j.tree.2015.03.013. [DOI] [PubMed] [Google Scholar]