Significance
This study develops and exploits an expanded single-molecule fluorescence technique to understand the molecular mechanism of assembly, translocation, and disassembly of a hexameric DNA motor, FtsK, which functions ubiquitously in bacterial chromosome segregation. Assembly of single hexamers on DNA and their subsequent rapid translocation were directly assayed. FtsK hexamers dissociated soon after encountering and activating XerCD-dif recombination complexes. This work contrasts with previously published reports, which suggested that FtsK can reverse spontaneously during translocation, or upon encountering XerCD-dif. Furthermore, in some previous assays, the readout was DNA looping; here, we show that looping does not occur with single hexamer translocation. The technique used provides a blueprint for mechanistic real-time studies of individual protein–nucleic acid complexes.
Keywords: tethered fluorophore motion, DNA translocation, site-specific DNA recombination, chromosome segregation, single-molecule FRET
Abstract
The FtsK dsDNA translocase functions in bacterial chromosome unlinking by activating XerCD-dif recombination in the replication terminus region. To analyze FtsK assembly and translocation, and the subsequent activation of XerCD-dif recombination, we extended the tethered fluorophore motion technique, using two spectrally distinct fluorophores to monitor two effective lengths along the same tethered DNA molecule. We observed that FtsK assembled stepwise on DNA into a single hexamer, and began translocation rapidly (∼0.25 s). Without extruding DNA loops, single FtsK hexamers approached XerCD-dif and resided there for ∼0.5 s irrespective of whether XerCD-dif was synapsed or unsynapsed. FtsK then dissociated, rather than reversing. Infrequently, FtsK activated XerCD-dif recombination when it encountered a preformed synaptic complex, and dissociated before the completion of recombination, consistent with each FtsK–XerCD-dif encounter activating only one round of recombination.
Understanding how molecular machines assemble and act requires a combination of biochemical, structural, and biophysical approaches. In recent years, single-molecule techniques have allowed the observation of biological reactions in real time, thereby avoiding the averaging of ensemble experiments. These single-molecule studies rely on information gained from existing biochemical characterization to set experimental parameters and to place their results in context. To follow complex multistep reactions involving multiple components, methods capable of tracking several observables simultaneously have been established (1–4). In this work, we push the boundaries of a recently developed technique, and apply it to further our understanding of the Escherichia coli XerCD-dif–FtsK molecular machine, which functions in chromosome segregation and coordinates it with cell division.
FtsK is a 1,329-aa DNA translocase, which assembles at the division septum and functions in segregating sister chromosomes during the late stages of the cell cycle in a wide range of bacteria (5–8) by activating site-specific recombination by XerCD at dif (9). The two related Tyr recombinases, XerC and XerD, bind the 28-bp dif site located within the terminus region, ter, on the E. coli chromosome. They unlink catenated chromosomes and resolve chromosome dimers formed by homologous recombination (10–14). Independent of its role in activating XerCD-dif recombination, FtsK appears to play a direct role in the segregation of ter (8). FtsK consists of three domains: an essential 179-aa N-terminal domain that anchors it to the division septum, an ∼500-aa C-terminal motor domain, and an ∼650-aa linker domain (6, 7, 15, 16). The motor domain, FtsKC, is composed of α-, β-, and γ-subdomains (17). The α- and β-subdomains form a dsDNA translocase, belonging to the RecA family of ATPases (17). The γ-subdomain plays a regulatory role in the recognition of the FtsK orientating polar sequence (KOPS) that guides FtsK translocation toward the dif site at ter (18–20), and in the activation of XerCD-dif recombination (9, 14, 21). Activation of recombination requires direct interaction between the γ-subdomain and XerD (22, 23).
Our previous work, using a single tethered fluorophore motion (TFM) reporter, in combination with Förster resonance energy transfer (FRET), determined the conformational transitions in the XerCD-dif complex that occurred as XerD mediated an initial strand exchange to form a Holliday junction (HJ), which was resolved by a subsequent XerC-mediated strand exchange (24). In that work, we indirectly inferred the presence of FtsKC, taking advantage of protein-induced fluorescence enhancement (PIFE) (25). Here, we have expanded TFM-FRET, using two spectrally distinct TFM reporters (one on FtsKC and one on DNA), to directly observe FtsKC as it assembles and translocates, and to correlate its behavior, upon arrival at XerCD, with the progress of the recombination reaction.
Previous studies of FtsKC assembly and translocation have used biochemical methods (26, 27) and single-molecule techniques, including magnetic and optical tweezers (18, 28–30); tethered particle motion (TPM) (18); and, more recently, DNA curtains (31, 32). Optical/magnetic tweezers and TPM experiments have relied on loop extrusion by FtsKC to observe its action (looping by FtsK shortens the length between two DNA ends, hence displacing the bead used in TPM or optical/magnetic tweezers). Many of the single-molecule assays involved the attachment of FtsKC to quantum dot (QD) labels or used derivatives that were known to aggregate, thereby potentially confounding the interpretation of data, because multiple motors could be present in the region of analysis. Experiments utilizing DNA curtains have revealed that FtsKC can push, evict, and bypass proteins bound to DNA as it translocates (32). However, FtsKC stops at least transiently and/or dissociates at XerCD bound to dif (27, 32). Reversals in translocation direction have been observed to occur spontaneously (18, 28, 30, 31) and in response to XerCD bound to dif (32). The use of a fluorophore label, along with singly tethered DNA, has allowed us to observe FtsKC without requiring any loop extrusion, and to observe its interaction with synaptic complexes of XerCD, where previous single-molecule work has only dealt with unsynapsed XerCD-dif (32). Using this approach, we have determined that FtsKC assembles on DNA as a single hexamer, and begins translocating rapidly (∼0.25 s), without extruding a loop of DNA. When it reached XerCD bound to dif, either in a synapsed or unsynapsed conformation, it resided briefly for ∼0.5 s and then dissociated without any evidence of reversal. FtsKC activated recombination when it met synapsed XerCD-dif complexes, and then dissociated faster than the completion of recombination by XerCD.
Results
Spectrally Separate Fluorophores Report on DNA Conformation and FtsKC Behavior.
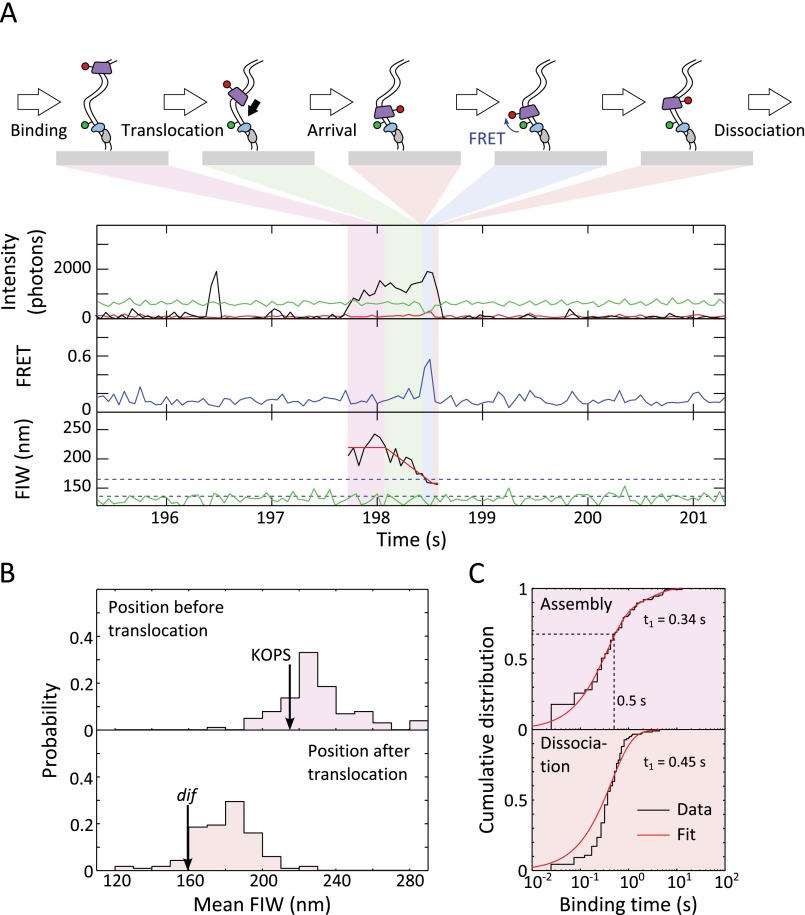
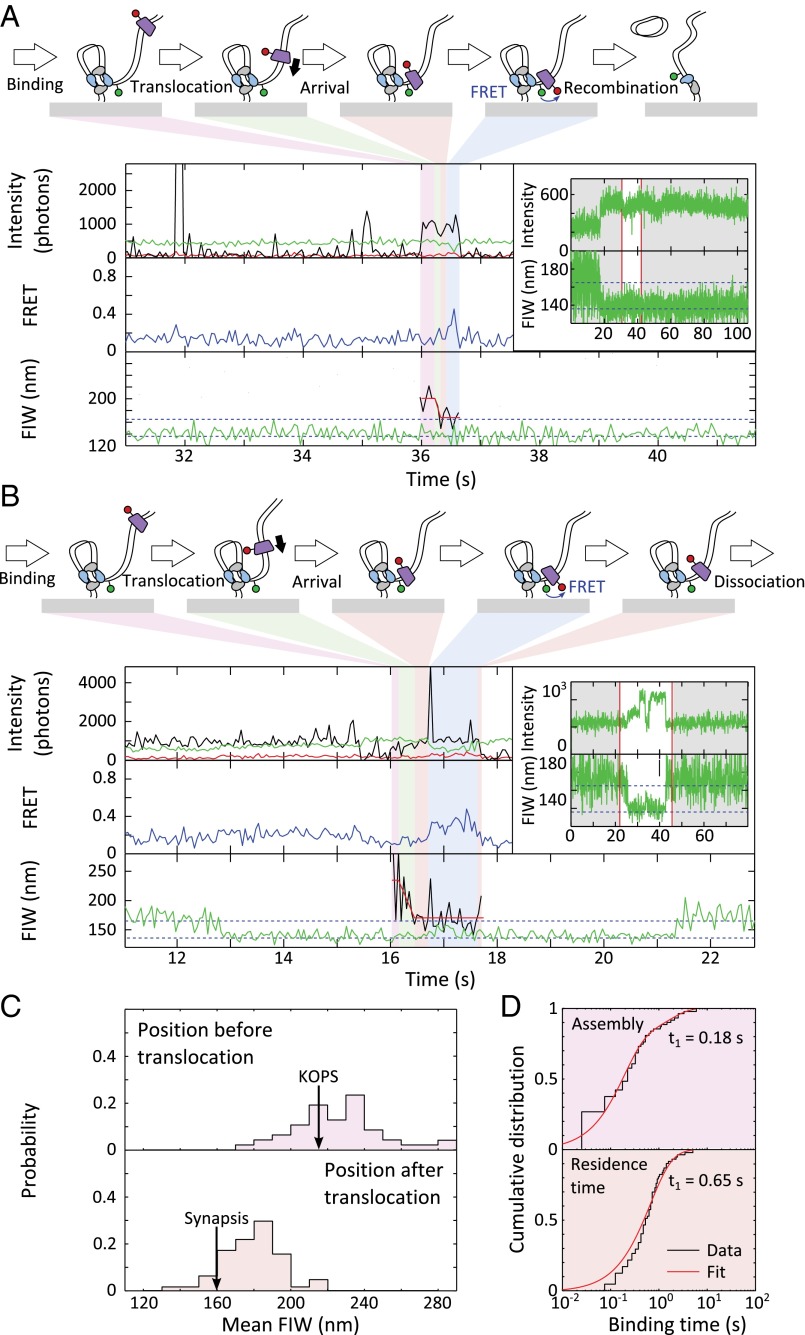
We adapted TFM-FRET (24, 33, 34) to follow the assembly, translocation, and behavior of FtsKC when it interacts with XerCD. We made a DNA substrate carrying two dif sites separated by a 1-kb spacer (Fig. 1A). The substrate was attached to the slide surface through a biotinylated 5′ end that was located 200 bp away from the first surface-proximal dif site, and the second surface-distal dif site was flanked by a 2.8-kb DNA segment containing a triple KOPS near its end. The KOPS was orientated toward the surface (Fig. 1A, Fig. S1A, and Methods). The DNA was labeled with Cy3B 1 bp away, in the direction of the free end, from the XerD binding sequence of the surface-distal dif site. TFM uses the width of the image of this single fluorophore to measure an effective distance along DNA; the fluorescence image width (FIW) decreases as the effective tether length decreases. Synapsis formation between the two dif sites, mediated by XerCD, was evident as a reduction in the green (Cy3B) FIW, and recombination was evident as a permanent reduction in the green FIW (Fig. 1A).
Fig. 1.
Experimental design. (A) Recombination is monitored using Cy3B. Synapsis transiently reduces the fluorescence image width (FIW), and recombination permanently reduces FIW. Blue dashed lines represent population average FIWs for the unsynapsed and synapsed/product DNA (165 nm and 136 nm, respectively). (B) Cy5 monitors FtsKC behavior. Translocation decreases or increases FIW. The red line represents LS fit, which was used to segment translocation. (C) FIW calibration. The SD from frame to frame over all of the molecules is shown as the shaded region. The red line represents a quadratic LS fit (n > 50 at each DNA length).
Fig. S1.
DNA substrate and Cy5-FtsK. (A) Sequence of DNA used throughout this study. Binding sites for XerC and XerD are indicated as boxes. The green Cy3B indicates the attachment of a Cy3B fluorophore via a C6-linker. (B) Absorption spectrum of the labeled FtsKC was used to estimate the labeling efficiency. The absorption wavelengths used for Cy5 and for the protein are indicated as dashed lines. (C) Cy5-FtsK was pulled down to the surface, in the presence of a 100-fold excess of unlabeled FtsKC trimer, using biotinylated anti-His antibody. The majority of time traces (87%) showed single-step photobleaching, but a minority (13%) displayed a photobleaching intermediate. (D) Photobleaching time for the surface-immobilized Cy5-FtsK was 28 ± 3 s, which is much longer than any of the FtsKC time scales reported in our experiments. (E) Correcting for the uneven illumination across our FOV by imaging a short DNA fragment carrying a single Cy5 (Cy5-DNA), we produced a histogram of single-molecule intensities for Cy5-FtsK. We find the majority (84%) of frames of Cy5-FtsK, pulled down in the presence of a 100-fold excess of unlabeled FtsKC, emission have an intensity 1.22-fold higher than 87-bp Cy5-DNA imaged at the same position and that a minority (16%) of frames have a relative intensity of 0.42, corresponding to the intensity of the observed photobleaching intermediate. The intensity increase relative to DNA is consistent with PIFE, commonly observed when particular dyes, including Cy5, are in close proximity to a protein. The width parameters (SDs) of the fitted intensity distributions were 0.21 and 0.28 relative intensity units for the minor and major populations, respectively. (F) Stoichiometry of Cy5-FtsK in solution. We used our His pull-down assay, without any excess of unlabeled FtsKC, and found that only 10% of molecules had an intensity corresponding to a doubly labeled hexamer of FtsKC (blue line). Given our labeling efficiency of ∼50%, we predict that if FtsKC formed hexamers in solution, we would have seen 33% of molecules with two Cy5 labels. Hence, we conclude that FtsKC predominantly existed, in solution, as trimers.
We used a covalent trimer of FtsKC (35), singly labeled with Cy5 at a surface Cys introduced in the motor β-subdomain of the middle monomer (Cy5-FtsK, with a labeling efficiency of ∼50%; Fig. S1 B–F and Methods). The trimer concentration, 5 nM, was close to the ∼40 nM in vivo monomer concentration (36). The binding of FtsKC was apparent as an appearance of signal in the red (Cy5) channel, and the position and behavior of the FtsKC were monitored via the red FIW (Fig. 1B). To convert between the red FIW and the position of FtsKC along DNA, we imaged dsDNA, with lengths between 87 and 4,000 bp, using our total internal reflection fluorescence (TIRF) microscope and standard conditions of 25-ms alternating laser excitation (ALEX) (37) (Fig. 1C and Methods). At around a 1-kb tether length, with the 400 photons per 25-ms frame observed using our illumination power (corresponding to a 16-kHz emission rate), we would need to measure for 15 frames (∼400 ms of observation time) to be able to distinguish with 95% confidence a 500-bp difference in effective tether length (Fig. 1C). The spatial and temporal resolution of TFM is discussed in our previous work (34).
In the absence of ATP and XerCD, we observed transient binding events that showed no obvious translocation (Fig. S2A); these binding events were distributed randomly along the DNA (Fig. S2B). The FtsKC dwell time was fit using three exponentials (Fig. S2C and Methods), recovering a major time of 0.84 ± 0.07 s (61 ± 5%) and two minor times of 4.0 ± 0.9 s (18 ± 5%) and 24 ± 3 s (19 ± 3%). We attribute the shortest time to transient association of a single Cy5-FtsK with DNA, the intermediate time to formation of a hexamer from two Cy5-FtsK molecules on DNA (because a hexamer would be more stable than a trimer), and the longest time to nonspecific sticking to the surface of the slide and subsequent photobleaching (photobleaching time for Cy5 at the surface was 28 ± 3 s) (Fig. S1D). These nonspecific sticking events were distinguished by their fixed narrow red FIW and were excluded from further analysis. Hence, around a quarter of DNA binding events had a dwell time that indicated hexamer assembly.
Fig. S2.
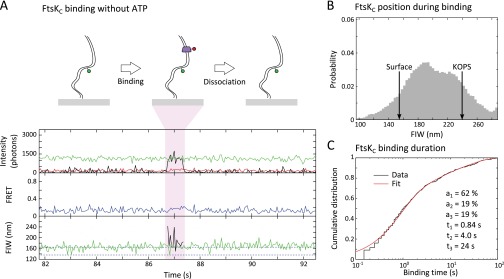
FtsKC binds DNA in the absence of ATP. (A) FtsKC binding is apparent as red fluorescence. (Top) Intensities recorded in the green emission channel under green excitation (DD, green line), red emission under green excitation (DA, red line), and red emission under red excitation (AA, black line). (Middle) Apparent FRET between the Cy3B and Cy5. Under these conditions, no FRET was observed. (Bottom) FIW in the green channel under green excitation (green) and in the red channel under red excitation (black). (B) FtsKC position during binding events is distributed along the DNA. The red FIW corresponding to binding at the surface and binding at the KOPS is indicated with arrows (n = 490 binding events). (C) Binding time distribution of FtsKC is well fit by three exponential dwell times, which suggests that there are different types of binding events as FtsKC assembles on DNA.
In the presence of ATP, in addition to transient binding events (Fig. S3A), we observed events with a decreasing red FIW, consistent with Cy5-FtsK translocations along DNA (Fig. 2A and Fig. S3B). We used a least-squares (LS) fit to the red FIW (Methods) to segment translocation events into three stages: assembly before translocation, translocation, and residence after translocation. The mean FIW before translocation, 220 nm, corresponded to the position of the KOPSs (FIW ≈ 230 nm), and the mean FIW after translocation, 161 nm, corresponded to close to the surface (FIW ≈ 155 nm), past the Cy3B (FIW ≈ 183 nm) (Figs. 1C and 2B), indicating that Cy3B does not impede FtsKC translocation. These mean FIWs suggest that the translocating FtsKC we observe assembles at or near the KOPSs and translocates past Cy3B, reaching the surface attachment point of the DNA. FRET, which reports the proximity of Cy5-FtsK to Cy3B within ∼10 nm [Förster radius, R0 ≈ 6.5 nm for these fluorophores (24)], was not observed under these conditions. Occasionally (n = 7), the red and green fluorescence disappeared simultaneously, which indicated that FtsKC had displaced the biotin–neutravidin interaction that tethered the DNA to the surface (Fig. S3C). This rare displacement is consistent with previously described behavior (38).
Fig. S3.
FtsKC binding and translocation in the presence of ATP. (A) Transient binding events with no obvious translocation are apparent in the presence of ATP, perhaps associated with incomplete FtsKC assembly or a translocation distance that is too short to be resolved in our assay. (B) Translocation event in the presence of ATP. The same color coding is used throughout this work. (C) Displacement of tethering biotin–neutravidin interaction by FtsKC (n = 7). (D) Potential translocation-dependent looping event apparent as a narrow green FIW at around 69.5 s (n = 8 events with FtsKC present during a transient narrowing of green FIW in the absence of XerCD).
Fig. 2.
Translocation events in the absence of XerCD. (A) Translocation event. (Top) Intensity: green emission under green excitation (DD, green), red emission under green excitation (DA, red), and red emission under red excitation (AA, black). (Middle) Apparent FRET. Under these conditions, no FRET was observed. (Bottom) FIW in the green channel (green) and in the red channel (black), and LS fit to the red FIW (red) used to segment the event into before (purple shading), during (green shading), and after (orange shading) translocation (n = 179 events). (B) Start and end positions of translocations. Black arrows represent positions of features on the DNA, obtained using the FIW calibration. (C) Dwell times. (Top) FtsKC assembly. (Bottom) Residence after translocation. Distributions were fit using a two-exponential process (Table S1 and Methods).
We determined the duration of FtsKC assembly, translocation, and residence using the LS fit to the red FIW. We found that dwell time distributions were in better agreement with two independent exponential processes rather than one, as judged using the Bayesian information criterion (BIC) (39) (ΔBIC > 10 in all cases, indicating strong evidence in favor of a two-exponential model). Hence, except where otherwise noted, we fit dwell time distributions with two exponentials and restrict our discussion to the major population (Table S1). Averaging across all fits to dwell times, the major population accounts for ∼90% of dwells, and we suggest that the minor population is a fit to background events. Using this method, the assembly time before translocation was 0.26 ± 0.12 s and the residence time after translocation was 2.1 ± 0.2 s (Fig. 2C, Table S1, and Methods). The mean translocation time was 0.33 ± 0.02 (±SEM), which corresponds to a translocation velocity of 12 ± 1 kb⋅s−1 at 21 °C (given that translocation events originated at the KOPS and ended close to the surface), which is a factor of two faster than previously measured at this temperature (30), but we note that our method did not require looping by FtsKC or the movement of a large bead with associated hydrodynamic drag to observe translocation. Hence, this velocity corresponds to the physiological translocation speed along unconstrained, flexible, “naked” dsDNA.
Table S1.
Maximum likelihood fits to dwell times presented in figures
| Description | a | t1, s | t2, s |
| Translocation on substrate without XerCD | |||
| Assembly | 80 (+20, −30) % | 0.26 ± 0.12 | 1.4 (+1.3, −0.6) |
| Residence | 88 ± 5% | 2.1 ± 0.2 | 14 ± 4 |
| Translocation toward single dif | |||
| Assembly | 90 ± 6% | 0.25 ± 0.03 | 2.7 ± 0.7 |
| Residence | 95 ± 2% | 0.50 ± 0.02 | 3.5 ± 0.6 |
| Translocation toward synapsis | |||
| Assembly | 77 ± 12% | 0.18 ± 0.04 | 1.6 ± 0.6 |
| Residence | 92 ± 9% | 0.65 ± 0.08 | 1.9 ± 0.5 |
| Translocation on product | |||
| Assembly | 82 ± 8% | 0.34 ± 0.05 | 2.9 ± 0.9 |
| Residence | 97 ± 2% | 0.45 ± 0.03 | 2.2 ± 0.5 |
| Recombination | |||
| Narrow FIW duration | 60 ± 6% | 13 ± 1 | 56 ± 5 |
| FtsK dwell after synapsis | 78 ± 9% | 0.9 ± 0.2 | 16 ± 6 |
| FRET events | |||
| Substrate FRET duration | 90 ± 3% | 0.30 ± 0.04 | 17 ± 6 |
| Product FRET duration | 91 ± 4% | 0.23 ± 0.02 | 1.7 ± 0.4 |
| Substrate dwell after FRET | 90 ± 5% | 0.09 ± 0.02 | 6 ± 4 |
| Product dwell after FRET | 92 ± 4% | 0.07 ± 0.01 | 5 ± 2 |
Errors are estimated as the SD of the parameter fit, using bootstrapping with 100 resamples.
A minority of events (n = 8, compared with 176 obvious translocations) showed a decrease in the green FIW, during the time that FtsKC was bound to DNA, which could be interpreted as the sticking of FtsKC to the surface or translocation-induced looping (Fig. S3D). These results establish that, in general, translocation does not induce DNA looping.
Stoichiometry of Translocating FtsKC.
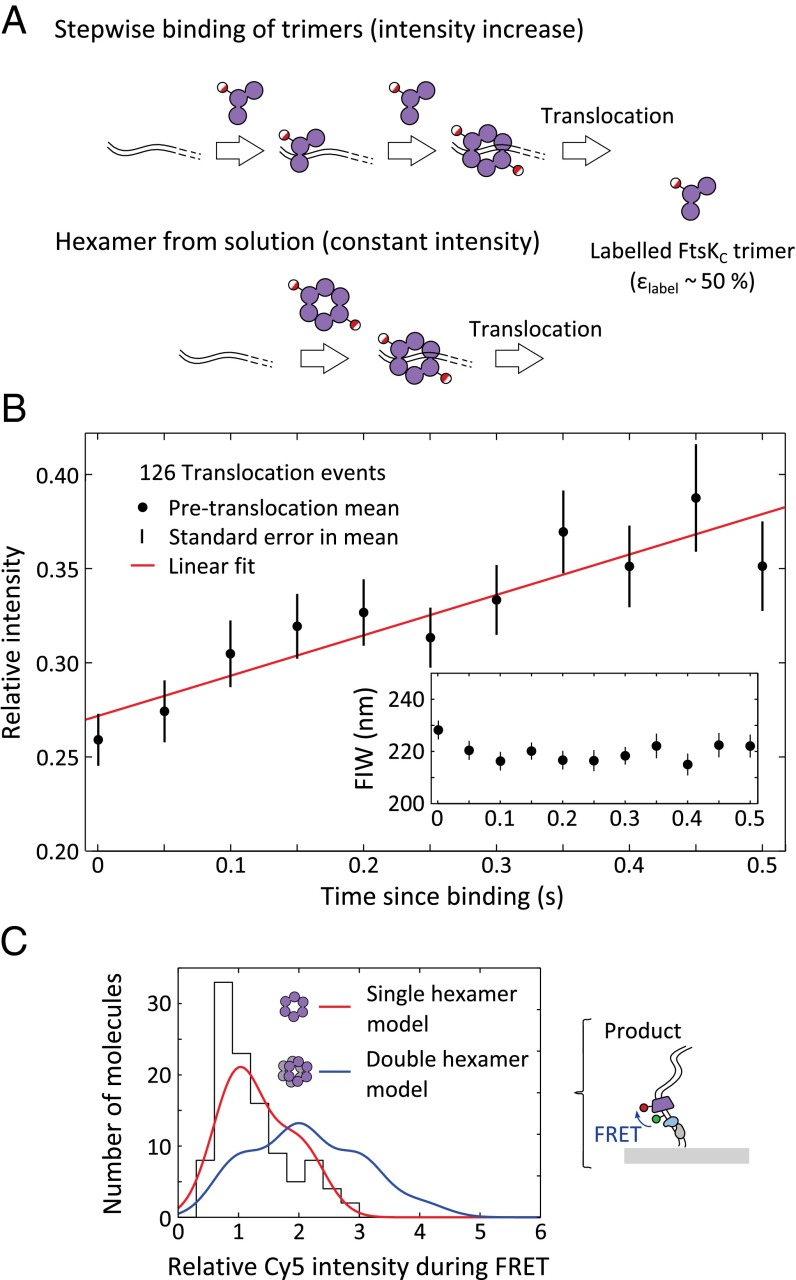
Previous biochemical analysis has suggested that the translocating unit of FtsKC assembles on DNA in six steps to form single hexamers (26). However, double-ring structures that could correspond to head-to-head hexamers on DNA have been observed in electron micrographs of Pseudomonas aeruginosa FtsKC lacking the γ-subdomain, and crystal structures of this protein contained dodecameric ring structures (17). To assess the stoichiometry of the active unit of FtsKC directly, we measured the intensities of Cy5-FtsK and counted the number of fluorophores present during assembly and during interactions with XerCD. A single hexamer, made up of two labeled trimers, would have between one and two fluorophores attached, and a double hexamer would have between one and four fluorophores attached.
For this analysis, we used the shorter, single dif-containing, DNA fragments formed by XerCD recombination at the surface (Fig. S4A), because all features of the DNA (dif site, KOPS, and Cy3B) were 1,000 bp closer to the surface than in the original substrate, and hence in a region of higher illumination intensity in the evanescent field of TIRF. This DNA recombinant product was identified as having a permanent narrow green FIW. Translocation events were segmented as before (Fig. S4B). If FtsKC preassembled in solution and then bound DNA fully assembled, we would predict no change in intensity between FtsKC binding and the start of translocation; however, if FtsKC assembled on DNA, the intensity should increase before translocation begins (Fig. 3A). The mean intensities during assembly, across all translocation events, were extracted (Fig. 3B). At a time 0.5 s after the appearance of red fluorescence signaling the presence of FtsKC, more than 66% of translocations had begun (Fig. S4C). We saw that on this time scale, there was a clear increase in the intensity of the red signal without any change in FIW (Fig. 3B), which implies that there was a stepwise assembly of Cy5-FtsK before translocation. Given our labeling efficiency of ∼50%, if FtsKC assembled into a single hexamer composed of two trimers, we would anticipate an average increase of 33%, and if it assembled into a double hexamer, we would anticipate an average increase of 110% before translocation (Methods). The observed 38.5% increase in intensity during the 0.5 s after binding is consistent with assembly into a single hexamer (Fig. 3B). The intensity increase suggests that FtsKC assembles on DNA rather than in solution. Using biotinylated anti-His antibodies, we pulled Cy5-FtsK out of solution, and determined that, at 5 nM and in the absence of DNA, the predominant stoichiometry was a single trimer (Fig. S1F), consistent with stepwise assembly on DNA before translocation.
Fig. S4.
FtsKC translocates on the product of recombination. (A) FtsKC translocation on the product of recombination. The product of recombination is a single dif site with a 2.7-kb tail, with the furthest part of the DNA 1 kb closer to the surface than in the DNA substrate (n = 126, with FRET apparent in 59). (B) Start and end positions of translocations. The mean starting FIW is 225 ± 2 nm, and the mean end FIW is 174 ± 2 nm (with the errors computed using bootstrapping with 1,000 resamples). (C) Dwell times before and after translocation. Distributions are fit using MATLAB (Table S1). (Top) At a time 0.5 s after binding (dashed line), more than 66% of translocations have begun.
Fig. 3.
FtsKC stoichiometry. (A) Intensity changes before translocation. If FtsKC assembles in a stepwise fashion before translocation, we expect an increase in intensity before translocation starts, whereas if fully assembled oligomers in solution bind and translocate, there would be no increase. (B) Mean relative intensity before translocation. A single Cy5 had a relative intensity of 0.2–0.4 between 2 and 3 kb along the DNA (Methods). Error bars: ±SEM. Least-squares fit (red) with an intensity increase of 38.5 ± 0.2% in the 0.5 s since binding (by which time more than 66% of translocations have begun). (Inset) Mean FIW for the same molecules. (C) Cy5 intensity during FRET. (Right) Corrected intensity during FRET events for the product DNA conformation were extracted and histogrammed (n = 108). (Left) Predicted intensity distribution is shown for a single hexamer (red) and a double hexamer (blue) of FtsKC. A single hexamer provides a better explanation of our data [single hexamer: sum of squared deviation (SSD) = 330; double hexamer: SSD = 1,230].
We also determined the stoichiometry of FtsKC during its interaction with XerCD complexes. In the presence of Cy5-FtsK, ATP, and XerCD, we observed FRET, signaling proximity between Cy5-FtsK and the Cy3B placed near the XerD binding site (Fig. S4A). We used the Cy5 intensity during FRET to extract the number of Cy5 fluorophores attached to a FtsKC complex while it interacted with XerCD-dif (Methods). This intensity was expressed relative to the calculated intensity for a single Cy5 at the same position (40) (Methods). Using this intensity for a single fluorophore, the labeling efficiency of Cy5-FtsK, and the width of the intensity distribution for a single fluorophore (Fig. S1E), we predicted the intensity distribution for a single hexamer and a double hexamer (Fig. 3C, Fig. S5, and Methods). Across all DNA conformations, the data agree much better with the single-hexamer prediction (Fig. 3C and Fig. S5). Hence, we conclude that, consistent with its assembly into a single hexamer before translocation, FtsKC interacts with XerCD as a single hexamer.
Fig. S5.
Stoichiometry of FtsKC approaching unsynapsed dif. During FRET, the Cy5 attached to FtsKC must be within ∼10 nm of the Cy3B attached to the DNA; hence, we know where in the evanescent field the Cy5 is (on average). (Right) Corrected intensity during FRET events for unsynapsed DNA conformations was extracted and histogrammed (n = 122). (Left) Given a labeling efficiency of 53%, a correction for the distance along the DNA in our evanescent excitation field, and the width of the intensity distribution for single Cy5-FtsK pulled down to the surface, we plot the predicted intensity distribution for a single hexamer (red) and a double hexamer (blue) of FtsKC. We find that a single hexamer provides a better explanation of our data [sum of squared deviation (SSD) = 230 for the single hexamer and SSD = 600 for the double hexamer].
Interactions Between Translocating FtsKC and XerCD-dif Complexes.
FtsK activates site-specific recombination by XerCD, and biochemical studies have shown that when FtsKC encounters XerD during translocation, it stops there (27). Moreover, QD-labeled FtsKC has been observed to pause and reverse translocation direction after meeting unsynapsed XerCD-dif on DNA curtains (32). In this work, we use singly Cy5-labeled FtsKC and observe its interaction with XerCD-dif in both unsynapsed and synapsed configurations.
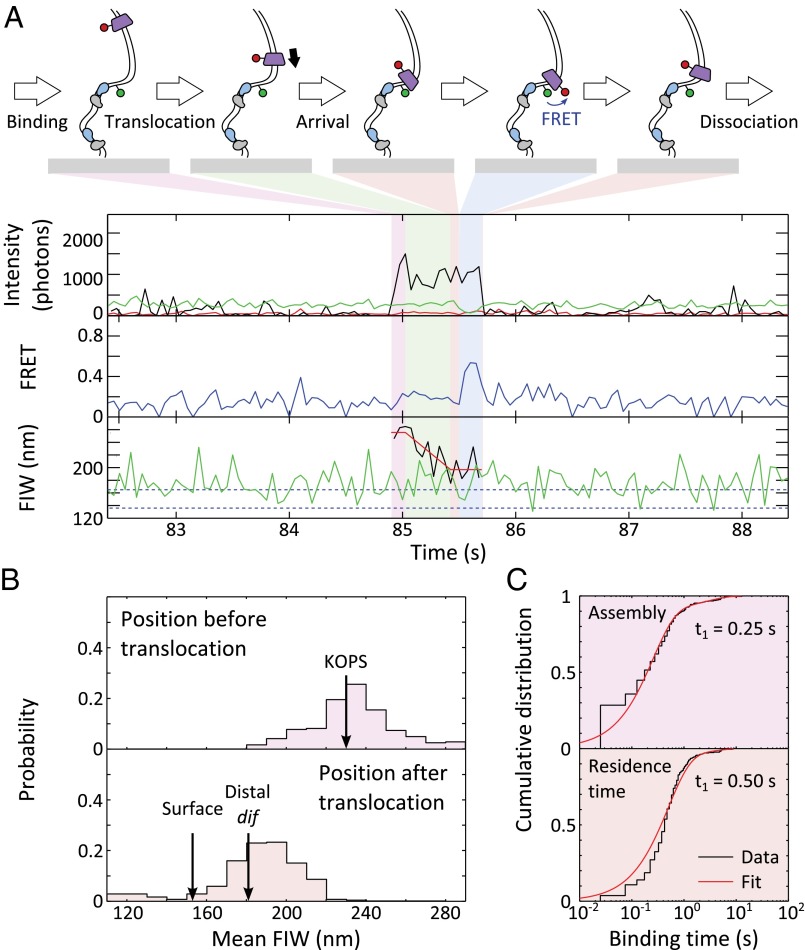
We imaged our double-dif tethered DNA substrate in the presence of Cy5-FtsK, ATP, and XerCD. We observed FtsKC translocations toward the surface when the DNA was in an unsynapsed conformation, as judged by the green FIW (n = 342; Fig. 4A and Fig. S6). In 30% of these events, at least one frame of FRET was apparent between the Cy3B attached to DNA and Cy5-FtsK, suggesting that FtsKC remained within ∼10 nm of the dif site for more than approximately the camera frame time (25 ms). Segmenting the translocation events as before, we observed that unsynapsed XerCD-dif acts as a roadblock for FtsKC, with the mean FIW after translocation, 181 nm, matching the location of the surface-distal dif site (FIW ≈ 183 nm) (Fig. 4B). The dwell times for assembly and residence after translocation were fit (Fig. 4C), recovering an assembly time of 0.25 ± 0.03 s and a residence time of 0.50 ± 0.02 s. The assembly times were independent of the presence of XerCD as anticipated, but the residence time after translocation decreased fourfold when FtsKC encountered XerCD bound to unsynapsed dif (compared with when it encountered the biotinylated DNA end), suggesting that interactions between XerD and FtsKC promote FtsKC dissociation. This decreased residence time gives us confidence that, under these conditions, it reflects interactions with XerCD bound to dif. Only a small minority (<0.5%, n = 1) of translocations toward unsynapsed XerCD resulted in a reversal by FtsKC, apparent as an increasing red FIW after residence. The lack of reversals in our data contradicts previous observations (32), where FtsKC reversed translocation direction after meeting XerCD bound to dif. The previous observations were made with QD-labeled FtsKC trimers, where there was the possibility of multiple FtsKC trimers tethered to the same QD. This multiple tethering could permit loading of a second FtsKC motor during the residence time of the original motor at XerCD, and may explain the presence of reversals in the previous observations. The Cy5-FtsK used in this work had only one trimer of FtsKC per fluorescent label, and hence overcomes this limitation. In the presence of XerCD, we recovered a mean translocation time of 0.18 ± 0.02 s (±SEM), which is shorter than the translocation time (0.33 ± 0.02 s) observed in the absence of XerCD, which, given the shorter translocation distance from the KOPS to XerD, corresponds to a similar mean translocation speed of 15 ± 1 kb⋅s−1.
Fig. 4.
Translocation stops at unsynapsed XerCD-dif. (A) Translocation. Blue shading highlights FRET. Translocation towards unsynapsed XerCD-dif is apparent in 342 events, of which 102 have at least a single frame of FRET accompanied by anticorrelated DD and DA changes. (B) Start and end positions of translocations. The mean starting FIW is 229 ± 1 nm (±SEM), and the mean end FIW is 181 ± 1 nm (±SEM). (C) Dwell times before and after translocation. Distributions are fit in MATLAB (MathWorks) (Table S1).
Fig. S6.
FtsKC interacts with XerCD after translocation. Translocation toward single XerCD-loaded dif (same conditions and similar event as in Fig. 4A) is shown.
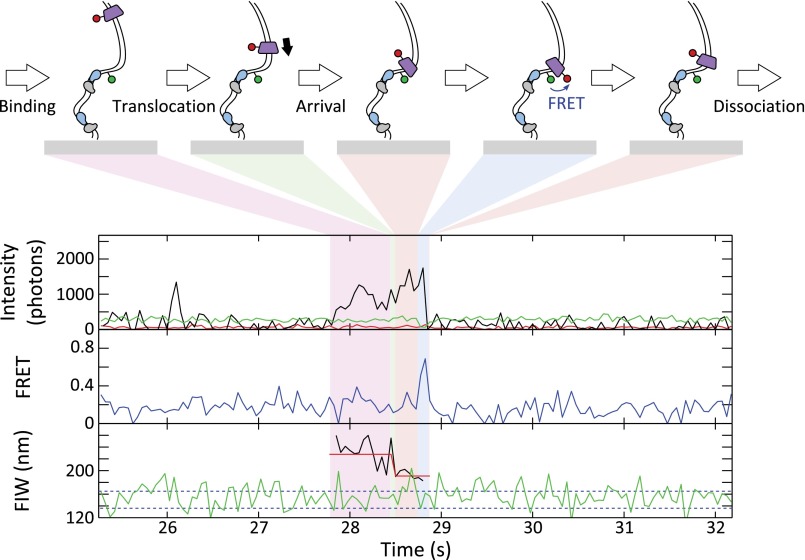
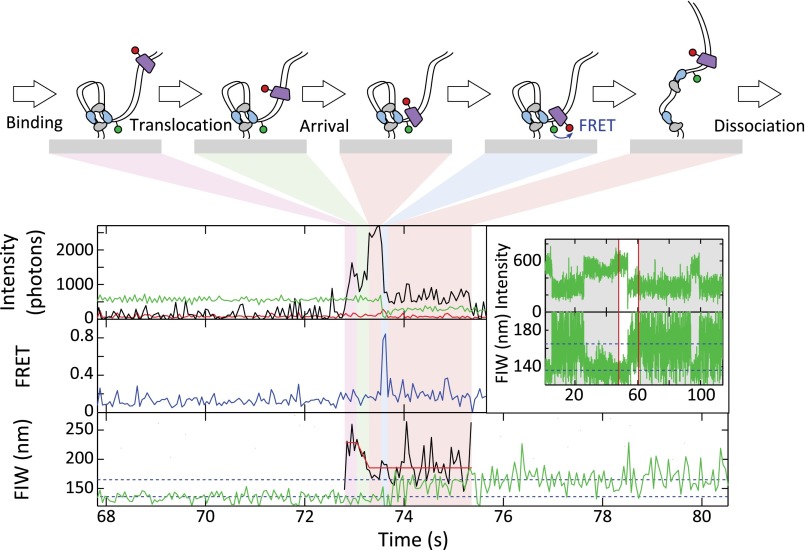
FtsKC activates XerCD recombination at dif, which was apparent as DNA molecules that transitioned to a persistent narrow green FIW (Fig. S7A). Of these events, 90% that had remained with a narrow FIW for at least 43 s had recombined (rather than being long-lived synapses; Methods), and we define a recombination event as meeting this criterion (n = 73). In 50% of recombination events (n = 39), FtsKC binding was apparent after synapsis formation (Fig. 5A). In 8% of recombination events (n = 6), obvious translocation (consisting of binding followed by a decreasing image width without any anomalous changes in intensity) accompanied recombination (Fig. 5A). Obvious translocations did not accompany all recombination events because translocations may not have initiated at the KOPS, and hence would have been too short to resolve in our assays, and because recombination could have been activated by the ∼25% of FtsKC complexes that would carry no fluorescent label. If translocation-induced looping (leading to formation of a synapsis) were a prerequisite for the activation of recombination, it would have been apparent as a FtsKC presence during synapsis formation. However, only 10% of recombination events (n = 8) followed this pattern (Fig. S7B), suggesting that translocation-induced looping between dif sites was not necessary for the activation of recombination. This suggestion is consistent with the previous result that the formation of the XerCD-dif synapsis is independent of FtsKC (24).
Fig. S7.
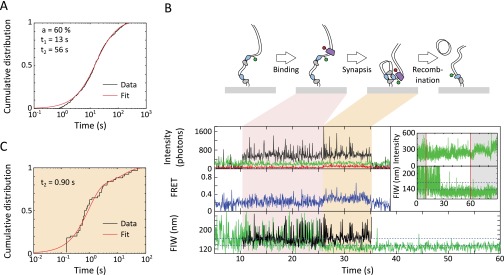
Recombination events. (A) Dwell times of recombination-like events, as distinguished by a narrow green FIW at the end of data acquisition. The dwell times were fit by two exponentials, recovering that 60 ± 6% of dwells had a lifetime of 12.5 ± 1.3 s (consistent with a synapsis lifetime of around 9 s) and that the remaining 40 ± 6% of events had a lifetime of 56 ± 5 s. The longer events correspond to true recombinations, and so a recombination-like event with a narrow FIW lasting 43 s or longer has a 90% chance of being a true recombination. (B) Recombination event with FtsKC present during synapsis formation. If FtsKC-induced looping was required for recombination, recombination events would have predominately followed this pattern. FtsKC binding is apparent during synapsis formation in eight of 73 recombination events. (C) Dwell time of first FtsKC to dissociate after synapsis formation in recombining events (n = 47 recombination events with FtsKC binding).
Fig. 5.
Arrival of FtsKC at XerCD-dif synapses. (A) FtsKC arrives after synapsis formation, leading to recombination. (Inset) Successful recombination reactions result in a permanent narrow green FIW. The highlighted region corresponds to the main figure. FtsKC binding is apparent after synapsis formation (but not during synapse formation) in 39 of 73 recombination events that meet the 90% confidence criterion. (B) FtsKC arrives at a synapsis and dissociates before the synapsis disassembles. (Inset) Synapsis formation and disassembly are apparent as a transiently narrowing FIW (n = 48, of which 20 show FRET). (C) Start and end positions of translocations. The mean starting FIW is 221 ± 4 nm (±SEM), and the mean end FIW is 175 ± 2 nm (±SEM). (D) Dwell times before and after translocation. Distributions are fit using MATLAB (Table S1).
FtsK can resolve catenated chromosomes produced by replication (41). We have previously suggested that FtsK may facilitate this decatenation by remaining in the vicinity of XerCD-dif after the activation of recombination and activating multiple rounds of recombination (12, 24). Taking the residence time (during synapsis) of the first FtsKC to dissociate after the synapsis formation that leads to recombination [i.e., the best candidate for having activated the recombination (Fig. S7C)], we recovered a residence time of 0.9 ± 0.2 s. This residence time is the total time from the first observation of binding (or synapsis formation) until FtsKC dissociates, and is hence an overestimation of the time FtsKC spends at XerCD-dif. Despite this overestimation, it is still shorter than the 1.6 s it takes XerCD to complete the recombination reaction (24), suggesting that FtsKC does not reside at XerCD long enough to activate multiple further rounds of recombination.
Not all translocations toward synapsed XerCD-dif resulted in recombination. We analyzed these nonproductive translocation events, distinguished by a transiently narrowing green FIW (Fig. 5B and Fig. S8); of 65 events, around a quarter showed FtsKC present during synapsis disassembly (Fig. S8). Again, XerCD acted as a roadblock for FtsKC, with the end of translocation approximately matching the position of the synapsis (Fig. 5C). FRET was apparent after translocation in 46% of events. For all these translocations toward a synapsis, the assembly time was 0.18 ± 0.4 s, consistent with the assembly time on unsynapsed DNA, and the posttranslocation residence time was 0.65 ± 0.07 s, again consistent with the residence time at unsynapsed XerCD-dif (Fig. 5D), as well as with the residence time of FtsKC during productive recombination events (0.9 ± 0.2 s). From this consistency, we infer that FtsKC does not reside for a significantly longer or shorter time depending on whether it encounters a single XerCD-dif or a XerCD-dif synaptic complex, or depending on whether it has activated recombination.
Fig. S8.
FtsKC present during synapsis disassembly. A translocating FtsKC meets a XerCD-dif synapsis, which disassembles, whereas FtsKC remains bound to DNA. The red and green intensities simultaneously decrease at around 73.5 s, consistent with FtsKC remaining at the distal dif site as the synapsis disassembles (n = 17, of which 10 show FRET).
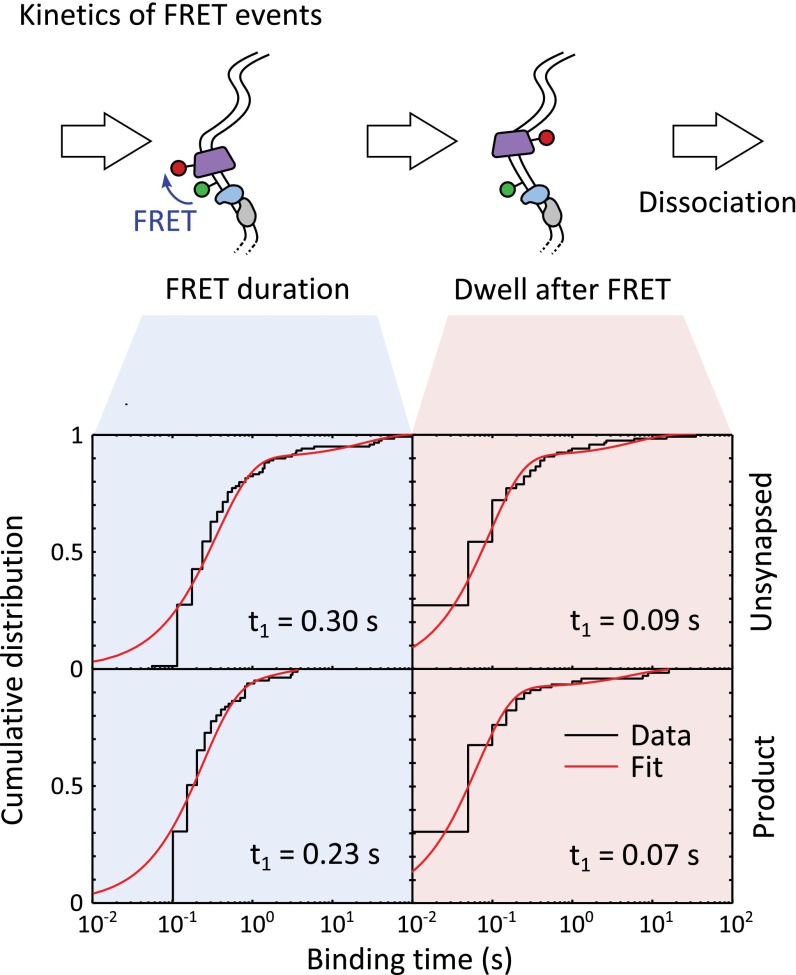
FRET may represent a transient association between Cy5-FtsK and XerCD-dif complexes. The automatic extraction of FRET events (Methods) required a minimum duration of 0.1 s (two frames), and so an upper bound on the duration of this association was estimated (Fig. S9). We find that FRET events had a dwell time of 0.30 ± 0.04 s and 0.23 ± 0.02 s for DNA in the substrate and product configurations, respectively. The dwell after FRET (before dissociation) was 0.09 ± 0.02 s and 0.07 ± 0.01 s for DNA in the substrate and product configurations, respectively. These dwells, between the end of FRET and dissociation, are shorter than the mean translocation time of 0.18 ± 0.02 s, suggesting there is no time for FtsKC to reverse and translocate after leaving the proximity of XerCD, consistent with the absence of an increasing FIW before dissociation, confirming that FtsKC dissociates, rather than reverses, after meeting XerCD-dif.
Fig. S9.
FRET reports on the proximity of FtsKC to the XerCD bound to dif. The dwell time distribution for automatically extracted FRET events (lasting a minimum of two frames) for unsynapsed (Top) and product (Bottom) DNA is fit using two exponentials (Table S1). The dwell times during and after FRET are similar for both DNA conformations, and the short time between the end of FRET and the disappearance of red fluorescence suggests that FtsKC dissociates after meeting XerCD bound to dif, rather than reversing translocation direction.
Discussion
FtsKC Translocates on DNA as a Single Hexamer.
In this work, using labeled FtsKC trimers, we addressed the question of whether FtsKC functions as a single or double hexamer. Double hexamers of FtsKC have been observed using EM and in crystal structures of FtsKC, and they have been implicated in the explanation of translocation reversals, and the extrusion of DNA loops, in optical tweezers single-molecule experiments (17, 28, 30). Our precise fluorescence measurements unequivocally show that FtsKC assembles on DNA into a single hexamer, rather than by forming a solution hexamer that then loads onto DNA and subsequently translocates as a single hexamer. Moreover, our data show that FtsKC activates XerCD-dif recombination as a single hexamer. We therefore conclude that double hexamers may not be physiologically relevant. Consistent with our observations, previous biochemical analysis using FtsK monomers led to the proposal that FtsKc assembles on DNA from monomers in a stepwise fashion (26). These results contrast with the demonstration that Bacillus subtilis SpoIIIE, a FtsK ortholog, forms hexamers in the absence of DNA at comparable concentrations to those concentrations used here (42, 43).
As revealed by a structural analysis, three FtsK γ-subdomains bind a KOPS (44). Hence, it seems likely that interaction of a single FtsK trimer with a KOPS will allow the engagement of the three γ-domains with DNA, supporting the physiological relevance of the transient association time, 0.84 ± 0.07 s, that we determined for a single trimer and DNA. The onset of hexamer translocation happens on a similar time scale (∼0.25 s; Fig. 6A), indicating that assembly of a second trimer into a functional hexamer and subsequent translocation are rapid, and will be dependent on the local concentration of FtsKC in the cell. A short initial association may serve to prevent FtsKC from assembling and translocating on DNA when its local concentration is low, which is the case during most of the cell cycle. In vivo, FtsK translocation occurs only when its concentration is high, at an invaginating septum (36, 45).
Fig. 6.
FtsKC assembly, stoichiometry, translocation, and activation of recombination. (A) FtsKC assembles stepwise on DNA into a single hexamer, which begins translocating around 0.25 s after the first binding. We note that in vivo FtsKC is expected to assemble from monomers rather than trimers. (B) When FtsKC arrives at a single dif site loaded with XerCD, it resides for around 0.5 s and then dissociates. (C) FtsKC arrives at a preformed XerCD-dif synapsis, resides for 0.5–1 s, and then dissociates. Frequently, the synapsis then disassembles, but, occasionally, recombination is activated.
Previous TPM and optical trapping experiments have required loop extrusion by FtsKC to observe its action at the single-molecule level. In some of these experiments, large visible FtsKC aggregates were observed to translocate at the same rate as species observed using loop extrusion in optical trapping (28). It has been suggested that loop extrusion arises from multiple points of contact on the same FtsKC, and that reversals by FtsKC require that it exists as more than a “single unidirectional motor” (13, 28, 30). Our work establishes that FtsKC assembles into a single hexamer before translocating, does not usually extrude loops of DNA, and dissociates after it encounters XerCD-dif. Hence, we suggest that the loop extrusion and reversals observed in TPM (18) and optical trapping experiments (28, 29) may have been introduced by the surface or by protein aggregates.
FtsKC Resides Briefly at XerCD-dif Before Dissociating.
When FtsKC encountered XerCD bound to dif, it resided there with a lifetime of 0.5–1 s, irrespective of whether it encountered an unsynapsed or synapsed XerCD-dif complex and independent of whether the encounter led to recombination (Figs. 4–6 B and C). This pause time is consistent with the pause time observed following collisions of QD-labeled FtsKC trimers with XerCD-dif or XerD-dif in studies utilizing DNA curtains (32). However, the outcome of the encounters we observed was different. Although 80% of encounters of QD-labeled FtsKC with XerCD-dif led to reversals of translocation direction, in our experiments, more than 99% of encounters of Cy5-FtsK led to dissociation. A possible explanation is that the Cy3B label, positioned 1 bp away from the surface-distal XerD binding site, influenced the behavior of FtsKC in our experiments. However, in the absence of XerCD, the translocation of Cy5-FtsK was not impeded by the Cy3B linkage on DNA, and when Cy5-FtsK encountered another obstruction, the biotin-neutravidin used for surface attachment, we did not observe reversals. We note that in our previous work, the activation of recombination was no less efficient when a fluorophore was placed in the position used here (24). It is also possible that our assay would not be able to resolve a rapid translocation away from XerCD-dif; however, using the time between the disappearance of FRET and the disappearance of fluorescence marking the disassociation of Cy5-FtsK, we have established that the average time of 0.08 ± 0.02 s that FtsKC appeared to remain on DNA after leaving the XerCD-dif complex was too short for it to have translocated off the “free end” of our DNA (which would have taken 0.18 ± 0.02 s at the measured translocation velocity). Because we had no more than one trimer of FtsKC per Cy5 fluorophore in our experiments, we suggest that QD-labeled FtsKC could have had other FtsKC trimers, attached to the same fluorescent probe, which might have assembled on DNA during residence at XerD. Because assembly before translocation took ∼0.25 s, and because the residence time at XerCD-dif was ∼0.5 s, there is sufficient time for the assembly of a second hexamer, attached to the same QD, while the first resides at XerCD-dif.
FtsKC Activates a Single Round of Recombination Before Dissociating from DNA.
Using single-color TFM-FRET, we previously determined that FtsKC activates recombination by inducing the remodeling of preexisting XerCD-dif synapses (24). In these experiments, we deduced the fate of FtsKC after activation by following the disappearance of PIFE, which resulted from the close proximity of FtsKC to the recombining complex. The PIFE signal was only observed up to isomerization of the HJ, which was generated by XerD-mediated DNA strand exchanges, which initiate FtsK-dependent recombination. This disappearance of PIFE, however, could not determine FtsKC dissociation from the complex, because the HJ isomerization could also lead to a change in conformation, which, in turn, could lead to the end of PIFE, because PIFE is only sensitive to distance changes in the range of 1–2 nm (25). Here, using a labeled trimer of FtsKC, we have followed precisely the presence of FtsKC at the recombining complex. Our results show that FtsKC predominantly dissociated 0.5–1 s after arriving at XerCD-dif. Because HJ formation and resolution by XerCD takes an average of 1.6 s (24), it seems unlikely that FtsK can activate subsequent rounds of recombination without dissociation, rebinding DNA, and encountering the XerCD-dif complex de novo. Therefore, multiple rounds of recombination would have to be mediated by multiple arrivals of FtsKC hexamers at XerCD-dif. The rapid dissociation of FtsKC, observed once it encounters XerCD-dif, may prevent a FtsKC hexamer from blocking access to XerCD-dif during these subsequent activation attempts by other FtsKC hexamers.
The vast majority of the events where FtsK successfully activated XerCD-dif recombination were not accompanied by any evidence of translocation-mediated looping. This lack of looping provides more evidence to support the conclusion that FtsK remodels existing initial synapses (24), rather than extruding a loop and forming an active synapsis or otherwise promoting active synapse assembly as suggested by TPM experiments using a purified γ-subdomain (13, 46, 47).
Most frequently, when FtsKC encountered a XerCD-dif synapsis, recombination was not activated. If FtsK behaves in a similar fashion in vivo, this low likelihood of activation would serve as a regulatory mechanism to help ensure it resolves, rather than produces, chromosome dimers. Only when a synapsis reformed several times, because chromosome topology was preventing segregation, would recombination, on average, be activated.
Two-Color TFM-FRET Follows Two Effective Lengths Along the Same DNA.
The extension of TFM, using two spectrally distinct fluorophores to track two positions simultaneously along the same DNA, has allowed us to correlate large-scale DNA conformation with the behavior of FtsKC as it assembled, translocated, and interacted with XerCD-dif. This method, performed on a standard fluorescence microscope, provides a blueprint for single-molecule experiments with increasing bandwidth, allowing the study of increasingly complicated protein–nucleic acid systems. This work has encoded information in the intensities, widths, and positions of the images of two fluorophores, realizing the possibility of six simultaneous fluorescence observables (increased further with the use of ALEX). TFM-FRET can be extended with the introduction of a third spectral channel (3, 48) and a third excitation laser, which would allow up to three simultaneous FRET distances, colocalization of three binding partners, and the simultaneous measurement of three distances along the same DNA molecule.
Methods
Standard methods were used for DNA and protein preparation, and they are described in detail in SI Text. Sample preparation and single-molecule experiments are described in SI Text. Data analysis, including event extraction, dwell time fitting, and the segmentation of translocation, is described in SI Text. The procedure for correcting Cy5 intensity and predicting the intensity distribution of labeled FtsK is described in SI Text.
SI Text
DNA Preparation.
The 4-kb DNA substrate (Fig. S1A) was prepared using PCR with one fluorescently labeled oligonucleotide and a plasmid template containing directly repeated dif sites separated by a 1-kb KmR gene cassette (pRB10) and Phusion High-Fidelity DNA polymerase (New England Biolabs). Oligonucleotides were synthesized and HPLC-purified by ATDBio Ltd. Cy3B labeling was performed as previously described (49). After the PCR assay, the product was digested with NcoI and ligated to a 200-bp fragment, produced by a separate PCR, which contained a 5′ biotin moiety. A 2.8-kb tail was ligated, following SalI digestion, and the 4-kb DNA produced was gel-purified.
Protein Preparation.
XerC, XerD, and unlabeled FtsKC were purified according to established procedures (17, 50). The mutant trimer of FtsKC was prepared using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). We mutated the Asx at position 953 in the middle subunit of the covalent trimer to a Cys. The oligonucleotides used were as follows: forward, 5′-ATCTGCGCGAAGTTTTGTGTAACGCCAAATTCCGC-3′; reverse, 5′-GCGGAATTTGGCGTTACACAAAACTTCGCGCAGAT-3′.
After purification, 1 mg of the covalent trimer was incubated for 20 min with 5 mM DTT. Next, the trimer was loaded onto a heparin column and washed extensively with buffer [25 mM Tris (pH 7.5), 5% (vol/vol) glycerol, 100 mM NaCl] to remove DTT. Subsequently, Cy5-maleimide was dissolved in 50 μL of DMSO, and 950 μL of buffer was then added. This dye solution was injected into the column containing the trimer; after 20 min of incubation on ice, unbound dye was washed away using buffer and the labeled trimer was eluted in a NaCl gradient. WT protein was reacted with Cy5-maleimide under the same conditions, and no Cy5 fluorescence was observed for the product, confirming that labeling targets the Cys introduced at the surface of the protein.
The labeling efficiency of Cy5-FtsK was determined using a Cary UV-Vis (Agilent) spectrophotometer (Fig. S1B). We used an absorption coefficient of 250,000 cm−1⋅M−1 for Cy5 and 519,870 cm−1⋅M−1 for our covalent trimer of FtsKC, determined from its primary sequence using an ExPASy ProtParam Tool (51), giving a labeling efficiency of 53 ± 5% (with the error quoted from the uncertainty in the protein absorption coefficient). To confirm that only one Cy5 label had been attached to each FtsKC trimer, we pulled down Cy5-FtsK to our slide surface by its His-tag, using a biotinylated anti-His antibody (Qiagen). We found that 86% of the 215 molecules analyzed showed single-step photobleaching (Fig. S1C), and that a minority displayed a photobleaching intermediate with an intensity of ∼40% of the intensity of a Cy5 attached to a short (87-bp) DNA at the same position in our field of view (FOV). This finding confirms that we have, at most, one label per Cy5-FtsK.
Instrumentation.
Single-molecule TIRF experiments were performed on a custom-built, objective-type TIRF microscope. A green laser (532-nm Cobolt Samba; Cobalt) and a red laser (635-nm CUBE; Coherent) were combined using a dichroic mirror and coupled into a fiberoptic cable. The output of the fiber was focused into the back focal plane of the objective (magnification of 100×, oil immersion, N.A. = 1.4, f/26.5; UPlanSApo, Olympus) and displaced perpendicular to the optical axis such that laser light was incident at the slide/solution interface at greater than the critical angle, creating an evanescent excitation field. Illumination powers were set to 2 mW, which corresponds to an approximate power density of 0.8 μW/μm2 incident on the 50 × 50-μm FOV. ALEX, with each laser on alternately for 25 ms, was implemented using an acousto-optic modulator (Isomet) for the green laser and direct modulation of the red laser. Fluorescence emission was collected by the objective and separated from the excitation light by a dichroic mirror (545 nm/650 nm; Semrock) and clean-up filters (545-nm long pass, Chroma; 633/25-nm notch filter, Semrock). The emission signal was focused on a rectangular slit to crop the image and then spectrally separated, using a dichroic mirror (630-nm long pass; Omega), into two emission channels that were focused side-by-side onto an electron multiplying charge coupled device (EMCCD) camera (iXon 897; Andor). The EMCCD was set to an EM gain of 300, corresponding to an approximate real gain of four counts per photon. Each pixel on the EMCCD corresponded to a 96 × 96-nm region in the imaging plane. A continuous reflective-interface feedback focus (ASI) autofocus system was used throughout the work to ensure focus stability over the course of data acquisition.
Sample Preparation.
DNA (∼200 pM) was incubated for 60 s at the surface of a polyethylene-glycol–passivated coverslip and attached to the coverslip through biotin–neutravidin interactions, which was sealed using a silicone gasket (Grace Biolaboratories) and a second coverslip. Imaging was performed in a buffer consisting of 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 100 μg/mL BSA, and 1 mM UV-treated Trolox (Sigma-Aldrich). An enzymatic oxygen scavenging system consisting of 1 mg/mL glucose oxidase, 40 μg/mL catalase, and 1.4% (wt/vol) glucose was added before sealing the sample before image acquisition. ATP was used at a concentration of 1 mM, and XerC and XerD were used at a concentration of 2 μM.
Data Analysis.
Extraction of fluorescence intensity signals from microscope images was performed using previously described TwoTone software (52). An apparent FRET efficiency was calculated from the extracted fluorescence emission ():
where is the fluorescence emission from the acceptor (Cy5) under donor excitation and is the emission from the donor (Cy3B). The FIW was obtained from the mean width of the fitted elliptical Gaussian.
Binding Event Extraction.
A 2D projective transformation between our emission channels was generated using the image of 100-nm TetraSpeck Microspheres (Life Technologies Ltd.) and the previously described TwoTone software (52).
Binding of Cy5-FtsK to DNA was automatically extracted using the following procedure. Candidate events started when the red intensity under red illumination () was at least 250 photons per frame and ended when the intensity fell below 100 photons for three consecutive frames. Of these candidates, binding events were identified as having at least two frames in duration, a mean binding location in the FOV within 150 nm of the moving average position of a green signal (averaged over the preceding 2.4 s), and at least a third of individual frame localizations within 150 nm of the green moving average (53).
FRET Event Extraction.
FRET was used to determine when Cy5-FtsK was within ∼10 nm of the distal dif site. Because we were interested in the red intensity under red illumination (AA) signal during FRET events, we looked for events with a minimum duration of two frames (50 ms), because we could be sure of the location of Cy5-FtsK for the red frames that came between the first and last frames of a FRET event. To search for FRET events within binding events, we corrected the apparent FRET efficiency for leakage of the Cy3B signal into the red detection channel and for direct excitation of Cy5 by the green laser using:
The proximity ratio, EPR, is the FRET efficiency corrected for leakage and direct excitation. The leakage and the direct excitation were determined by fitting the 2D surface defined by:
using linear LS in MATLAB (MathWorks). For our purposes, , the signal when no FRET is occurring, was well approximated by our whole dataset, because FRET occurred in a vanishingly small proportion of frames. We recovered and , consistent with our previous measurements for these fluorophores on our microscope (24, 40).
FRET events were considered to begin when was above 0.2 and to end when dropped below 0.1 for more than one consecutive frame. Single-frame events were discarded for the purpose of dwell time and stoichiometry analysis. The red intensity during FRET was the median intensity for the red frames that fell between the start and the end of the automatically extracted FRET event within a binding event.
Dwell Time Fitting.
Dwell times were fit with the sum of two exponentials, with the model:
Fits were performed using maximum likelihood estimation in MATLAB, and errors were estimated as the SD in each estimated parameter using bootstrap resampling with 100 resamples.
The three-exponential fit, to all FtsKC dwell times on DNA, was performed using the same procedure with the model:
Translocation Segmentation.
Translocation events were segmented using an ordinary LS fit to the red FIW, , with the model:
where and are the FIWs before and after translocation, and and are the start and end times of translocation, which are constrained to be exactly coincident with a frame for computational efficiency. When extracting image widths and intensities, the frames associated with and were discarded to mitigate the effect of the finite camera integration time.
Likelihood of Recombination.
Recombination was apparent as a transition to a fixed, narrow green FIW. However, some proportion of time traces that ended in a narrow FIW was due to synapsis formation (which did not proceed to recombination), with the transition back to a broad green FIW not seen due to photobleaching or the end of a movie. Because synapsis has an exponentially distributed dwell time with a mean time of 9 ± 1 s (24), and recombination leads to a permanent narrowing of FIW, “true” recombinations can be distinguished with some degree of certainty based on the time for which the FIW has remained narrow.
We extracted the Cy3B bleaching time by fitting the mean intensity over time for all our experiments, and recovered a time of 320 ± 20 s. Using this bleaching time, the length of our movies (205 s), the lifetime of synapsis [8.6 ± 0.6 s (24)], and the observation that 50% of our DNAs imaged in the presence of XerCD show some synaptic activity, we determined a dwell time between synapsis formation of ∼190 s by matching the fraction showing synaptic activity in a Markov chain Monte Carlo simulation. Using this simulation of synapsis-only time traces (where recombination was not possible), we found the dwell time for synaptic events that end in a narrow FIW (due to photobleaching or the end of a movie) was 9 ± 1 s (with the uncertainty dominated by the uncertainty in the synapsis lifetime).
Extracting the dwell times for events that end in a narrow FIW from our experimental data, and using our two-exponential fit (Fig. S7A), we find that 60 ± 6% of our dwell times come from a distribution with a dwell time of 13 ± 1 s, which is broadly consistent with the synaptic event lifetime determined above, and that the remaining 40 ± 6% of the dwell times come from a distribution with a dwell time of 56 ± 5 s, which we attribute to true recombination events. Given the relative proportions of the populations and their time scales, we determine that 90% of time traces that end in a narrow green FIW, which has been narrow for 43 s or longer, are true recombinations.
Relative Cy5 Intensities.
In our experiments, the detected fluorescence intensity from a single Cy5 fluorophore depended on its position within the microscope FOV, its binding position along DNA, and what biomolecule it was attached to. The dependency on FOV position was due to the nonuniform excitation field across the FOV. We followed our established method for determining this dependency (40), modifying the procedure to use a reference standard DNA. Our reference DNA consisted of an internally labeled Cy5 separated from the 5′ biotin moiety, used for surface attachment, by 87 bp. After setting laser powers and the TIRF angle, we imaged many (∼100) FOVs of this Cy5-DNA standard. We excluded molecules that photobleached, or displayed photoblinking, by filtering those molecules whose intensities deviated by more than twice the median absolute deviation for three or more frames. The mean intensity and the localization position were fit with a 2D elliptical Gaussian, using nonlinear LS fitting in MATLAB, which recovered a predicted intensity for a single Cy5-DNA at any position within the FOV.
We corrected for the particular single-molecule intensity of Cy5-FtsK using our anti-His pull-down data. We extracted the frame-by-frame intensities of Cy5-FtsK pulled down to our surface using a biotinylated anti-His antibody, and we fit two Gaussian distributions, using nonlinear LS in MATLAB (Fig. S1E). We extracted a relative intensity for Cy5-FtsK 1.22-fold higher than for the Cy5-DNA standard used. The minor population (∼16% of frames) corresponds to the rare photobleaching intermediate observed in 13% of pull-down time traces (Fig. S1C).
We also applied a correction for the evanescent TIRF field in our experiments, because the further along DNA that Cy5-FtsK bound, the dimmer it appeared. To estimate the TIRF decay length in our experiments, we compared the mean intensity of synapsed and product DNAs with the mean intensity of unsynapsed DNAs. We found that synapsed and product DNA had 210 ± 10% of the brightness of unsynapsed DNA (with the error quoted as the SD between sample wells). Assuming the position of the fluorophore above the coverslip is linearly proportional to its length along DNA, this corresponds to a TIRF decay length in the green channel of 1,400 bp, corresponding to a decay length of 1,700 bp in the red channel, given that both green and red lasers are incident on the coverslip at the same angle but differ in wavelength (for a given incident angle, the TIRF decay length is inversely proportional to the wavelength used). Using this estimated red channel decay length, we predict that Cy5 attached to DNA at the position of the distal dif site in DNA in the unsynapsed configuration (∼1,300 bp) has 50% of the intensity of our Cy5-DNA standard and that Cy5 attached to DNA at the position of a synapsis, or the dif site in the DNA product of recombination (∼230 bp), should have 92% of the intensity of our Cy5-DNA standard. Using these three correction factors (for nonuniform illumination, the single-molecule intensity of Cy5-FtsK, and the TIRF field), we could predict the intensity of a single Cy5 attached to FtsKC at any position within our FOV and bound anywhere along DNA.
Predicted Intensity Distributions.
Given the labeling efficiency and the intensity of Cy5-FtsK, we predicted the range of intensities expected for a single hexamer of FtsKC and a double hexamer of FtsKC. For an assembly of trimers (two for a single hexamer and four for a double hexamer), with a labeling efficiency , the probability of observing a complex with labels () is:
Hence, the predicted intensity distribution is given by:
The intensity refers to the predicted intensity for a single fluorophore ( when we are expressing relative intensities), and is the width of the intensity distribution, which can be estimated from the intensity distribution for His pull-down Cy5-FtsK as 0.28 intensity units relative to the intensity of Cy5-DNA at the surface.
The average number of fluorophores per assembly is given by:
For a labeling efficiency of ∼50%, we get an average of 1.33 fluorophores per single hexamer and an average of 2.13 per double hexamer. Hence, we predict that a single hexamer would show a 33% increase in intensity, on average, after the first labeled FtsKC trimer binds to DNA and that a double hexamer would show a 110% increase.
Acknowledgments
P.F.J.M. was supported by MathWorks (Cambridge, UK). P.Z. was supported by the Leverhulme Trust (Grant RP2013-K-017). Work in the D.J.S. laboratory was supported by a Wellcome Trust Senior Investigator Award (Grant 099204/Z/12Z). Work in the A.N.K. laboratory was supported by the European Commission Seventh Framework Program (Grant FP7/2007-2013 HEALTH-F4-2008-201418), the Biotechnology and Biological Research Council (Grant BB/H01795X/1), and the European Research Council (Starter Grant 261227).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1510814112/-/DCSupplemental.
References
- 1.Hohng S, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318(5848):279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms GS, et al. Probing conformational changes of gramicidin ion channels by single-molecule patch-clamp fluorescence microscopy. Biophys J. 2003;85(3):1826–1838. doi: 10.1016/S0006-3495(03)74611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S, Lee J, Hohng S. Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS One. 2010;5(8):e12270. doi: 10.1371/journal.pone.0012270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J, et al. Single-molecule four-color FRET. Angew Chem Int Ed Engl. 2010;49(51):9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigot S, Sivanathan V, Possoz C, Barre F-X, Cornet F. FtsK, a literate chromosome segregation machine. Mol Microbiol. 2007;64(6):1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 6.Barre F-X. FtsK and SpoIIIE: The tale of the conserved tails. Mol Microbiol. 2007;66(5):1051–1055. doi: 10.1111/j.1365-2958.2007.05981.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaimer C, Graumann PL. Players between the worlds: Multifunctional DNA translocases. Curr Opin Microbiol. 2011;14(6):719–725. doi: 10.1016/j.mib.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Stouf M, Meile J-C, Cornet F. FtsK actively segregates sister chromosomes in Escherichia coli. Proc Natl Acad Sci USA. 2013;110(27):11157–11162. doi: 10.1073/pnas.1304080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aussel L, et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108(2):195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 10.Hallet B, Sherratt DJ. Transposition and site-specific recombination: Adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21(2):157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 11.Sherratt DJ, et al. Site-specific recombination and circular chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 1995;347(1319):37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 12.Grainge I, et al. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26(19):4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip SCY, Bregu M, Barre FX, Sherratt DJ. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 2003;22(23):6399–6407. doi: 10.1093/emboj/cdg589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barre F-X, Sherratt DJ. Chromosome dimer resolution. In: Higgins NP, editor. The Bacterial Chromosome. American Society for Microbiology; Washington, DC: 2005. pp. 513–524. [Google Scholar]
- 15.Draper GC, McLennan N, Begg K, Masters M, Donachie WD. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180(17):4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu XC, Weihe EK, Margolin W. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol. 1998;180(23):6424–6428. doi: 10.1128/jb.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Löwe J. Double-stranded DNA translocation: Structure and mechanism of hexameric FtsK. Mol Cell. 2006;23(4):457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Ptacin JL, Nöllmann M, Bustamante C, Cozzarelli NR. Identification of the FtsK sequence-recognition domain. Nat Struct Mol Biol. 2006;13(11):1023–1025. doi: 10.1038/nsmb1157. [DOI] [PubMed] [Google Scholar]
- 19.Bigot S, et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24(21):3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sivanathan V, et al. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13(11):965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigot S, Corre J, Louarn J-M, Cornet F, Barre F-X. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol Microbiol. 2004;54(4):876–886. doi: 10.1111/j.1365-2958.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- 22.Massey TH, Aussel L, Barre F-X, Sherratt DJ. Asymmetric activation of Xer site-specific recombination by FtsK. EMBO Rep. 2004;5(4):399–404. doi: 10.1038/sj.embor.7400116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yates J, et al. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol Microbiol. 2006;59(6):1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zawadzki P, et al. Conformational transitions during FtsK translocase activation of individual XerCD-dif recombination complexes. Proc Natl Acad Sci USA. 2013;110(43):17302–17307. doi: 10.1073/pnas.1311065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang H, Kim H, Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci USA. 2011;108(18):7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham JE, Sherratt DJ, Szczelkun MD. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc Natl Acad Sci USA. 2010;107(47):20263–20268. doi: 10.1073/pnas.1007518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham JE, Sivanathan V, Sherratt DJ, Arciszewska LK. FtsK translocation on DNA stops at XerCD-dif. Nucleic Acids Res. 2010;38(1):72–81. doi: 10.1093/nar/gkp843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pease PJ, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307(5709):586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 29.Levy O, et al. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci USA. 2005;102(49):17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saleh OA, Pérals C, Barre FX, Allemand JF. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23(12):2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Finkelstein IJ, Crozat E, Sherratt DJ, Greene EC. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc Natl Acad Sci USA. 2012;109(17):6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Finkelstein IJ, Arciszewska LK, Sherratt DJ, Greene EC. Single-molecule imaging of FtsK translocation reveals mechanistic features of protein-protein collisions on DNA. Mol Cell. 2014;54(5):832–843. doi: 10.1016/j.molcel.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinkney JNM, et al. Capturing reaction paths and intermediates in Cre-loxP recombination using single-molecule fluorescence. Proc Natl Acad Sci USA. 2012;109(51):20871–20876. doi: 10.1073/pnas.1211922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.May PFJ, et al. Tethered fluorophore motion: Studying large DNA conformational changes by single-fluorophore imaging. Biophys J. 2014;107(5):1205–1216. doi: 10.1016/j.bpj.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crozat E, et al. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J. 2010;29(8):1423–1433. doi: 10.1038/emboj.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bisicchia P, Steel B, Mariam Debela MH, Löwe J, Sherratt D. The N-terminal membrane-spanning domain of the Escherichia coli DNA translocase FtsK hexamerizes at midcell. MBio. 2013;4(6):e00800–e00813. doi: 10.1128/mBio.00800-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101(24):8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chivers CE, et al. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat Methods. 2010;7(5):391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90(420):773–795. [Google Scholar]
- 40.Periz J, et al. Rotavirus mRNAS are released by transcript-specific channels in the double-layered viral capsid. Proc Natl Acad Sci USA. 2013;110(29):12042–12047. doi: 10.1073/pnas.1220345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimokawa K, Ishihara K, Grainge I, Sherratt DJ, Vazquez M. FtsK-dependent XerCD-dif recombination unlinks replication catenanes in a stepwise manner. Proc Natl Acad Sci USA. 2013;110(52):20906–20911. doi: 10.1073/pnas.1308450110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cattoni DI, et al. Structure and DNA-binding properties of the Bacillus subtilis SpoIIIE DNA translocase revealed by single-molecule and electron microscopies. Nucleic Acids Res. 2014;42(4):2624–2636. doi: 10.1093/nar/gkt1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cattoni DI, et al. SpoIIIE mechanism of directional translocation involves target search coupled to sequence-dependent motor stimulation. EMBO Rep. 2013;14(5):473–479. doi: 10.1038/embor.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Löwe J, et al. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol Cell. 2008;31(4):498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hale CA, de Boer PAJ. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J Bacteriol. 2002;184(9):2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diagne CT, et al. TPM analyses reveal that FtsK contributes both to the assembly and the activation of the XerCD-dif recombination synapse. Nucleic Acids Res. 2014;42(3):1721–1732. doi: 10.1093/nar/gkt1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Besprozvannaya M, Burton BM. Do the same traffic rules apply? Directional chromosome segregation by SpoIIIE and FtsK. Mol Microbiol. 2014;93(4):599–608. doi: 10.1111/mmi.12708. [DOI] [PubMed] [Google Scholar]
- 48.Hohng S, Joo C, Ha T. Single-molecule three-color FRET. Biophys J. 2004;87(2):1328–1337. doi: 10.1529/biophysj.104.043935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapanidis AN. Alternating-laser excitation of single molecules. In: Selvin PR, Ha T, editors. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 2008. pp. 85–119. [Google Scholar]
- 50.Ferreira H, Sherratt D, Arciszewska L. Switching catalytic activity in the XerCD site-specific recombination machine. J Mol Biol. 2001;312(1):45–57. doi: 10.1006/jmbi.2001.4940. [DOI] [PubMed] [Google Scholar]
- 51.Gasteiger E, et al. Protein identification and analysis tools on the ExPASY server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- 52.Holden SJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J. 2010;99(9):3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans GW, Hohlbein J, Craggs T, Aigrain L, Kapanidis AN. Real-time single-molecule studies of the motions of DNA polymerase fingers illuminate DNA synthesis mechanisms. Nucleic Acids Res. 2015;43(12):5998–6008. doi: 10.1093/nar/gkv547. [DOI] [PMC free article] [PubMed] [Google Scholar]